Abstract
Image analysis is used in numerous studies of root system architecture (RSA). To date, fully automatic procedures have not been good enough to completely replace alternative manual methods. DART (Data Analysis of Root Tracings) is freeware based on human vision to identify roots, particularly across time-series. Each root is described by a series of ordered links encapsulating specific information and is connected to other roots. The population of links constitutes the RSA. DART creates a comprehensive dataset ready for individual or global analyses and this can display root growth sequences along time. We exemplify here individual tomato root growth response to shortfall in solar radiation and we analyse the global distribution of the inter-root branching distances. DART helps in studying RSA and in producing structured and flexible datasets of individual root growth parameters. It is written in JAVA and relies on manual procedures to minimize the risks of errors and biases in datasets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent methods for studying plant root development utilize images (Smit et al. 2000). Indeed, growing roots are fine and fragile organs, which require absolute protection against direct touching during measurements. Furthermore, the dynamic methods of root observation are often based on 2D systems in which all or part of the root system is forced to develop on a plane. This is, for example, the case for flat containers with transparent-walls (Devienne-Baret et al. 2006; Iijima et al. 1998) or soil-located rhizotrons using windows or transparent tubes (for review Johnson et al. 2001; Pagès and Bengough 1997; Vamerali et al. 1999). The plane image emerges therefore as a convenient tool to capture and measure several parameters of root system architecture (RSA). This approach is further strengthened by the rapid development of low cost cameras and scanners capable of acquiring digital images of high definition/resolution.
Following this acquisition step, biologists are given raw datasets of pixel characteristics, which must be converted into interesting information such as root length, branching densities, diameters of apices, etc. The domain of digital image processing is under rapid expansion as assessed by the development of automatic algorithms for root discrimination (Kimura et al. 1999; Zeng et al. 2008) and the appearance of various image analysis packages, such as freeware and open-source software either making a general and versatile job (ImageJ http://rsb.info.nih.gov/ij/; NIH-Image http://rsb.info.nih.gov/nih-image/; Object-Image http://simon.bio.uva.nl/Object-Image/object-image.html; ObjectJ http://simon.bio.uva.nl/objectj/index.html) or offering specialized features (EZ-Rhizo http://www.ez-rhizo.psrg.org.uk/; Root Image Analyzer http://rootimage.msu.edu/root_images/new), mainly serving minirhizotron image analysis (RMS from Ingram and Leers 2001; MR-RIPL 2.0 & 3.0 http://rootimage.msu.edu/; MSU ROOTs Tracer http://roots.rsgis.msu.edu/; Rootfly http://www.ces.clemson.edu/~stb/rootfly/; RootView http://www.mv.helsinki.fi/aphalo/RootView.html; WR-RIPL 2.0 http://rootimage.msu.edu/) and commercial proprietary software designed for RSA measurements (Delta-T-Scan http://www.delta-t.co.uk/groups.html?group2005092301354; RooTracker www.biology.duke.edu/rootracker/; WinRHIZO™ www.regentinstruments.com). These software make use of mouse-clicks and more or less automated procedures such as edge detection, segmentation and thresholding, and skeletonization, to acquire global variables (i.e. root order, total root length, mean root diameter, number of apices, colour, branching angle, etc) and eventually supply elaborated variables (typically areas and volumes) from specific assumptions. Barring their cost, these programs are requisite tools for root researchers as they provide user-friendly procedures making it feasible to rapidly capture datasets and (in theory) without significant mental fatigue.
