Abstract
Background and aims
Rice (Oryza sativa L.) and pearl millet (Pennisetum glaucum L.) biofortification breeding programs require accurate and convenient methods to identify nutrient dense genotypes. The aim of this study was to investigate energy-dispersive X-ray fluorescence spectrometry (EDXRF) for the measurement of zinc (Zn) and iron (Fe) concentration in whole grain rice and pearl millet.
Methods
Grain samples were obtained from existing biofortification breeding programs. Reference Zn and Fe concentrations obtained by inductively-coupled plasma-optical emission spectroscopy (ICP-OES) were used to calibrate the EDXRF instrument. Calibration was performed with 24 samples and separate calibrations were developed for rice and pearl millet. To validate calibrations, EDXRF analyses were conducted on an additional 40 samples of each species.
Results
EDXRF results were highly correlated with ICP-OES values for both Zn and Fe in both species (r2 = 0.79 to 0.98). EDXRF predicted Zn and Fe in rice to within 1.9 and 1.6 mg kg−1 of ICP-OES values, and Zn and Fe in pearl millet to within 7.6 and 12.5 mg kg−1 of ICP-OES values, at a 95% confidence level.
Conclusion
EDXRF offers a convenient, economical tool for screening Zn and Fe concentration in rice and pearl millet biofortification breeding programs.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Considerable effort is being made around the world to breed zinc (Zn)- and iron (Fe)-dense grain crops for improved human nutrition. Within the HarvestPlus biofortification program (Nestel et al. 2006; Pfeiffer and McClafferty 2007), a major focus is to breed rice (Oryza sativa L.) containing more Zn, with high Fe rice an important secondary objective. Another area of focus for HarvestPlus is to breed high Fe pearl millet (Pennisetum glaucum L.), with high Zn genotypes an important secondary objective. Reliable methods to analyse Zn and Fe in rice and pearl millet grain samples are fundamental to these efforts. In recent years, the main method used for elemental analysis of plant tissues has been inductively coupled plasma-optical emission spectrometry (ICP-OES) (Zarcinas et al. 1987). This form of analysis requires expensive equipment, highly trained analysts, contamination free reagents and extensive sample preparation. It has not proven practicable for most biofortification breeding programs to conduct their analysis ‘in-house’, particularly where work is conducted in less-developed countries. As a consequence, most HarvestPlus breeding programs have had their samples analysed abroad (e.g., Velu et al. 2011). While quality of results has been highly satisfactory for the breeding programs, the expense of the analysis and quarantine issues associated with the international transport of grain have represented major constraints. Alternative, colorimetric methods have been developed for Zn and Fe analysis in grain (Prom-u-thai et al. 2003; Ozturk et al. 2006; Choi et al. 2007). Though simpler to undertake than ICP-OES, these methods are only semi-quantitative and, when thousands of samples are to be analysed, too time consuming.
X-ray fluorescence analysis (XRF) has previously been used in quantitative elemental analysis of a wide range of organic and inorganic samples (for reviews, see Arai 2006; West et al. 2009; West et al. 2010). The basis for the technique is that all elements emit secondary (‘fluorescent’) X-rays of characteristic energy when exposed to X-rays of appropriate higher energy, with energy and intensity of emitted X-rays used to determine elemental composition. In general, the heavier the element being analysed, the higher the energy of X-rays required to elicit fluorescence, the higher the energy of fluorescence, and the easier it is to detect fluorescence. The lightest elements found in biological samples (e.g. H, B, C, N, O) are not generally detectable by XRF, while elements such as Na, Mg, P, S, Cl, K, Ca are detectable, but only at high concentrations or with highly specialized conditions, and heavier elements such as the trace metals Mn, Fe, Cu and Zn or toxic heavy metals are readily analyzed, even at trace levels. Major advantages of XRF over ‘wet chemistry’ methods are that analyses are non-destructive, use no noxious chemicals and produce no toxic wastes, and can be made on solid samples. Since XRF signal is obtained from transitions among inner shell electrons, not bonding electrons, XRF also has the advantage that signal is independent of chemical form.
