Abstract
Plant growth promoting rhizobacteria (PGPR) confer disease resistance in many agricultural crops. In the case of Bacillus subtilis (UFLA285) isolated from the cotton producing state of Mato Grosso (Brazil), in addition to inducing foliar and root growth, disease resistance against damping-off caused by Rhizoctonia solani was observed. The aim of this cotton study was to identify gene transcriptional events altered with exposure to the PGPR strain UFLA285 in infected plants. Global gene transcription was profiled using a commercially-available cotton gene chip; cotton plants with and without UFLA285-seed treatment were infected with R. solani 9-days after planting and harvested on day14. Microarray data of stem tissue revealed 247 genes differentially regulated in infected plants, seed treated versus untreated with UFLA285. Transcripts encoding disease resistance proteins via jasmonate/ethylene signaling as well as osmotic regulation via proline synthesis genes were differentially expressed with UFLA285 induction. Consistent with transcriptional regulation, UFLA285 increased plant-proline accumulation and dry weight. This study has identified transcriptional changes in cotton, induced by the beneficial soil bacterium UFLA285 and associated with disease control.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil microbes adapt to particular rhizosphere conditions during as well as between successive growing seasons for crops such as cotton that often are grown for several years without crop rotation (Malik et al. 2005; Hulugalle and Scott 2008). Hence a cotton pathogen such as Rhizoctonia solani that is able to overwinter as dormant sclerotia, clamidospores or in a saprofic form (Manian and Manibhushanrao 1990) can cause substantial harvest losses as soil-pathogen titers increase (Kirkpatrick and Rothrock 2001). Such a plant pathogen interferes with xylem flow, simulate water-stress conditions and result in greater plant susceptibility to drought or salt stress (Dowd et al. 2004). While sustained efforts have been underway to breed for abiotic stress tolerance in cotton (Parida et al. 2008), few programs have targeted soil-borne pathogen resistance (Lopez-Lavalle et al. 2007).
A complex and integrated set of plant responses are mobilized in cotton with pathogen attack including constitutive and induced defenses (Martinez et al. 2000). Characterized plant signals that activate such defense responses include ethylene, salicylic acid, jasmonic acid and reactive oxygen species; downstream defenses responses such as the production of phenolics, phytoalexins, antimicrobial peptides and physical barriers have also been characterized. Priming of plant defenses has been shown to be an effective means for the tailoring of induced resistance against plant pathogens (Yi et al. 2009). Local resistance (LR) and systemic acquired resistance (SAR) are generally activated by plant-pathogen interactions and accompanied by elevated levels of endogenous salicylic acid (SA) (Dorey et al. 1997). Induced systemic resistance (ISR) is also a heightened plant defense state although exogenous signaling is via non-pathogenic/beneficial bacteria with elevated endogenous jasmonic acid (JA) and ethylene often reported as a secondary signal (Pieterse et al. 2002, 2007). While such bacteria can directly function as antibiosis agents in the rhizosphere, root colonization can also trigger changes in plant transcript profiles associated with indirect disease suppression (Dowd et al. 2004).
Many plant growth promoting rhizobacteria (PGPR) reduce disease symptoms in a wide range of agricultural crops (Mondal and Verma 2002). Model systems have been employed to unravel mechanisms of plant inducible biotic and abiotic stress-tolerance activated by PGPR. In the case of Pseudomonas fluorescens root colonization in Arabidopsis, disease suppression against P. syringae DC3000 is observed with up—(95) and down—(105) regulation for transcripts encoding metabolism, signal transduction, and stress response proteins (Wang et al. 2005). With PGPR-induced salt tolerance, the sodium uptake transporter HKT1 has been shown to be organ specifically regulated in Arabidopsis by B. subtilis resulting in reduced endogenous sodium when plants are grown with elevated salt (Zhang et al. 2008). For growth promotion, PGPR have been linked with auxin redistribution from leaves to roots that in turn is associated with foliar cell expansion and lateral root proliferation in Arabidopsis (Zhang et al. 2007; Xie et al. 2009).