Although automated procedures represent an ideal ultimate solution, they still present different shortcomings such as algorithm misinterpretation leading to miscalculation of variables and they cannot overcome several difficulties more or less specific to studies with root systems (Armengaud et al. 2009; Russ 1999; Ryser 2006; Vamerali et al. 2003; Zobel 2008). Indeed, large differences remain between digital image analysis and direct interpretation by human vision. Image processing often requires that the objects be well defined by their edges, a uniform brightness and contrast (Russ 1999). This is particularly important for roots, which are sometimes difficult to distinguish from their background (Zeng et al. 2008). Due to their transparency, fine roots may have the same colour than soil particles, and if root staining has proven useful (Bouma et al. 2000; Costa et al. 2001; Ortiz-Ribbing and Eastburn 2003), it also forbids temporal studies. From an image processing perspective, root tracking is a challenging task in which the ability to discern root branching from crossings controls the number of errors introduced in datasets, both for individual root length and final branching densities (Armengaud et al. 2009). The challenge is strengthened further in time series images, by the requirement to recognize automatically and unambiguously, evolving structures such as growing or necrotizing roots and their associated colour or diameter changes (Ryser 2006; Zeng et al. 2008). Thus, present-day technological developments allow us to consider that automated procedures of root image analysis are capable of estimating global criteria of branched systems (i.e. total length, mean diameter, number of apices, fractal dimension), with the confidence in the measurements being dependent mainly on the image quality (Himmelbauer et al. 2004; Ortiz-Ribbing and Eastburn 2003; Zobel 2003). As a consequence, numerous studies continue to keep faith with manual analysis of images (Danjon and Reubens 2008), particularly to assess the time-evolving topology of entire root systems, including fast growing annual species or young trees, for which minirhizotrons are not suited. Moreover, it is recognized that automated analyses of dense root systems with commercial software are prone to errors (Ortiz-Ribbing and Eastburn 2003; note on the web site of WinRHIZO™ www.regentinstruments.com).
In addition to global variables (such as total root length, or total length per diameter class, or fractal dimension) topological criteria have been shown important to characterize functional aspects of root systems (Fitter 1986; 1987; Fitter et al. 1991; Taub and Goldberg 1996) and they are necessary for modeling root system development (Pagès et al. 2004). As a starting point, it requires an image depicting the integral connectivity of the links relating different parts of the root system (Berntson 1997). In order to extract from this image a practical meaning in terms of root development, several well-identified structures must be located precisely, such as apices (clearly distinguished from ending parts of broken roots, or roots vanishing in the soil) and branching points. Furthermore, when dealing with time-series images, it is particularly important to identify the homologous structures, which evolve with time, in order to measure the rate of change in their properties. Thus, successive images should not be processed independently of their related links. This requirement is not satisfied in most existing software packages.
In this context, due to the growing importance of image analysis in the root research domain, we have developed specialized software, called DART (Data Analysis of Root Tracings), capable of conveying supplementary (i.e. topological) meaning to the information extracted from the measurements of root images. It uses a mix of human vision expertise and the capacity of the computer to rearrange the information into an exploitable form. Indeed, while automated procedures fail to differentiate between the pertinent structures of the developing root systems (i.e. branching—crossings, apices), human vision stands out in this task. Thus, we designed the software so that the image is the support for this expertise and the dataset is properly organized in order to provide the researchers with parameter estimates directly connected with the RSA analysis. This particularly concerns the kinetics of individual root emergence, growth rates and branching densities along axes, etc. In this paper we detail two examples of typical raw images that we captured in past experiments and describe the problems associated with their processing. We discuss the choices made in the software to cope with these difficulties. The software has been written in the Java programming language in order to take advantage of its object-oriented methodology and its platform independence resulting from its ability to be executed on multiple operating systems (Mac OS X, Linux, Microsoft Windows). DART is distributed as free-software which can be downloaded from (http://www.avignon.inra.fr/psh/outils/dart), thereby helping those researchers who cannot afford the cost of available proprietary-software. DART has emerged following the work of Colin-Belgrand et al. (1989), novel ideas, our own expertise in the domain of RSA analysis and the progress made in computer tools. Finally, we will illustrate the usefulness of the method from preliminary results taken from the selected examples.
Materials and methods
Typical images and associated problems
In situ analysis of root architecture is a difficult task, revealing several problems, particularly when seeking dynamic and non-destructive data over long periods of time (weeks to months). It is not the purpose of the paper to cover all these problems. Nevertheless we will describe here two examples of typical datasets obtained during experiments conducted on root systems.