Of the various types of X-ray spectrometry available, laboratory ‘bench-top’ Energy-Dispersive X-ray Fluorescence (EDXRF) is the form most commonly used for routine analysis of large numbers of samples. In EDXRF, detectors are used which discriminate X-rays based on energy, enabling the simultaneous detection of multiple elements. Samples are mostly presented for analysis in cuvettes on a sample carousel, enabling multiple samples to be analysed in a single run. The other major form of XRF currently used is Wavelength-Dispersive XRF (WD-XRF), which discriminates emissions after diffraction through a crystal. The major advantage of EDXRF over WD-XRF is that equipment is simpler and cheaper with lower output X-ray tubes, albeit with lower resolving power and higher background. Portable XRF devices have also been developed which have similar analytical performance to EDXRF, but lack multiple sample capability (Melquiades and Appolini 2004). In the area of plant and food science, applications have been found for EDXRF in the determination of iodine and calcium in bread improver (Ekinci et al. 2002), minerals in milk powder (Perring and Andrey 2003; Perring and Blanc 2008), Fe, Cu and Zn in food premixes (Perring et al. 2005; Perring and Blanc 2007), and phosphorus in potato starch (Noda et al. 2006).
Calibration of an EDXRF can be achieved using either of two methods. In situations where general purpose calibrations are required that cover a large variety of sample types, Fundamental Parameters (FP) approaches can be used, which use complex mathematical algorithms to model the response of the EDXRF to pure elements in a given sample type (Rousseau et al. 1996). In applications where the range of analyte concentrations is limited, and where sets of standards can be developed that are similar in composition and morphology to samples of interest, it is more common to use the empirical calibration approach. Here, elemental concentrations are measured in a set of calibration standards using an established ‘reference’ method (such as ICP-OES), and these values are related to the intensity of X-ray emissions for these samples. Such calibrations are then validated by comparing EDXRF and reference method values on an independent set of samples. The studies of Perring and Andrey (2003), Perring et al. (2005), Perring and Blanc (2007) and Perring and Blanc (2008) all utilized the empirical approach to calibration, and sets of calibration and validation samples in which reference values were determined by duplicate ICP-OES. In all these studies, EDXRF reliably determined concentrations of the target elements.
It is accepted that the greatest source of error in EDXRF is specimen inhomogeneity (Blank and Eksperiandova 1998; Injuk et al. 2006). Accordingly, analyses are usually conducted on ground samples. These can be analysed as loose powder poured into a specimen holder sealed at the bottom with a Mylar window or, in most cases with biological samples, powdered specimens are compressed into pellets to overcome effects arising from differences in particle size or sample density. The EDXRF protocols published so far for the analysis of trace metals in food-stuffs and cereal powders have all involved sample grinding, then compression of 4–10 g of powder into discs of 30–40 mm diameter. For the analysis of trace metals in pressed samples, the analytical signal is obtained from only the surface few millimeters of sample (depending on the analyte and sample density). Consequently, sample thickness is typically increased until there is no increase in signal intensity, i.e., until the sample is ‘infinitely thick’.
Despite previous studies demonstrating the utility and economy of EDXRF in the analysis of Zn and Fe in food-stuffs and cereals, the technique has never been applied by plant breeders, either because large scale breeding programs are relatively recent or because of the requirement of existing protocols for samples to be ground and pelletized. We postulated that EDXRF on whole grain rice and pearl millet could provide a convenient, reliable screening tool in biofortification breeding programs. This hypothesis was tested by developing a calibration relationship between EDXRF counts and ICP-OES-determined values for Zn and Fe concentration in rice and pearl millet grain. We then tested EDXRF calibrations on validation sets selected at random from rice and pearl millet biofortification breeding populations.