With cotton, a microarray chip was originally developed based on infected plant ESTs to study the interaction of cotton plant and Fusarium oxysporum f.sp. vasinfectum infection (Dowd et al. 2004). The authors observed defense- as well as drought tolerance-related genes to be up-regulated. Recently, an enhanced microarray chip was generated containing all deposited Gossypium spp ESTs (Udall et al. 2007) and this enlarged data set has been utilized here to study the tritrophic interaction between cotton, the common cotton pathogen R. solani AG4 and the newly reported beneficial rhizobacterium B. subtilis (UFLA285) isolated from the cotton rhizosphere (Medeiros et al. 2008). To probe plant-signaling pathways activated by UFLA285 seed treatment in combination with damping-off disease, overall transcript changes have been characterized using the Gossypium spp microarray chip. This global gene profiling was employed to identify defense- as well as stress-related genes that are differentially regulated in pathogen infected cotton with versus without UFLA285 seed treatment.
Material and methods
Bacterial cultures
Bacillus subtilis UFLA285 previously selected for the control of cotton seed-borne diseases (Medeiros et al. 2008) was streaked from −80°C preserved slants to LB agar; 24 h post-incubation at 28°C, isolated colonies were cultured in liquid LB media. To separate cells from the media, the suspension was centrifuged at 10,000 g for 5 min. Cells were re-suspended in saline buffer (0.85% NaCl) and the concentration set to 109 cfu/mL based on optical density measurements inferred from a standard curve (Jeger and Viljanen-Rollinson 2001).
Fungal cultures
The pathogen Rhizoctonia solani AG4 was initially frozen at −80°C as sclerotia in 40% aqueous glycerol, transferred to potato dextrose agar (Difco Laboratories, Detroit, MI) and after 7 days at 24°C, 5 mycelium plugs from the colony extremity were inoculated into V8 medium (200 mL commercial V8 juice with 3 g/L CaCO3). This suspension was incubated at 24°C and 150 rpm for 4 days and the mycelium was then harvested by centrifugation, re-suspended in water and blended to give a homogenous suspension. The threshold concentration of 102 cfu/mL required for wilting in all plants 4 days after inoculation was empirically determined; this concentration was then employed in all trials where pathogen exposure was provided. Unless otherwise stated, 9 days after planting, stems were infected with the pathogen suspension (250 μL, 102 cfu/mL) by deposition at the root/stem inter-phase in direct contact although without plant wounding.
Plant material
Gossypium hirsutum seeds cv Deltapine Acala 90 were surface sterilized in sodium hypochloride (0.5% active chloride), washed thoroughly with sterilized distilled water, air dried and then treated with the bacterial suspension (2 mL 109 cfu/mL/g of seed), saline (2 mL saline buffer 0.85% NaCl (w/w)/g of seed) or fungicide (triadimenol 10 μL active ingredient/g of seed). Seeds were sown into 2 L pots containing 400 g of the potting mix Sunshine® All-Purpose Planting Mix (Sun Gro Horticulture, Vancouver, CA), fertilized with 5 g of osmocote fertilizer (Scotts-Sierra Horticulture, Marysville, OH) and irrigated daily to field capacity. Plants (4/pot) were kept in a growth room under controlled temperature (25°C ± 4), relative humidity 40 ± 10% and light (200 μmol m−2 s−1) by using a combination of metal halide and high sodium pressure lamps set for 14 h/day.
Disease assessment
At 4–10 days after germination, plants were assessed daily for disease severity according to a 1–5 numerical scale as previously described (Keinath et al. 2000), where (1) represents no visible symptoms, (2) less than 10 pinpoint lesions on diffuse discolored areas, (3) distinct necrotic lesions, (4) girdling lesions and (5) damped-off or killed seedling. The obtained data were analyzed collectively by an area under the disease progress curve (Shaner and Finney 1977).
