Abstract
Reed canary grass (Phalaris arundinacea, L.) invasion of wetlands is an ecological issue that has received attention, but its impact on soil microbial diversity is not well documented. The present study assessed the size (substrate-induced respiration), catabolic diversity (CLPP, community level physiological profiles) and composition (selective inhibition) of the soil microbial community in invaded (>95% P. arundinacea cover) and in non-invaded areas of a wetland occupied by native species grown either as a mixed assemblage (22 species) or as quasi-monotypic stands of Scirpus cyperinus (74% cover). The study also tested the hypothesis that decomposition of lignin- and phenolics-rich plant tissues would be fastest in soils exhibiting high catabolic diversity. Results showed that soil respiration, microbial biomass and diversity were significantly higher (P < 0.03; 1.5 to 3 fold) in P. arundinacea-invaded soils than in soils supporting native plant species. Fungal to bacterial ratios were also higher in invaded (0.6) than in non-invaded (0.4) plots. Further, canonical discriminant analysis of CLPP data showed distinct communities of soil decomposers associated with each plant community. However, these differences in microbial attributes had no effect on decomposition of plant biomass which was primarily controlled by its chemical composition. While P. arundinacea invasion has substantially reduced plant diversity, this study found no parallel decline in the size and diversity of the soil microbial community in the invaded areas.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The invasion of terrestrial ecosystems by exotic species often results in the replacement of taxonomically diverse communities of plants by monotypic stands. The impact of such shifts in vegetation composition on soil functions is an ecological issue that has been the focus of several recent studies (Hook et al. 2004; Wolfe and Klironomos 2005; Li et al. 2007; Liao et al. 2007; Rothman and Bouchard 2007; Marchante et al. 2008), but important questions remain with respect to process-level understanding of the factors associated with soil responses to plant invasion. Such responses include alteration of the soil microbial community which in turn could affect important soil processes such as litter decomposition. A limited number of studies have examined the linkage between soil microbial community and soil functions in terrestrial ecosystems undergoing vegetation shift, and some recent reports (Wardle 1998; Meier and Bowman 2008; also see review byWolfe and Klironomos 2005) have discussed the difficulties in establishing these linkages. Meier and Bowman (2008) argued that relationships between plant diversity and soil microbial functions are more strongly mediated by the heterogeneity of litter chemistry than by the number of plant species present in an ecosystem because some of the plant species may produce chemically similar litter.
Wetland soils are important carbon sinks but, throughout the United States and Canada, these ecosystems have experienced invasion by reed canary grass (Phalaris arundinacea, L.), a Eurasian perennial grass that was introduced in North America for forage production and soil erosion control (Lavergne and Molofsky 2004). Variable effects (positive, Liao et al. 2007; neutral, Rothman and Bouchard 2007; negative, Hook et al. 2004) of plant invasion on soil organic C (SOC) stocks have been reported. At a south-central Indiana wetland, larger SOC pools were recorded in P. arundinacea-invaded areas than under native vegetation, but plant biomass production and composition could not explain the results (Bills 2008).
In terrestrial ecosystems, SOC stocks reflect the amount and quality of plant litter returned to soils as well as the composition and activity of the soil microbial community. Plant biomass contains a wide variety of biochemical constituents ranging from readily biodegradable molecules (e.g. carbohydrates and amino acids) to more recalcitrant molecules including structural polymers such as lignin and defense compounds such as phenolics (Fog 1988; Heal et al. 1997). These compositional variations in plant biomass have been integrated into a residue quality index (RQI; Tian et al. 1995), a parameter that is inversely related to the C/N ratio, lignin and phenolics content of plant biomass. Because the abundance of these molecules in plant tissue varies with species and growth physiology (Tian et al. 1995; Heal et al. 1997), it is expected that residue quality could provide the link between soil C cycling and plant invasion. Plant tissues rich in lignin and phenolics for example would be expected to be more recalcitrant than biomass rich in easily decomposable components such as simple carbohydrates. However, Fog (1988) cautioned that the composition of the soil decomposer community may be more important to decomposition of plant materials than biomass quality. Noting that no single microbial species has the enzymatic capability to decompose the wide variety of compounds present in plant litter, others (Wardle 1998; Sinsabaugh et al. 2002) have further proposed that the composition and catabolic diversity of soil microbes determine their ability to degrade plant matter. In light of the foregoing, it is hypothesized that the decomposition of lignin- and phenolics-rich plant biomass (low RQI) would be faster in soils exhibiting high level of microbial diversity than in low-diversity soils.