Materials and methods
Trial samples
Given the unavailability of whole grain reference samples, it was necessary to develop a set of calibration and validation standards for this study. Rice and pearl millet samples were sourced from breeding programs connected to HarvestPlus. Rice was obtained from the International Rice Research Institute, Philippines, and the Bangladesh Rice Research Institute, Bangladesh. All rice samples had been polished, as this is the form in which rice samples are typically eaten, and consequently the form in which micronutrient content is assessed. Pearl millet was sourced from the International Center for Agricultural Research in the Dry Areas, India. Samples were sterilized by irradiation at 50 kGray (5 Mrad) upon entry to Australia. Reference values were determined by duplicate ICP-OES analysis using a closed-tube nitric acid/hydrogen peroxide digestion method. This method has been shown to give full recovery of Zn and Fe from plant tissues, with variability of 2–6% relative standard deviation (RSD) (Wheal et al. 2011). Approximately 0.8 g (45 rice grains, or 60 pearl millet grains) was used for each ICP-OES analysis. All samples contained < 4 mg kg−1 Al; according to HarvestPlus internal standards, this indicated samples could be generally regarded as being free of soil contamination.
EDXRF
EDXRF was performed using an Oxford Instruments X-Supreme 8000 fitted with a 10 place autosampler. Measurement conditions were as recommended by the manufacturer for analysis of Zn and Fe in a cellulose matrix (see Table 1). Total analysis time for each sample was 186 s, which included 60 s acquisition times for the separate Zn and Fe conditions as well as 66 s ‘dead time’ during which the EDXRF established each measurement condition. Scans were conducted in sample cups assembled from 21 mm diameter Al cups combined with polypropylene inner cups sealed at one end with 4 μm Poly-4 XRF sample film. Cups containing samples were gently shaken to evenly distribute grains. Preliminary studies showed that a sample depth of ≥ 6 mm was required for maximum recovery of Zn signal in both rice and pearl millet, with ≥ 3 mm required for Fe. The difference between Zn and Fe was expected since Zn emissions are of higher energy and therefore more penetrative than Fe emissions. In 21 mm cups, minimum depths equated to ≥ 4 g of grain for Zn analysis, and ≥ 2 g of grain for Fe analysis, so sample mass was fixed at 4 g. According to the manufacturer, the X-Supreme 8000 scans a circle of 21 mm diameter with the sample spinner on. All scans in this study were performed in this mode, so the scanned area was 346 mm2. Sampling volumes were therefore ~2.1 cm3 for Zn and ~1.0 cm3 for Fe analysis. For Zn and Fe these volumes equated to about 100 and 50 rice grains, respectively, or 120 and 60 pearl millet grains.
Statistical definitions
Statistical definitions and calculations were based on the methods of Perring and Andrey (2003).
Reference method (ICP-OES) | y i | |
Alternative method (EDXRF) | \( {\hat{y}_i} \) | |
Number of coefficients used in the calibration equations | p | |
Bias | \( \frac{{\Sigma_{i = 1}^n\left( {{{\hat{y}}_i} - {y_i}} \right)}}{n} \) | |
Standard error of calibration (SEC) | \( \sqrt {{ \frac{{\Sigma_{i = 1}^n{{\left( {{{\hat{y}}_i} - {y_i}} \right)}^2}}}{{n - p - 1}}}} \) | |
Standard error of prediction (SEP) | \( \sqrt {{\frac{{\Sigma_{i = 1}^n{{\left( {{{\hat{y}}_i} - {y_i}} \right)}^2}}}{n}}} \) | |
Limits of quantification (LOQ) were calculated according to the following (Perring and Andrey 2003):
where N = noise of instrument, S = signal, and [analyte] = concentration of the analyte (mg kg−1).
Calibration and validation
The range of Zn and Fe concentrations occurring in the rice and pearl millet samples either did not overlap (Fe), or scarcely overlapped (Zn). Based on the principle that the samples used for empirical calibration should be as similar as possible to test samples in their composition, morphology and concentration range, separate calibrations were conducted for rice and pearl millet. Twenty-four calibration samples were used in each case. Samples were chosen to ensure a wide range of Zn and Fe concentrations was represented for each species and that there was an approximately even spread of concentrations for both elements. All calibration samples contained Zn and Fe at or above the LOQ.