Plant sampling and RNA extraction
At 10, 11, 12 and 13 days after planting, stems and roots were harvested, separately water rinsed, and frozen in liquid nitrogen. Plant material was stored at −80°C until time of RNA extraction in which tissue (0.2 g) was mortar-and-pestle ground with liquid nitrogen and extracted by a hot borate protocol (Wan and Wilkins 1994). Total RNA was purified using an RNEasy MinElute Cleanup kit (Qiagen, Valencia, CA) including the RNase-free DNAse treatment step from the same manufacturer. RNA was quantified and stored at −80°C.
RT-PCR analysis
First strand cDNA was synthesized from 5 μg of total RNA following a previously described protocol (Zhang et al. 2007) and PCR was performed using (5′-3′) primers, designed based on deposited sequences (Table 1). Agarose gel electrophoresis images were taken by Kodak Gel Logic 100 Imaging System (Fisher Scientific, Houston, TX) and band intensity quantified by Image J 1.33u (http://rsb.info.nih.gov/ij/, National Institute of Health).
Microarray analysis
Microarray hybridizations consisted of four biological replicates (AG4 infected plants with or without UFLA285 as a biological control treatment) with one dye swap. Extracted RNA was transcribed to aRNA in a three step transcription using amino-allyl aRNA amplification kit (Ambion, Austin, TX) and labeled with NHS dyes Cy3 or Cy5 (Amersham Biosciences, Little Chalfont Buckinghamshire, UK) according to the manufacturer’s protocol. Slide pre-hybridization was performed according to the manufacturer’s instructions, whereas hybridization and post-hybridization followed an Arabidopsis protocol (Zhang et al. 2007). Arrays were scanned using a GenePix 4100 array scanner (Axon Instruments, Sunnyvale, CA). Spot statistical analysis was performed according to the manufacturer’s guidelines (Gene-Spring 7.0; Silicon Genetics, Redwood, CA). A 40% change, either up- or down-regulation, in the expression level compared with the control was designated as the threshold for a gene to be classified as altered in response to rhizobacterium treatment. Only genes that passed the flag filtering, identified as present (Gene-Spring 7.0), and passed the t-test p-values 0.10 were considered differentially regulated with UFLA285 treatment.
Proline quantification
For endogenous proline measurements, shoot tissue was extracted in 3% aqueous sulfosalicilic acid and vortexed for 1 min; the solution was then centrifuged (10,000 g) for 10 min and the supernatant (0.5 ml) diluted 1:1 with water following the procedure of Bates et al. (1973). Extracted proline was stained with Ninhydrin reagent (Sigma-Aldrich, St. Louis, MO). Ninhydrin (1.25 g) was dissolved in glacial acetic acid (30 mL) and 6 M phosphoric acid at 40 C until complete dissolved, heated to 100°C for 1 h in darkness, ice-bath cooled and extracted with toluene. The organic phase was measured at 520 nm and quantification is based on a proline-ninhydrin standard curve.
Statistical analysis
Whenever applicable, plots were composed of four plants. Those plants were either pooled for RNA extraction or the measured severity and photosynthesis averaged and each experiment was composed in order to reduce errors and the experiments encompassed three biological replicates. The obtained data was submitted to variance analysis ANOVA and for significant effects (p < 0.05), means were compared according to Turkey’s test using the SAS software (SAS Institute, Cary, NC).
Results
Damping-off resistance kinetics
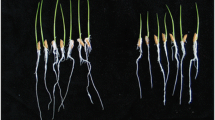
To establish the incubation time required for UFLA285-associated plant protection against R. solani, cotton seedlings were pathogen inoculated at different times after planting (Fig. 1a). At 9 days after planting, pathogen inoculated seedlings in combination with UFLA285 seed treatment resulted in lower disease symptoms compared to water control. Pathogen inoculated seedlings in combination with fungicide seed treatment resulted in lower disease symptoms at all days that were assayed (9–12 days) (Fig. 1b). As expected, light brown stem lesions, typical of R. solani infection (Kirkpatrick and Rothrock 2001), were observed 4 days after inoculation (Fig. 1c) and used to determine the area under the disease progress curve (AUDPC). The plant-infected fungus was positively identified by plating the root/shoot inter-phase on potato dextrose agar amended with streptomycin (100 ppm). All subsequent experiments were pathogen inoculated 9 days after planting.