Bacteria and fungi represent the largest subsets of the soil microbial community. Metabolic differences between these two groups are fairly well documented with generally greater C-use efficiency and higher rates of SOC accumulation in fungal- than in bacterial-dominated soil ecosystems (Frey et al. 1999). The catabolic diversity of soil microbes in invaded ecosystems has been investigated, but few studies have concurrently examined the composition of the soil microbial community in response to plant invasion. Despite similarities between Typha spp. and Phragmites australis biomass composition, Rothman and Bouchard (2007) observed differences in C utilization patterns in wetland soils invaded by these two plant species. Marchante et al. (2008) attributed alterations in soil microbial diversity following Acacia longifolia invasion to deposition of N-rich litter by the invading legume. Li et al. (2007) noted a progressive increase in soil microbial biomass carbon (MBC) and basal soil respiration (BSR) with increased level of forestland invasion by the vine Mikania micrantha. In a study comparing P. arundinacea to other forage crops, Drury et al. (1991) measured significantly greater MBC in soils where P. arundinacea was grown but information on microbial diversity and composition was not provided.
Thus, the objectives of this research were to (1) compare the size, composition and functional diversity of the soil microbial community in non-invaded and P. arundinacea-invaded sections of a wetland, and (2) examine the relative control of residue quality and soil microbial attributes on plant biomass decomposition.
Materials and methods
Sampling site and vegetation communities
Soil and plant biomass used in the study were collected from Beanblossom Bottoms, a wetland complex in south-central Indiana (39° 34′ 43″ N, 86° 34′ 22″ W) established on land which, until 1992, was used as cropland. Soils at the site are clay-loam classified as Bonnie (Typic Fluvaquents), Stendal (Fluventic Endoaqepts), and Zipp (Endoaquepts). Based on vegetation survey and site hydrology, four vegetation communities (separated by 50–500 m) were included in the study: (i) Community A, a mixed native plant assemblage comprising 22 plant species with Carex lurida, Juncus effusus, Juncus interior, and Solidago canadensis being the most abundant , (ii) Community B, dominated (74% cover) by the native sedge, Scirpus cyperinus (wool grass); Community C, a monotypic stand of P. arundinacea inundated during most of the growing season, and (iv) Community D, also a pure stand of P. arundinacea with the surface soil layer generally saturated (not ponded). Further description of the hydrology, soil properties and composition of plant communities at the study site is available elsewhere (Bills 2008). In each plant community, triplicate study plots (10 m × 10 m) were established.
To carry out the microbial assay described below, composite soil samples (0–10 cm; both underneath and between plants) were collected on August 8, 2008 from each study plot (6 sampling points per plot), transported to the laboratory in plastic bags and stored at 4°C. Dry soil subsamples were used for determination of soil chemical properties. Soil pH was measured with an Orion pH-meter (1:2 soil to water ratio). Exchangeable bases were extracted with 1 M ammonium acetate (pH 7) and quantified using inductively coupled plasma atomic emission spectroscopy (Leeman Labs, Hudson, NH). Soil organic C and N were determined by dry combustion (850°C) using CHNS analyzer (Thermo Electron, Waltham, MA).
Soil microbial biomass, composition and metabolic profile
The relative bacterial and fungal contribution to the soil microflora was determined using the selective inhibition technique (Anderson and Domsch 1975). Preliminary tests were conducted to determine the optimum amount of glucose (6 mg g−1 soil), anti-fungal (15 mg cycloheximide g−1 soil), and anti-bacterial (3 mg streptomycin g−1 soil) to use in the assays. For each soil sample, twelve (10 g moist soil, 0.32 ± 0.04 g water g−1 soil) sub-samples were weighed into glass beakers and distributed in triplicate among the following four treatments: glucose only; glucose plus cycloheximide; glucose plus streptomycin; and glucose plus cycloheximide and streptomycin. Beakers containing amended soils were placed into glass jars (450 mL) and incubated (18°C, 6 h). Air samples were withdrawn for jar headspace and analyzed for CO2 using a Varian CP-3800 gas chromatograph (Varian Inc., Palo Alto, CA). The relative contribution of fungal and bacterial biomass to soil respiration was determined (Anderson and Domsch 1975). The rate of CO2 production in the glucose-only treatment was used to determine microbial biomass C (MBC) following the computation approach described in Kaiser et al. (1992).