To calibrate the EDXRF instrument, reference (i.e., ICP-OES-determined) concentrations were entered into the machine before each sample was scanned. Clean Poly-4 film was used for each sample. The relationship between X-ray fluorescence and reference values was then established using the EDXRF calibrate function and a simple linear model. To validate calibrations, a further 40 rice and pearl millet samples were randomly selected from breeder’s populations and scanned once using EDXRF. Correlations between EDXRF and reference values were then examined, along with bias and standard errors of prediction (SEP), based on data from this single EDXRF scan. To investigate EDXRF precision, each validation sample was analysed a second time, allowing standard deviations and RSD to be calculated from the duplicate data of each sample. For comparison with ICP-OES performance, the same calculations were performed on duplicate ICP-OES values.
Results
Limits of quantification
The estimated LOQ for Zn and Fe in rice were ~12 and ~3 mg kg−1, respectively, while the corresponding LOQ in pearl millet were ~16 and ~5 mg kg−1, respectively. For Zn and Fe in rice, and for Zn in pearl millet, samples used to determine the LOQ values contained the analyte of interest at approximately the same concentration as the LOQ; for Fe in pearl millet, the sample used to estimate LOQ contained Fe at a higher level (18 mg kg−1), as this was the lowest Fe sample available.
Calibration
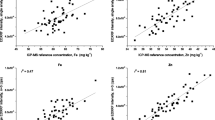
Strong correlations were observed between EDXRF counts and Zn and Fe concentration in both rice and pearl millet, with all calibration plots displaying coefficients of determination (r2) ≥ 0.85 (Table 2 and Fig. 1). The values of r2 were smaller for rice than pearl millet, presumably because of the smaller Zn and Fe concentrations in rice. Standard errors of calibration (SEC), which are based on the differences between each reference value and those calculated using the calibration model, were 1.3 and 0.6 mg kg−1 for Zn and Fe in rice, and 3.6 and 6.3 mg kg−1 for Zn and Fe in pearl millet. The lower SEC in rice were presumably due to the lower concentrations of Zn and Fe present. The strength of calibration relationships was similar to those seen in the EDXRF analysis of Zn and Fe in pressed milk powders and cereal samples (Perring and Andrey 2003). This, and the small SEC relative to the range of concentrations present, indicated validation steps were warranted for all four calibrations.
Because grain morphology and Zn and Fe concentration ranges differ between rice and pearl millet, calibrations were kept ‘species-specific’ in this study. However, to investigate the possibility of using a single calibration to analyse rice and pearl millet grain, calibration plots of the two species were overlaid (Fig. 2). For both Zn and Fe, the relationship between X-ray counts and reference values appeared subtly different between the two species.
Validation
The range of concentrations of Zn and Fe present in the validation population was similar to that used for calibration (Table 3). Similarly strong correlations were observed between EDXRF and reference results as in the calibration set, with all r2 ≥ 0.79 (Fig. 3). Uncertainty statistics for Zn and Fe (in this case, SEP, based on the differences between EDXRF and reference values) were similar to those seen in calibration, namely at 1.0 and 0.8 mg kg−1 for rice, and 3.9 and 6.4 mg kg−1 for pearl millet. Ninety-five percent confidence intervals, calculated from SEP, indicate that rice EDXRF results can be expected to be within ± 1.9 mg kg−1 of ICP-OES-Zn, and ± 1.6 mg kg−1 of ICP-OES-Fe, while pearl millet EDXRF results can be expected to be within ± 7.6 mg kg−1 of ICP-OES-Zn, and ± 12.5 mg kg−1 of ICP-OES-Fe.
Bias was estimated for each validation set by subtracting each EDXRF value from the corresponding ICP-OES value, and averaging these differences (Table 3). Bias values were all < 1 mg kg−1 and, according to paired t tests, were not significantly different from zero.
To investigate EDXRF precision, scans were repeated on both rice and pearl millet validation sets. This allowed standard deviations (SD) and RSD to be calculated for each sample (Table 3): SD averaged 0.4 and 0.3 mg kg−1 for Zn and Fe in rice, and 1.9 and 1.8 mg kg−1 for Zn and Fe in pearl millet, equating to average RSD values for Zn and Fe of 2.5 and 6.2% in rice, and 2.8 and 1.9% in pearl millet. The same calculation performed on duplicate ICP-OES results for the validation set revealed Zn and Fe SD to be 0.4 and 0.2 mg kg−1 in rice, and 2.7 and 2.5 mg kg−1 in pearl millet, equating to RSD of 2.7% and 4.6% for Zn and Fe in rice, and 3.7 and 2.8% for Zn and Fe in pearl millet.