Bacillus subtilis (UFLA285) incubation requirement for induced cotton protection against Rhizoctonia solani (AG4). Seeds were treated at the time of planting and germinated seedlings were inoculated with AG4 (+ pathogen) at 5, 6, 7, 8 or 9 days after planting (a). Disease severity was quantified based on an area under the disease progress curve (AUDPC) (Shaner and Finney 1977) with seeds treatments of water (white bars), UFLA 285 (black bars) or the commercial fungicide triadimenol (gray bars). Bars represent the mean ± SE with n = 4; bars marked with different letters within a given time period are statistically different based on a Turkey’s test p ≤ 0.05 (b). Seventeen day old cotton plants infected 9 days after planting exhibit brownish necrotic lesions (see arrow) at the root/shoot inter-phase typical of (AG4) damage (c)
Defense-related gene expression
Expression of established defense-related genes was up-regulated in an organ and time-dependent manner with UFLA seed treatment versus untreated controls (Fig. 2). A rapid ethylene-inducible protein, CD486177, was activated ca. 5 fold in shoots at 10 days after planting, while root expression was induced ca. 3 fold only at 13 days after planting. In contrast, peroxidase, induction was observed at day 13 with ca. 3-fold increase in both shoots and roots. Chitinase also exhibited a delayed induction, observed on day 12 and 13, for shoot and root, respectively.
Bacillus subtilis (UFLA285) induction of defense-related genes in cotton shoots and roots. Bars represent the transcriptional ratios between UFLA285-treated and untreated plants with regard to the defense related genes: ethylene inducible protein (black bar), peroxidase (gray bar) and chitinase (white bar); shown is the mean ± SE with n = 4
UFLA285 gene induction in R. solani infected cotton
To provide insight into underlying mechanisms responsible for rhizobacterium-mediated cotton disease protection, cDNA microarray experiments were performed with pathogen-exposed plants treated with UFLA285 or water. A total of 247 genes were differentially expressed with UFLA285 treatment and putatively identified genes were divided into primary (Table 2) and secondary metabolism (Table 3). For each category, responses were subdivided by functional groupings with primary metabolism including: lipid-, carbohydrate-, protein-metabolism, gene replication, transport, respiration, flowering, photosynthesis, and unidentified genes. Genes associated with defense encompassed the largest group of secondary metabolites and were preliminarily divided into the groupings: cell-wall modulators, stress-related signals, signal transduction activators and transcription factors.
To validate microarray results, five genes that were differentially expressed with UFLA285 treatment were assayed by RT-PCR and similar fold changes were observed for both microarray and RT-PCR analysis (p > 0.05) (Fig. 3).
Expression level comparison for RT-PCR (black bars) and microarray (white bars) analysis of 13-day-old cotton shoots, AG4-infected on day 9, with and without UFLA285 seed treatment Selected genes include: lipoxygenase (LOX), cytochrome P450 (P450), xyloglucan endoglycosylase (XTH), aquaporin (AQUA), heat shock protein (HS), and ethylene response factor (ET); bars represent the mean ± SE with n = 4 (A). Representative RT-PCR gel band images are shown including ubiquitin (UBI) (b)
Genes associated with growth and development
Several genes associated with DNA replication were up-regulated (22 genes 95% of which were up-regulated) including: mini-chromosome maintenance protein (DNA replication initiation), unwinding of nucleic acid (helicase), ribonuclease III and helicase activities (ribonuclease), alternative splicing as well as single-stranded RNA binding (RNA recognition protein), double stranded RNA binding (C-terminal domain phosphatase-like 1), acetylation (histone), RNA replication (RNA polymerase), DNA polymerase (reverse transcriptase), ligase (ARIADNE-like protein), guanine tetraphosphate metabolic process (rel A/spo T homologous protein RSH2), and nucleic acid binding (small nuclear ribonucleoprotein).