Community level physiological profiles (CLPP) were assessed using BIOLOG Eco microplates (Biolog Inc., Hayward, CA, USA). Each microplate contains 31 carbon sources comprising 6 categories of substrates: amines, amino acids, carbohydrates, carboxylic acids, phenolics and polymers. Bottles and solution used in this assay were autoclaved. For each soil sample, duplicate 1 g sub-samples were suspended in 100 mL of phosphate buffer (10 mM, pH 7.4) and the suspension vortexed for several minutes. After settling briefly, 1 ml of that suspension was diluted in another 100 ml of phosphate buffer solution and vortexed. Finally, 150 µl aliquots of that final dilution (10−4) were added to each of the 96 microplate wells. Plates were incubated at 18°C and absorbance read (λ: 595 nm) at 24, 72, 96, and 120 h using a VersaMax microplate reader (Molecular Devices, Sunnyvale, CA). Shannon’s diversity index (H) and substrate richness (S, number of different substrates used by the microbial community) were computed:
where, pi is the ratio of absorbance for a given substrate to the sum of absorbance of all wells in a micro-plate. Average absorbance for each substrate category was also computed.
Plant biomass decomposition
An incubation study was conducted to assess the decomposition of plant biomass in relation to biochemical properties of soils. Subsamples of plant biomass (collected in July 2007) were analyzed for lignin, cellulose (Gessner 2005), phenolics (Bärlocher and Graça 2005), and total C and N (CHNS analyzer, Thermo Scientific, Waltham, MA). Chemical composition is reported in Table 1.
The incubation experiment involved four (4) soil types (soil from communities A, B, C and D) and 3 types of plant matter (mixed native vegetation, S. cypernicus and P. arundinacea from community C). For each soil type, a control (with no plant biomass added) was included. Each treatment was run in triplicate. Approximately 25 g of fresh soil was thoroughly mixed with 0.5 g of leaves (shred to 2 to 3.44 mm size) and incubated (900 mL glass jars, 19°C) for 21 days. Jar headspace was periodically sampled to determine CO2 concentration. The rate of CO2 production in the controls was used as a measure of basal soil respiration (BSR). The amount of CO2 attributable to the decomposition of added plant biomass was computed as CO2 produced in the treatment minus that produced in corresponding controls. Cumulative CO2 (Ct) produced was fitted to the non-linear model Ct = Co (1−exp−kt) where k is the rate constant (d−1) and Co is pool of readily decomposable C in the plant biomass (mg C g−1 plant biomass).
Statistical analyses
Garland (1996) cautioned that differences in initial inoculum cell density could affect well color development and introduce biases in CLPP data. In accord with approaches suggested by Garland (1996) and Grayston et al. (1998), well absorbance readings were normalized by dividing the absorbance of each well by the average color of all wells in a plate. The 96-h readings were analyzed using canonical discriminant analysis (CVA) using the CANDISC procedure (SAS 2003). This form of multivariate analysis was selected instead of principal component analysis (PCA) because, as Grayston et al. (1998) have also found, it provided a better differentiation among the vegetation communities than did PCA. The non-linear regression (NLIN) procedure of SAS (2003) was used to fit CO2 produced during the incubation study, and the values of Co derived were submitted to analysis of variance (ANOVA using the GLM procedure in SAS) to assess the effect of soil type and plant species on biomass decomposition. When a statistically significant effect was detected, Fisher’s protected LSD test was used for comparison of the means. Unless otherwise noted, statistical significance was determined at the 95% confidence level.
Results
Soil microbial biomass and composition
Significant differences (P < 0.001) among plant communities (Table 2) were detected with respect to soil microbial biomass (MBC). In the P. arundinacea-invaded communities, MBC was nearly 3-fold higher (mean: 182 mg kg−1) than in the native plant communities (62 mg kg−1). Although differences were not significant, the fungal to bacterial (F:B) ratios were generally higher in soils under P. arundinacea (0.63) than in soils under the native plant species (0.39). Using the total MBC and F:B ratios, an estimate of fungal biomass was found to be 4-times larger in P. arundinacea -invaded soils (70.2 mg kg−1) than in soils from areas occupied by native plant species (17.3 mg kg−1). Basal respiration (mg CO2-C kg−1 d−1) was also significantly (P < 0.03) higher in soils from P. arundinacea (15.4) than in soils from either the mixed native (7.7) or the S. cypernicus-dominated (4.9) community (Table 2). The metabolic quotient of soil respiration (qCO2, rate of respiration per unit of MBC) was similar among tested plant communities averaging 0.094 ± 0.017 mg CO2-C mg−1 MBC d−1. No significant effect of hydroperiod on any of the soil biological attributes was detected (Table 2) as similar levels were recorded in both the saturated (community D) and the inundated (community C) plots supporting P. arundinacea.