Discussion
The calibration relationships between X-ray counts and ICP-OES concentrations for Zn and Fe in whole grain rice and pearl millet were strong and justified validation. Validation showed that a single EDXRF scan predicts ICP-OES-Zn and Fe concentration to within 1.0 and 0.8 mg kg−1 for rice, and within 3.9 and 6.4 mg kg−1 for pearl millet. These uncertainties equated to 95% confidence intervals of ± 1.9 and ± 1.6 mg kg−1 for Zn and Fe in rice, and ± 7.6 and ± 12.5 mg kg−1 in pearl millet. LOQ values across rice and pearl millet were 12–16 mg kg−1 for Zn and 3–5 mg kg−1 for Fe. The micronutrient concentrations typically encountered in rice biofortification programs are in the range 15 to 27 mg kg−1 for Zn and 4 to 10 mg kg−1 for Fe, while in pearl millet programs the range is from around 50 to 110 mg kg−1 for Zn and 60 to 160 mg kg−1 for Fe [see Pfeiffer and McClafferty (2007), as well as the validation populations of this study]. The performance of EDXRF is therefore clearly sufficient to allow high micronutrient lines to be separated from low nutrient lines in these breeding populations. As an example of how EDXRF may be used, rice breeders seeking high Zn lines could use EDXRF to select for the approximately 10% of lines with Zn > 20 mg kg−1, with the expectation that > 95% of these lines would have Zn > 18 mg kg−1. Similarly, pearl millet breeders seeking high Fe genotypes could use EDXRF to screen for the 10% of lines with Fe > 120 mg kg−1, and have confidence that > 95% of these would lines contain Fe at > 106 mg kg−1. Only the selected nutrient dense lines would then be retained for further study. For extra confidence, high nutrient lines and checks could be re-tested by ICP-OES. In practice, accuracy of selection would probably be greater than outlined here since breeders make their decisions based on averaged data from replicated trials at multiple locations.
The current cost of ICP-OES analysis to rice and pearl millet breeding programs is in the range $US 7–15 per sample, depending on the laboratory used, and many thousands of samples are tested annually. The recommended retail price for the EDXRF used here is in the range $US 70,000–80,000, and the cost of consumables is negligible (approx. $US 0.15 per sample if film is changed for each sample). Analytical time was 31 min for each batch of ten samples, equating to a daily throughput of about 160 samples in an 8 h day. Thus a machine analyzing just 10,000 samples (possible in a few months in a single breeding season) would save enough money in ICP-OES analytical costs to pay for itself. Another general advantage of EDXRF is that technicians can be trained to conduct this form of analysis in a matter of days, as the technology requires considerably less analytical skill, and less sample preparation, than ICP-OES. The technology is therefore well suited to less developed settings. A final advantage of in-house EDXRF to HarvestPlus biofortification programs is that samples would no longer need be sent abroad, avoiding lengthy delays (often months) in quarantine and processing.
A disadvantage of the method presented here, relative to ICP-OES analysis, is that the elements typically used to check for soil contamination in grain, Al, Ti and Cr, are either too light (Al), or present at levels too low to be detected by EDXRF (Ti, Cr). This is not a major concern for Zn analysis, since soil and dust do not usually contain practically significant amounts of Zn, but may be of concern for Fe analysis, since soil and dust often contain enough Fe to compromise screening programs. It is therefore likely that most programs using EDXRF will still utilize some level of ICP-OES analysis to check for soil contamination, as well as to confirm lines predicted by EDXRF to have high Zn and/or Fe concentrations are indeed nutrient-dense.
The strategy of keeping calibrations ‘species-specific’ in this study was used to maximize the chance of developing bias-free calibrations and allow performance statistics to be examined separately in the two species. The strategy was further vindicated by the finding that Zn and Fe calibration relationships appeared subtly different in the two species. This suggests that whatever differences exist between rice and pearl millet in grain size and nutrient distribution only led to minor differences in Zn and Fe fluorescence. Nonetheless, it appears that whole grain calibrations for measuring Zn and Fe in rice and pearl millet should remain species-specific.