A second set of genes encoding for the transport of amino acids (peptide transport and permease), chloride, potassium or sulfate ions were up-regulated with UFLA285 treatment while proton and copper transporters were down-regulated. Other proteins involved in trans membrane transport selective for lysine amino acids (coatomer protein complex epsin-like protein) (Holstein and Oliviusson 2005), exocytosis (exocyst dubunit EX070 family protein E1), ABC transporter involved in the import of molecules through the membrane envelope (importin beta2 subunit family protein), Golgi complex internal transport (got1-like family protein), endoplasmic reticulum and Golgi complex-mediated transport (sedlin, transport component particle) (Muthusamya et al. 2009) were differentially regulated. In cases where transporters were closely associated with components of primary metabolism, the transporter was grouped with the process: transporters that take part in mitochondrial respiration (e.g. mitochondrial phosphate translocator and mitochondrial carrier protein-like), replication of genetic material (e.g. purine permease) and transporters of heterocyclic nitrogen compounds (e.g. ureide permease 1).
Putative secondary metabolism associated responses
Among the genes with altered expression levels, defense responses were the largest number (12% of total) with all members except aquaporin and dehydroascorbate peroxidase up-regulated (fold change >1.4 and p-value ≤ 0.05). Defense genes were sub-grouped as anti-oxidant/scavengers, PR-proteins, jasmonic acid biosynthesis, phenylpropanoid pathway and osmoregulators (Table 3). Defense-related metabolism such as those belonging to the PR-protein group or part of the phenylpropanoid pathway are commonly reported as part of a plant’s induced systemic resistance response (Hammerschmidt and Kuc 1995).
Six genes encoding cell wall modulation including up-regulation of cell wall reinforcement (transferase, callose synthase and lipid transfer protein) and down regulation of cell wall loosening (xyloglucan hydrolase) were observed with UFLA285 treatment.
Genes encoding detoxifying enzymes were selectively up-regulated with UFLA285 elicitation including alcohol dehydrogenase, heat shock proteins, luminal binding proteins, and disulfide isomerase. After pathogen penetration, plants respond either by producing enzymes targeting the pathogen and/or toxic pathogen metabolites (Buchanan et al. 2000). Alcohol dehydrogenase catalyzes the two-way reaction from acetaldehyde to ethanol, which is produced under anoxic conditions, frequently observed on necrotrophic pathogen causing tissue maceration (Kronstad 2000).
Signaling events associated with plant defense were among the largest group of genes to be up-regulated (13% of total genes). This group was divided into five sub-groups: hormone-related, calcium-dependent, leucine rich repeat, lectin repeat, and serine/threonine kinases. Transcription factors were grouped in the sub-classes: hormone-related, WRKY, MYB, WD-40 and miscellaneous (Table 3).
Inducible proline accumulation
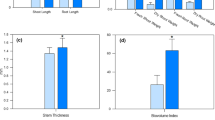
Inducible proline accumulation was examined with the observed differential regulation of a proline-synthesis enzyme with UFLA285 seed treatment (Table 3). With pathogen infection, a 1.5 times increase in proline accumulation was observed with UFLA285 (71 ± 9 μg/g shoot) versus untreated seeds (48 ± 4 μg/g shoot). Pathogen infected plants also had 1.9 times greater dry weight with UFLA285 treatment (0.93 ± 0.03 g/plant) than from untreated cotton seed (0.43 ± 0.03 g/plant).
Discussion
In a previous report, over 300 endospore-forming bacterial strains were screened for control of two important seed-borne cotton diseases: bacterial blight (Xanthomonas axonopodis pv. malvacearum) and damping-off/ramulose (Colletotrichum gossypii var. cephalosporioides) (Medeiros et al. 2008). Bacillus subtilis UFLA285 was obtained which has broad-spectrum activity against cotton pathogens. This rhizobacterium reduces post-emergence damping-off in the field and reduces seed-borne associated fungi. In the present study, control of damping off in cotton caused by R. solani AG4 is shown with UFLA285 seed inoculation (Fig. 1). Treatment was effective when plants were inoculated 9 days after planting that is also the incubation time necessary for chemical elicitation with benzothiadiazole, an activator of SAR (Jabaji-Hare and Neate 2005). While UFLA285 does produce metabolites with antibiotic activity against AG4 (data not shown) and the time necessary for the onset of disease control is sufficient for UFLA285 to cause direct disease suppress, the up-regulation of jasmonate and ethylene related transcripts suggests that UFLA285 may also participate in induced systemic resistance (ISR).