Functional diversity of the soil microbial biomass
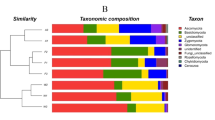
Marked differences were detected between soils (P < 0.007) from the P. arundinacea plots and soils supporting native vegetation with regard to C utilization patterns (Figs. 1 and 2). In many instances, microbes present in soils from P. arundinacea plots exhibited a greater ability to metabolize more C sources (as indicated by higher substrate richness) than microbes in soils from native plant communities (Figs. 1 and 2). Consequently, higher levels of microbial diversity were measured in soils from P. arundinacea than in soils underneath the native plant species after 96 and 120 h (Fig. 3). Explaining 90% of the variance, canonical discriminant analysis separated soil microbial communities into three distinct groups (Fig. 4). A net segregation of the soil microbial community from the S. cypernicus plots along the CV1 axis (74% of the explained variance) was observed. The soil microbial community in the mixed native plots was separated from that of the S. cypernicus and P. arundinacea monocultures along the CV2 axis (18%). There was no separation between the soil microbial communities from the two P. arundinacea-invaded areas (Fig. 4). The soil microbial parameters assumed strikingly similar values in P. arundinacea-invaded areas irrespective of whether the topsoil was frequently inundated or was merely saturated. Among the C sources, the ability to metabolize carbohydrates (especially mannitol, galactonic acid α-lactone, and N-acetyl-glucosamine), amino acids and polymers contributed the most to soil microbial community separation (Figs. 1 and 2). Soil microbes from the S. cypernicus plots showed almost no capacity to metabolize these C sources.
Decomposition of plant materials
Carbon dioxide produced during biomass decomposition (Fig. 5) was fitted (non-linear fitting) to the first order exponential model (r2: 0.88–0.99, P < 0.002) to derive the pool of readily decomposable C (Co) in plant biomass. Using Co values as response variables, ANOVA showed significant (P < 0.001) effect of plant species on decomposition of plant biomass (Table 3). The effect of soil type and soil type x plant species interaction was not significant. Regardless of soil type (hence microbial diversity), the decomposition of P. arundinacea biomass was consistently the highest and that of S. cypernicus biomass was always the lowest. Across soil types, the pool of readily decomposable C in P. arundinacea, mixed native and S. cypernicus biomass C averaged 71, 55, 33 mg C g−1 plant biomass, respectively (Table 3). P. arundinacea biomass had lower C/N ratio (21 vs 31), contained less lignin (7 vs 15.4%) and less phenolics (1.2 vs 2.2%) than S. cyperinus biomass and decomposed faster regardless of wetland soil attributes. Contrary to the study hypothesis, these results indicated that the biochemical composition of plant biomass was more important than soil microbial diversity in affecting decomposition.
Carbon dioxide production during decomposition of plant biomass incubated in different types of wetland soils. Each data point is the mean of 6 experimental units. Graph panels labeled a, b, c and d indicate that plant biomass was incubated in soils collected from plant community A (mixed native), community B (S. cypernicus), community C (P. arundinacea, inundated) and community D (P. arundinaea, saturated), respectively. For each treatment, the best fit of the data to the exponential model is represented by a solid line
Discussion
Plant diversity versus soil microbial diversity
Several studies (Kowalchuk et al. 2002; Benizri and Amiaud 2005) have reported strong correlations between plant diversity and functional diversity of soil decomposers. Elevated soil microbial diversity in taxonomically diverse plant communities has generally been thought as being the result of exposure of soil microbes to litter of diverse chemical composition (Kowalchuk et al. 2002; Benizri and Amiaud 2005). Thus, this line of reasoning suggests a direct link between plant species diversity and soil microbial diversity. Applying these considerations to our study, it was hypothesized that litter returned to soil in community A would be chemically diverse (22 different plant species) and that would in turn elicit the evolution of a soil microbial community functionally more diverse than found in monotypic stands of either S. cypernicus or P. arundinacea. However, our results did not provide support for that hypothesis. Although soil microbial parameters were higher in soil from community A (mixed native species) than in soil where a quasi-monotypic stand of the native S. cypernicus has established, differences were generally not significant (Table 2 and Figs. 1, 2 and 3). Moreover, all soil microbial parameters were significantly greater in areas invaded by P. arundinacea compared to areas occupied by the mixed native plant species.