There are a range of sources of error in EDXRF analysis which contribute to measurement uncertainty and overall SEC and SEP, including errors associated with sample preparation (Blank and Eksperiandova 1998; Injuk et al. 2006), as well as less significant ‘machine errors’ (e.g. variation in sensitivity and machine drift). Since samples were analysed as whole grains in this study, sample inhomogeneity was expected to be a major source of error. However, EDXRF repeatability was found to be high (EDXRF produced RSD values of 3–6%, of similar magnitude to the RSD values found for ICP-OES). Further, the overall SEC and SEP observed in this study were similar to those seen previously in the EDXRF analysis of pressed milk-powders (where SEP were approximately 3 mg kg−1 for both Zn and Fe, see Perring and Andrey 2003). Thus, the combined effects of sample and machine error in this study were typical of those seen when ground, pressed samples are analysed, and whole grain samples can be concluded to be homogenous enough to give reliable EDXRF results. The small grain size of rice and pearl millet, and the fact some 50–100 grains contribute to Zn and Fe X-ray signal in EDXRF of these species (see Methods), would have been major factors that helped overcome sample inhomogeneity.
To minimize error in the reference values of this study, ICP-OES analyses were conducted in duplicate. This same approach was used to minimize reference value error in the standards developed by Perring and Andrey (2003), Perring et al. (2005), Perring and Blanc (2007) and Perring and Blanc (2008). However, minor errors would still have existed in reference values due to sample inhomogeneity and ICP-OES uncertainty. The main effect of these errors in this study would have been to inflate SEC and SEP statistics. Indeed, given the SD of reference values was similar in magnitude to standard errors of calibration and validation, it is likely that reference data uncertainty was a major contributor to SEC and SEP statistics. Thus the true SEC and SEP of Zn and Fe EDXRF, and the derived 95% CI, are likely to be smaller than reported here.
Given the various uncertainties described here, a number of different avenues could be explored to improve EDXRF performance, including analysis of ground and pressed samples, replicating EDXRF analyses rather than relying on a single scan, and conducting more replicates of ICP-OES analysis to improve reference values. Another possibility to reduce machine error would be to increase scanning time beyond 60s to reduce counting errors in X-ray detection. However, all these approaches would reduce analytical throughput. An alternative approach that may be taken to increase throughput, albeit with increased measurement uncertainty, would be to reduce scanning time from 60 s to 30 s. This approach reduces analytical time from 186 s to 126 s (i.e., from about 160 samples in an 8 h day to 230) and, in the authors’ hands, only increases SEP by < 0.2 mg kg−1 for Zn and Fe in rice, and by 0.2 mg kg−1 for Zn and 1.4 mg kg−1 for Fe in pearl millet. Perring and Monard (2010) also found that EDXRF analysis of Zn and Fe in powdered foodstuffs could be conducted at 30 s rather than 60 s without appreciable loss of performance. For programs only interested in a single micronutrient (i.e., Zn or Fe, but not both), another approach to increasing throughput would be to program the EDXRF to scan for just the nutrient of interest (i.e. using just one set of analytical conditions), reducing analytical time by half. This would allow the analysis of 320 samples in an 8 h day at 60 s scanning time, or 460 samples a day at 30 s. Though clean cups and film was used for each sample in this study, a final approach breeders could use to increase throughput and reduce labour is to use cup and film assemblies multiple times. Provided samples are relatively clean, and cups are tapped sharply up-side-down on a clean surface after each sample to remove dust, we have found it possible to re-use assembled cups up to ten times without detectable cross-contamination.