UFLA285 induced transcripts encoding phenylpropanoid metabolism included: phenylalanine ammonia lyase, 2-hydroxyisoflavone reductase, dihydroflavonol-4-reductase, caffeic acid O-methyltransferase, and cinnamoyl CoA reductase, which catalyze the production of undifferentiated phenylpropenoids, phytoalexins, catechins/anthocyanins, and phenolics/lignin, respectively (Zabala et al. 2006). The oxidation products of catechins and tannins are the main polyphenols in cotton and are produced in high amounts in response to R. solani infection (Kirkpatrick and Rothrock 2001). Since tannins can inhibit fungal polygalacturonases, responsible for the tissue maceration (Kirkpatrick and Rothrock 2001) over-expression of tannin synthesis may well serve in plant defense. Lignin, also a product of the phenylpropanoid pathway can provide a physical barrier against additional microbial colonization (Vorwerk et al. 2004). Moreover other cell wall reinforcement strategies including down-regulation of transcripts that participate in cell wall loosening (Zhang et al. 2007) and up regulation of callose synthase genes coding for callose deposition have been associated with the thwarting further fungal colonization (Tiburzy and Reisener 1990; Taiz and Zeiger 1991). A down-regulation of xyloglucan endoglycosyl transferase aids in reducing natural openings for fungal invasion (Zhang et al. 2007).
Cotton defense responses induced with a combination of UFLA285 cotton seed treatment and AG4 infection including pathogenesis-related genes and genes encoding lignin biosynthesis have been previously observed when cotton hypocotyls were infected with Fusarium oxysporum f. sp. vasinfectum (Dowd et al. 2004). The repression of drought-responsive proteins such as aquaporins has also been observed previously (Dowd et al. 2004) which may be specific to vascular wilt diseases.
Necrotrofic infections such as produced by R. solani as well as drought can generate reactive oxygen species (ROS) that are in turn scavenged by plant peroxidases and glutathione-S-transferases (Able 2003; Ramanjulu and Bartels 2002). Other detoxifying strategies may also be operative in plant-pathogen interactions. To either eliminate the toxin from inside the cell or degrade such metabolites to non-toxic components, multidrug and toxin extrusion (MATE) transport proteins and cytochrome P450/endo-beta-N-acetylglucosaminidase are induced in host plant tissue (Eckardt 2001; Ralston et al. 2001), as observed with UFLA285 treatment.
Although R. solani has not been reported as being a xylem colonizer (Kirkpatrick and Rothrock 2001) from the initial infection, cushions form on the stem epidermis, the mycelium reach and damage the tracheary elements reducing the water conductivity (data not presented) and thus, with normal water transpiration, a negative water balance occurs leading irreversible plant wilting. On treated plants, in spite of the brownish necrotic lesions (Fig. 1b) no wilting symptoms were observed possibly due to a lower extent of pathogen internal tissue colonization due to many possible factors including cell wall reinforcement (callose and lignin deposition), as shown for the binucleate Rhizoctonia mediated biological control of damping-off (Cardoso and Echandi 1987).
For the first time, PGPR treatment resulting in cotton protection against a soil-borne pathogen has been proposed. Future experiments will verify the role of drought tolerance in the disease protection and enzyme activity as well as physical barriers predicted to be operative form the microarray data. It is expected that Bacillus subtilis UFLA285 will provide cotton growers with a new, environmentally innocuous, biological agent allowing for improved plant protection against biotic- and abiotic-stress and in turn, superior crop performance.