Although inconsistent with these past studies (Kowalchuk et al. 2002; Benizri and Amiaud 2005), the lack of correspondence between plant diversity and soil microbial diversity observed in the present work is well supported by several recent investigations (Li et al. 2007; Rodriguez-Loinaz et al. 2008). Li et al. (2007) reported increased catabolic diversity of soil following M. micrantha invasion of forestland. Rodriguez-Loinaz et al. (2008) noted that the relationships between plant diversity and soil microbial diversity vary with plant species; strong under ferns and herbaceous plants, but non-existent under shrubs and trees.
The present study clearly shows significant shifts in soil biological attributes in areas where P. arundinacea has established, but the available data does not allow elucidation of operating mechanisms. There are at least two physiological traits of P. arundinacea that can be invoked as an attempt to explain these results. One such trait is the high root/shoot ratio of P. arundinacea (Lavergne and Molofsky 2004) which, in the present study, was found to be 2–4 times that of the native plant species (Table 1). This may have contributed to the study results given reported relationships between root mass, root exudation and rhizospheric MBC (Grayston et al. 1998; Lu et al. 2002). Another well documented trait of P. arundinacea is its ability to store photosynthetic products, mostly nonstructural carbohydrates (e.g. fructose, glucose, sucrose, and starch) in its rhizomes during the dormant season allowing for an early resumption of growth during the spring (Tamura and Moriyama 2001). It is conceivable that some of these stored carbohydrates may leak into the root zone and become available to soil microbes. Lending credence to this interpretation is the fact that our soil sampling occurred in August, right at the end of the growing season. Further, CLPP analysis showed that carbohydrates were one of the categories of organic substrates that most clearly distinguished soil microbes in P. arundinacea-invaded plots from the microbial community in non-invaded areas (Fig. 1).
Soil microbial attributes and decomposition
Several authors (Wardle 1998; Sinsabaugh et al. 2002) proposed that diversity and composition of soil microbial communities significantly affect their ability to decompose plant tissues. Thus, in this study, a direct relationship was hypothesized between the decomposition of low RQI biomass and soil microbial diversity. More specifically, low RQI biomass was expected to decompose much faster in high diversity soils than in soils characterized by low diversity microbial population. The more diverse the soil microbes, the more likely that the enzyme systems needed to degrade the recalcitrant C fractions would be present. Our results, however, showed no statistically significant effect of soil microbial diversity on the decomposition of plant biomass (Table 3 and Fig. 5). Several past studies (Degens 1998; Sall et al. 2006) have also failed to establish correlations between catabolic diversity of soils and decomposition of plant materials.
Several factors may have contributed to these results. First, because decomposition of biomass-C can be carried out by widely diverse groups of microbes, functional diversity of decomposers may not be as important a determining factor as it would be for processes involving only a limited subset of the soil micro-flora. Second, the BIOLOG method may have favored the activity of fast-growing micro-organisms that may not be as active when exposed to phenol- and lignin-rich plant materials. Third, by stimulating the growth of soil microbes, addition of fresh plant biomass may nullify initial differences in catabolic diversity that may exist among soil types. This interpretation is consistent with the work of Bending et al. (2002) that showed a progressive convergence of the microbial diversity index of soils following amendment with various types of crop residue. Since fresh plant tissues (even low RQI) contain a certain amount of labile organic compounds, weathered plant biomass (depleted in labile C) could be more appropriate as starting material to investigate the effect of soil microbial diversity on decomposition. In designing future experiments, steps should be taken to overcome these methodological shortcomings.
P. arundinacea invasion and soil C cycling
As several past studies assessing the effect of plant invasion on total SOC stocks have been inconclusive, it has been suggested (Hook et al. 2004) that rapidly-cycling soil C fractions are more sensitive to vegetation shifts and thus could be a better indicator of soil C dynamics in ecosystems experiencing exotic plant invasion. Indeed, we measured significantly higher microbial biomass, respiratory activity and diversity in soils invaded by P. arundinacea. In addition to demonstrating the sensitivity of soil biological indexes to vegetation change, these findings further confirm the results obtained in our assessment of total SOC pools at the site (Bills 2008).