The demonstration here that EDXRF has great potential for Zn and Fe analysis in whole grain rice and pearl millet raises the possibility that the technique could be used on grains of other species, or for other mineral elements. For whole grain EDXRF to work, an analyte’s XRF signal must be both detectable and generated from enough of the sample to be representative. It is therefore expected that other cereal species with similar sized seeds to rice and pearl millet, and similar Zn and Fe concentration ranges [e.g., wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.)], will be as suited to whole grain EDXRF as rice and pearl millet. For crops with larger grains, such as maize (Zea mays L.) and common bean (Phaseolus vulgaris L.), it is expected samples would need to be ground prior to analysis. Preliminary data for wheat grain and bean flour support this hypothesis. Regarding other elements, it has already been shown that different forms of XRF may be used for the analysis of a range of plant essential elements beside Zn and Fe in ground, pressed samples, including Ca, Cl, Cu, K, Mg, Mn, P, S and Si (Kocman et al. 1991; Frank et al. 1992; Clark et al. 1992). For elements lighter than Fe, success with EDXRF of whole grains could not be assumed, since signal from those lighter elements would be weaker and may only come from the outer layers of grains, necessitating grinding. Chances of success with whole grains would be greater with Cu and Mn, since these elements have similar atomic weight to Fe and are present at similar or higher concentrations. Finally, for heavier elements of interest in the biofortification sector, e.g. Se and I, whole grain EDXRF may be viable but experiments are needed to determine whether the concentrations present in biofortified samples are above the limits of quantification.
Conclusions
The results show that EDXRF offers a convenient, economical tool for screening Zn and Fe concentration in rice and pearl millet biofortification breeding programs. Because of these advantages, EDXRF is expected to largely replace ICP-based and colorimetric methods in the large scale screening steps of these programs. We also envisage EDXRF-based screens will allow rice and pearl millet biofortification breeding programs to expand to include more lines for faster genetic gains, and allow the wider inclusion of Zn and Fe density as traits of interest in other rice and pearl millet breeding programs. EDXRF analysis of Zn and Fe may also find increased use in the crop sciences outside biofortification programs, in other situations where high throughput is required, for example, in the mapping of Zn and Fe density traits in different crops.
It is highly likely that the method described here will work equally well in whole grain wheat and barley, and that analysis of flour, if not grain, will be feasible in larger grained crops such as maize and beans. Thus, we predict that EDXRF will be a significant enabling technology in the global Zn and Fe biofortification effort and that this role will be enhanced as instruments become cheaper and more sensitive in future. Further work is needed to investigate which other elements can be analysed in whole grain samples of different crops, but the approach is likely to work best for elements with atomic weights similar to or heavier than Zn and Fe.
References
Arai T (2006) Introduction. In: Beckhoff B, Kanngießer B, Langhoff N, Wedell R, Wolff H (eds) Handbook of practical X-ray fluorescence analysis. Springer, Berlin Heidelberg, pp 1–31
Blank AB, Eksperiandova LP (1998) Specimen preparation in X-ray fluorescence analysis of materials and natural objects. X-ray Spectrom 27:147–160
Choi EY, Graham R, Stangoulis J (2007) Semi-quantitative analysis for selecting Fe- and Zn-dense genotypes of staple food crops. J Food Compos Anal 20:496–505
Clark RB, Frank KD, Zaifnejad M, Denning J (1992) X-ray fluorescence analysis of small leaf samples mixed with cellulose or boric acid. Commun Soil Sci Plant Anal 23:569–583
Ekinci N, Ekinci R, Sahin Y (2002) Determination of iodine and calcium concentrations in the bread improver using ED-XRF. J Quant Spectrosc Radiat Transf 74:783–787
Frank KD, Burch J, Denning J (1992) Mineral analysis of corn leaves by X-ray fluorescence on ground versus unground leaf samples. Commun Soil Sci Plant Anal 23:2415–2424