References
Able AJ (2003) Role of reactive oxygen species in the response of barley to necrotrophic pathogens. Protoplasma 221:137–143
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water stress studies. Plant Soil 39:205–207
Buchanan BB, Gruissem W, Jones RL (2000) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville
Cardoso JE, Echandi E (1987) Nature of protection of bean seedlings from Rhizoctonia root rot by a binucleate Rhizoctonia-like fungus. Phytopathology 77:1548–1551
Dorey S, Baillieul F, Pierrel MA, Saindrenan P, Fritig B, Kauffmann S (1997) Spatial and temporal induction of cell death, defense genes, and accumulation of salicylic acid in tobacco leaves reacting hypersensitively to a fungal glycoprotein elicitor. Mol Plant Microbe Interact 10:646–655
Dowd C, Wilson IW, McFadden H (2004) Gene expression profile changes in cotton root and hypocotyl tissues in response to infection with Fusarium oxysporum f.sp. vasinfectum. Mol Plant Microbe Interact 17:654–667
Eckardt NA (2001) Move it on out with MATEs. Plant Cell 13:1477–1480
Hammerschmidt R, Kuc J (1995) Induced resistance to disease in plants: developments in plant pathology. Kluwer Academic, Dordrech
Holstein SHE, Oliviusson P (2005) Sequence analysis of Arabidopsis thaliana E/ANTH-domain-containing proteins: membrane tethers of the claritin-dependent vesicle budding machinery. Protoplasma 226:13–21
Hulugalle NR, Scott F (2008) A review of the changes in soil quality and profitability accomplished by sowing rotation crops after cotton in Australian Vertosols from 1970 to 2006. Australian J Soil Res 46:173–190
Jabaji-Hare S, Neate SM (2005) Nonpathogenic binucleate Rhizoctonia spp. and benzothiadiazole protect cotton seedlings against Rhizoctonia damping-off and Alternaria leaf spot in cotton. Phytopathology 95:1030–1036
Jeger MJ, Viljanen-Rollinson SLH (2001) The use of the area under the disease-progress curve (AUDPC) to assess quantitative disease resistance in crop cultivars. Theor Appl Genet 102:32–40
Keinath AP, Batson WE Jr, Caceres J, Elliott ML, Sumner DR, Brannen PM, Rothrock CS, Huber DM, Benson DM, Conway KE, Schneider RN, Motsenbocker CE, Cubeta MA, Ownley BH, Canaday CH, Adams PD, Backman PA, Fajardo J (2000) Evaluation of biological and chemical seed treatments to improve stand of snap bean across the southern United States. Crop Protect 19:501–509
Kirkpatrick TL, Rothrock CS (2001) Compendium of cotton diseases, 2 ed. American Phytopathological Society, Saint Paul
Kronstad JW (2000) Fungal pathology. Academic Publishers, Kluwer The Netherlands
Lopez-Lavalle LB, McFadden H, Brubaker CL (2007) The effect of Gossypium C-genome chromosomes on resistance to Fusarium wilt in allotetraploid cotton. Theoretical App Gene 115:477–488
Malik KA, Hafeez FY, Miirza FS, Hameed GR, Bilal R (2005) Rhizospheric plant-microbe interactions for sustainable agriculture. In: Wang YP, Lin M, Tian ZX, Elmerich C, Newton WE (eds) Biological nitrogen fixation, sustainable agriculture and the environment, 1st edn. Springer, Dorchdrech, pp 257–260
Manian S, Manibhushanrao K (1990) Influence of some factors on the survival of Rhizoctonia solani in soil. Tropical Agric 67:207–208
Martinez C, Baccou JC, Bresson E, Baissac Y, Daniel JF, Jalloul A, Montillet JL, Geiger JP, Assigbetse K, Nicole M (2000) Salicylic acid mediated by the oxidative burst is a key molecule in local and systemic responses of cotton challenged by an avirulent race of Xanthomonas campestris pv malvacearum. Plant Physiol 122:757–766
Medeiros FH, Souza RM, Ferro HM, Medeiros FC, Pomella AW, Machado JC, Santos Neto H, Soares DA, Zanotto E, Paré PW (2008) Bacillus spp. to manage seed-born Colletotrichum gossypii var. cephalosporioides damping-off. Phytopathology 98:S102