In a study involving P. arundinacea and other forage crops, Drury et al. (1991) observed parallel increases in both MBC and soil aggregation in experimental plots seeded with P. arundinacea, and speculated that P. arundinacea may have induced an increase in fungal biomass which, in turn, had led to the formation of stable soil aggregates. Three years after P. arundinacea establishment, Chantigny et al. (1997) also noted significant increases in glucosamine content and mean-weight diameter of soil aggregates. As glucosamine is an amino sugar present in most fungal cell walls, these results were interpreted as indication that fungi played a dominant role in the formation of soil macro-aggregates. The physical entanglement of soil particles by fungal hyphae additionally contributes to SOC accumulation in fungal-dominated soil ecosystems (Frey et al. 1999). Higher soil F:B ratio in P. arundinacea-invaded areas (Table 2) could have important implications for nutrient cycling and have contributed to the higher SOC stock measured at these locations compared to plots vegetated by native plant species (Bills 2008).
Conclusions
Microorganisms are essential for the functioning of soil ecosystems. With the understanding that the chemical composition of litter and root exudates differ among plant species, concurrent change is generally expected in vegetation composition and the attributes of the soil decomposer communities. With these underlying assumptions, the present study was conducted to evaluate alterations in the catabolic diversity of the soil microbial community in a wetland ecosystem experiencing P. arundinacea encroachment. The data collected showed distinct effects of P. arundinacea invasion on soil respiration, and on the size, composition and diversity of the soil microbial community. However, these differences in soil microbial attributes did not affect the rate at which plant tissues decomposed in the wetland soils. Decomposition was primarily controlled by the chemical composition of plant biomass and not by soil microbial attributes. Overall, this study found increased soil microbial diversity and activity in wetland areas invaded by P. arundinacea, but further work is needed to elucidate ecosystem-level responses to these alterations in the soil microbial community.
Abbreviations
- BSR:
-
basal soil respiration
- CLPP:
-
community level physiological profiles
- MBC:
-
microbial biomass carbon
- RQI:
-
residue quality index
- SOC:
-
soil organic carbon
References
Anderson JPE, Domsch KH (1975) Measurement of bacterial and fungal contributions to respiration of selected agricultural and forest soils. Can J Microbiol 21:314–322
Bärlocher F, Graça MAS (2005) Total phenolics. In: Graça MAS, Barlocher F, Gessner MO (eds) Methods to study litter decomposition: a practical guide. Springer, Dordrecht, pp 97–99
Bending GD, Turner MK, Jones JE (2002) Interactions between crop residue and soil organic matter quality and the functional diversity of soil microbial communities. Soil Biol Biochem 34:1073–1082
Benizri E, Amiaud B (2005) Relationship between plants and soil microbial communities in fertilized grasslands. Soil Biol Biochem 37:2055–2064
Bills JS (2008) Invasive reed canary grass (Phalaris arundinacea) and carbon sequestration in a wetland complex. M.S. Thesis, Indiana University
Chantigny MH, Angers DA, Prevost D, Vezina LP, Chalifour FP (1997) Soil aggregation and fungal and bacterial biomass under annual and perennial cropping systems. Soil Sci Soc Am J 61:262–267
Degens BP (1998) Decreases in microbial functional diversity do not result in corresponding changes in decomposition under different moisture conditions. Soil Biol Biochem 30:1989–2000
Drury CF, Stone JA, Findlay WI (1991) Microbial biomass and soil structure associated with corn, grasses and legumes. Soil Sci Soc Am J 55:805–811
Fog K (1988) The effect of added nitrogen on the rate of decomposition of organic matter. Biol Rev 63:433–462
Frey SD, Elliott ET, Paustian K (1999) Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biol Biochem 31:573–585
Garland JL (1996) Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol Biochem 28:213–221