Injuk J, Van Grieken R, Blank A, Eksperiandova L, Buhrke V (2006) Specimen preparation. In: Beckhoff B, Kanngießer B, Langhoff N, Wedell R, Wolff H (eds) Handbook of practical X-ray fluorescence analysis. Springer, Berlin Heidelberg, pp 411–432
Kocman V, Peel TE, Tomlinson GH (1991) Rapid analysis of macro and micro nutrients in leaves and vegetation by automated x-ray-fluorescence spectrometry (a case study of an acid-rain affected forest). Commun Soil Sci Plant Anal 22:2063–2075
Melquiades FL, Appolini CR (2004) Application of XRF for environmental analysis. J Radioanal Nucl Chem 262:533–541
Nestel P, Bouis HE, Meenakshi JV, Pfeiffer W (2006) Biofortification of staple food crops. J Nutr 136:1064–1067
Noda T, Tsuda S, Mori M, Takigawa S, Matsuura-Endo C, Kim S-J, Hashimoto N, Yamauchi H (2006) Determination of the phosphorus content in potato starch using an energy-dispersive X-ray fluorescence method. Food Chem 95:632–637
Ozturk L, Yazici MA, Yucel C, Torun A, Cekic C, Bagci A, Ozkan H, Braun HJ, Sayers Z, Cakmak I (2006) Concentration and localization of zinc during seed development and germination in wheat. Physiol Plant 128:144–152
Perring L, Andrey D (2003) ED-XRF as a tool for rapid minerals control in milk-based products. J Agric Food Chem 51:4207–4212
Perring L, Andrey D, Basic-Dvorzak M, Hammer D (2005) Rapid quantification of iron, copper and zinc in food premixes using energy dispersive X-ray fluorescence. J Food Compos Anal 18:655–663
Perring L, Blanc J (2007) EDXRF determination of iron during infant cereals production and its fitness for purpose. Int J Food Sci Technol 42:551–555
Perring L, Blanc J (2008) Validation of quick measurement of mineral nutrients in milk powders: comparison of energy dispersive X-ray fluorescence with inductively coupled plasma-optical emission spectroscopy and potentiometry reference methods. Sens Instrum Food Qual 2:254–261
Perring L, Monard F (2010) Improvement of Energy Dispersive X-Ray Fluorescence throughput: influence of measuring times and number of replicates on validation performance characteristics. Food Anal Methods 3:104–115
Pfeiffer WH, McClafferty B (2007) Biofortification: Breeding micronutrient-dense crops. In: Kang MS, Priyadarshan PM (eds) Breeding major food staples. Blackwell, Ames, pp 61–91
Prom-u-thai C, Dell B, Thomson G, Rerkssem B (2003) Easy and rapid detection of iron in rice seed. Sci Asia 29:314–317
Rousseau RM, Willis JP, Duncan AR (1996) Practical XRF calibration procedures for major and trace elements. X-ray Spectrom 25:179–189
Velu G, Ortiz-Monasterio I, Singh RP, Payne T (2011) Variation for grain micronutrients concentration in wheat core-collection accessions of diverse origin. Asian J Crop Sci 3:43–48
West M, Ellis AT, Potts PJ, Streli C, Vanhoof C, Wegrzynek D, Wobrauschek P (2009) Atomic spectrometry update. X-Ray fluorescence spectrometry. J Anal At Spectrom 24:1289–1326
West M, Ellis AT, Potts PJ, Streli C, Vanhoof C, Wegrzynek D, Wobrauschek P (2010) Atomic spectrometry update–X-Ray fluorescence spectrometry. J Anal At Spectrom 25:1503–1545
Wheal MS, Fowles TO, Palmer LT (2011) A cost-effective acid digestion method using closed polypropylene tubes for inductively coupled plasma optical emission spectrometry (ICP-OES) analysis of plant essential elements. Anal Methods (in press) doi:10.1039/c1ay05430a
Zarcinas BA, Cartwright B, Spouncer LR (1987) Nitric acid digestion and multi-element analysis of plant material by inductively coupled plasma spectrometry. Commun Soil Sci Plant Anal 18:131–146
Acknowledgements
Funding for this work was provided by HarvestPlus. We thank Oxford Instruments and Neal Robson for help identifying optimal Zn and Fe EDXRF conditions, Parminder Virk and Alamgir Hossain for supplying rice samples, Kedar Rai for supplying pearl millet samples, Waite Analytical Services for ICP-OES analysis, Zarina Yasmin for technical assistance, and Robin Graham and Wendy Telfer for helpful suggestions on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Ismail Cakmak.
Rights and permissions
About this article
Cite this article
Paltridge, N.G., Palmer, L.J., Milham, P.J. et al. Energy-dispersive X-ray fluorescence analysis of zinc and iron concentration in rice and pearl millet grain. Plant Soil 361, 251–260 (2012). https://doi.org/10.1007/s11104-011-1104-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-011-1104-4