Mondal KK, Verma JP (2002) Biological control of cotton diseases. In: Gnanamanickam SS (ed) Biological control of crop diseases. M.Dekker, New York, pp 96–119
Muthusamya BP, Natarajana P, Zhoua X, Graham TR (2009) Linking phospholipid flippases to vesicle-mediated protein transport. Biochim Biophys Acta Mol Cell Biol Lipid 1791:612–619
Parida A, Dagaonkar V, Phalak M, Aurangabadkarl L (2008) Differential responses of the enzymes involved in proline biosyntheis and degradation in drought tolerant and sensitive cotton genotypes during drought stress and recovery. Acta Physiol Plantarum 30:619–627
Pieterse CMJ, Van Wees SCM, Ton J, Van Pelt JA, Van Loon LC (2002) Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biology 4:535–544
Pieterse CMJ, Van Der Ent S, Van Pelt JA, Van Loon LC (2007) The role of ethylene in rhizobacteria-induced systemic resistance (ISR). In: Ramina A et al (eds) Advances in plant ethylene research, 7th edn. Springer, Dorchdrech, pp 325–331
Ralston L, Kwon ST, Schoenbeck M, Ralston J, Schenk DJ, Coates RM, Chappell J (2001) Cloning heterologous expression and functional characterization of 5-epi-aristolochene-1,3-dihydrolase from tobacco (Nicotiana tabacum). Archives Biochem Biophysics 393:222–235
Ramanjulu S, Bartels D (2002) Drought and desication-induced modulation of gene expression in plants. Plant Cell Environment 25:141–151
Shaner G, Finney RE (1977) The effect of nitrogen fertilization on the expression of slow-mildewing resistance in Knox wheat. Phytopathology 67:1051–1056
Taiz L, Zeiger E (1991) Plant physiology. The Benjamin/Cummings Publishing Co., Inc., Redwood City
Tiburzy R, Reisener HJ (1990) Resistance of wheat to Puccinia graminis f. sp. tritici: association of the hypersensitive reaction with the cellular accumulation of lignin-like material and callose. Physiol Mol Plant Path 36:109–120
Udall JA, Flagel LE, Cheung F, Woodward AW, Hovav R, Rapp RA, Swanson JM, Lee JJ, Gingle AR, Nettleton D, Town CD, Chen ZJ, Wendel JF (2007) Spotted cotton oligonucleotide microarrays for gene expression analysis. BMC Genomics 8:81
Vorwerk S, Somerville S, Somerville C (2004) The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci 9:203–209
Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223:7–12
Wang Y, Ohara Y, Nakayashiki H, Tosa Y, Mayama S (2005) Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol Plant Microbe Interact 18:385–396
Xie X, Zhang H, Pare PW (2009) Sustained growth promotion in Arabidopsis with long-term exposure to the beneficial soil bacterium Bacillus substilis (GB03). Plant Signal Behav 4:948–953
Yi HS, Heil M, Adame-Alvarez RM, Ballhorn DJ, Ryu CM (2009) Airborne induction and priming of plant defenses against a bacterial pathogen. Plant Physiol 151:2152–2161
Zabala G, Zou J, Tuteja J, Gonzalez D, Clough SJ, Vodkin LO (2006) Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biology 6:26
Zhang H, Kim MS, Krishnamachari V, Payton P, Sun Y, Grimson M, Farag MA, Ryu CM, Allen R, Melo IS, Pare PW (2007) Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226:839–851
Zhang H, Kim MS, Sun Y, Dowd SE, Shi H, Pare PW (2008) Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol Plant Microbe Interact 21:737–744
Acknowledgements
Financial assistance was provided in part by a grant from the Robert Welch Foundation (D-1478) and comments on an earlier draft of this manuscript were provided by Dr. Huazhong Shi.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Jesus Mercado-Blanco.
Rights and permissions
About this article
Cite this article
Medeiros, F.H.V., Souza, R.M., Medeiros, F.C.L. et al. Transcriptional profiling in cotton associated with Bacillus subtilis (UFLA285) induced biotic-stress tolerance. Plant Soil 347, 327–337 (2011). https://doi.org/10.1007/s11104-011-0852-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-011-0852-5