Gessner MO (2005) Proximate lignin and cellulose. In: Graça MAS, Barlocher F, Gessner MO (eds) Methods to study litter decomposition: a practical guide. Springer, Dordrecht, Netherlands, pp 115–120
Grayston SJ, Wang SQ, Campbell CD, Edwards AC (1998) Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem 30:369–378
Heal OW, Anderson JM, Swift MJ (1997) Plant litter quality and decomposition: an historical overview. In: Cadisch G, Giller KE (eds) Driven by nature - plant litter quality and decomposition. CAB International, Wallingford, pp 3–10
Hook PB, Olsen BE, Wraith JM (2004) Effects of the invasive forb Centaura maculosa on grassland carbon and nitrogen pools in Montana, USA. Ecosystems 7:686–694
Kaiser EA, Mueller T, Joergensen RG, Insam H, Heinemeyer O (1992) Evaluation of methods to estimate the soil microbial biomass and the relationship with soil texture and organic matter. Soil Biol Biochem 24:675–683
Kowalchuk GA, Buma DS, de Boer W, Klinkhamer PGL, van Veen JA (2002) Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie van Leeuwenhoek J Microbiol 81:509–520
Lavergne S, Molofsky J (2004) Reed canary grass (Phalaris arundinacea) as a biological model in the study of plant invasions. Crit Rev Plant Sci 23:415–429
Li WH, Zhang C, Gao G, Zan Q, Yang Z (2007) Relationship between Mikania micrantha invasion and soil microbial biomass, respiration and functional diversity. Plant Soil 296:197–207
Liao C, Luo Y, Jiang L, Zhou X, Wu X, Fang C, Cheng J, Li B (2007) Invasion of Spartina alterniflora enhanced ecosystem carbon and nitrogen stocks in the Yangtze Estuary, China. Ecosystems 10:1351–1361
Lu YH, Watanabe A, Kimura M (2002) Contribution of plant-derived carbon to soil microbial biomass dynamics in a paddy rice microcosm. Biol Fert Soils 36:136–142
Marchante E, Kjoller A, Struwe S, Freitas H (2008) Invasive Acacia longifolia induce changes in the microbial catabolic diversity of sand dunes. Soil Biol Biochem 40:2563–2568
Meier CL, Bowman WD (2008) Links between plant litter chemistry, species diversity, and below-ground ecosystem function. Proc Natl Acad Sci USA 105:19780–19785
Rodriguez-Loinaz G, Onaindia M, Amezaga I, Mijangos I, Grabisu C (2008) Relationship between vegetation diversity and soil functional diversity in native mixed-oak forests. Soil Biol Biochem 40:49–60
Rothman E, Bouchard V (2007) Regulation of carbon processes by macrophyte species in a Great Lakes coastal wetland. Wetlands 27:1134–1143
Sall SN, Masse D, Ndour NYB, Chotte JL (2006) Does cropping modify the decomposition function and the diversity of the soil microbial community of tropical fallow soil? Appl Soil Ecol 31:211–219
SAS (2003) SAS System for Windows, Version 9.1. SAS Institute Inc., Cary
Sinsabaugh RL, Carreiro MM, Repert DA (2002) Allocation of extracellular enzymatic activity in relation to litter composition, N deposition, and mass loss. Biogeochemistry 60:1–24
Tamura Y, Moriyama M (2001) Nonstructural carbohydrate reserves in roots and the ability of temperate perennial grasses to overwinter in early growth stages. Plant Prod Sci 4:56–61
Tian G, Brussard L, Kang BT (1995) An index for assessing the quality of plant residues and evaluating their effects on soil and crop in the (sub-) humid tropics. Appl Soil Ecol 2:25–32
Wardle DA (1998) Controls of temporal variability of the soil microbial biomass: a global-scale synthesis. Soil Biol Biochem 30:1627–1637
Wolfe BE, Klironomos JN (2005) Breaking new ground: soil communities and exotic plant invasion. Bioscience 55:477–487
Acknowledgements
The authors thank the Sycamore Land Trust for providing access to the study site; Andrew Mertz, of Indy Parks, for his help with the vegetation survey; Vince Hernly and Bob Hall for their support with field reconnaissance and well installation; and several interns from the Center for Earth and Environmental Science (IUPUI) who helped with field sampling.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Harsh P. Bais.
Rights and permissions
About this article
Cite this article
Jacinthe, PA., Bills, J.S. & Tedesco, L.P. Size, activity and catabolic diversity of the soil microbial biomass in a wetland complex invaded by reed canary grass. Plant Soil 329, 227–238 (2010). https://doi.org/10.1007/s11104-009-0147-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-009-0147-2