Example 1
This digital image (Fig. 1a) was acquired on a common flatbed scanner from a hand drawn root system. In this experiment, young oak seedlings (Quercus pubescens Willd.) were grown in a rhizobox, i.e. a flat container with transparent walls. Periodically, a record of the architecture was taken by transfer drawing all individual roots on a cellulose acetate sheet, using coloured ink pens, a unique colour being specific to the date of observation. With this method, the lines code for spatial information and the colours for temporal ones. This method appears common in the international community as it represents a simple means to observe RSA (Cahn et al. 1989; Cheng et al. 1991; Devienne-Baret et al. 2006; Iijima et al. 1998).
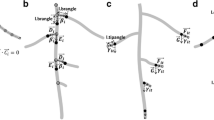
(a) Scanned image of a drawing made of an oak (Quercus pubescens L.) root system growing in a vertical rhizobox, using the technique of periodic (1-2 days) transfers on to an acetate sheet. The drawing was made using ink pens of different colour. Each colour is typical of the date at which the root segments have been observed. (b-d) Scaled digital images (montage of azimuthal views) of a single tomato (Solanum lycopersicum L.) root system shot 15 (b), 19 (c) and 28 (d) days after sowing (das) in a horizontal hydroponic rhizobox of 0.55 m width and 1.50 m length. In (b-c) the bottom parts of the rhizobox have been erased during image processing, in order to keep only the areas covered by the roots
Example 2
This is a time-series composite image (Fig. 1b–d) of a tomato (Solanum lycopersicum L.) root system shot by a digital camera. The original images were captured at 24 h intervals and analysed by DART accordingly, but only selected frames are shown here. The greenhouse tomato plants were raised hydroponically in horizontal flat boxes, using a modified recycling NFT (nutrient film technique) system in which the solution temperature was regulated (23 ± 1°C). The root images were mounted as a stack since they were taken from the same position using the same focal length, thus they are frames in a movie loop. The technique is similar to that described on rhizotrons or minirhizotrons set in laboratory or field conditions (Iijima et al. 1998; Walter et al. 2002).
In both examples, the image and the stack contain three types of information that must be extracted and assembled within a common dataset. These are (1) spatial data described by the Cartesian co-ordinates of the relevant pixels attached to all visible objects in the images, (2) connection data defining how roots are connected together within the network and (3) temporal data which allow the dating of the state of each object described in the image. In the first example, the latter are given by the colour code while in the second they are given by the rank of the frame within the stack. In the second example, it must also be noticed that the roots are not immobile, due to the substrate-less nature of their cultural system, implying that the roots of two successive frames may not coincide. This can create problems in time-series image analyses if the interpreted data cannot be moved, accordingly. In this case, different software become inoperative. This specific problem has, however, a broader importance, because soil-grown roots and apices also undergo slight irregular motion (circumnutation) either due to the plant root functioning (Shabala and Newman 1997) or associated with soil movements.
Concerning the spatial information, several objects are of prime importance and deserve a precise description. Among these are the positions of the apices between two successive frames (Example 2) and those of the ending points of all segments coded with the same colour (Example 1). These positions are informative of the length of individual roots throughout the time of the experiment. Similarly, from the connection viewpoint, the branching points (1) define the root filiations through the linkage between each individual root and its parents within the system and (2) determine the calculation of the branching density along the axes.
Based on these principles, and considering the relevant level of organization sought in our experiments, we propose to model the root system architecture as a set of “links”, each being defined as an object (within an object-oriented programming language) encapsulating the following information:
-
initial point (2D co-ordinates of origin);
-
observation number coding for the date at which the link was seen for the first time;
-
branching state (Boolean, indicative of the nascent or continuative nature of the link) where true denotes a branching link while false is a continuing link;
-
pointer on (i.e. address of) the father link which is the unique object where the actual link has emerged or of which it is the prolongation;
-
pointer on the list of children links. Indeed each link may carry from (0 to n) children links, ordered from proximal to distal.
Thus, hereafter, the link represents the atomic part of the root system, inferring that a root is defined by the linear series of its links standing from proximal to distal, the root system being determined by the entire link population. Moreover, the temporal level of information emerges when observation numbers are transcribed into the dates at which the observations were made. These allow calculation of the date of root emergence and time intervals between successive observations, which are required to compute individual root growth rates.
Principle of data acquisition and software features
Data acquisition is made manually within a graphical user interface (Fig. 2) comprising two windows, a command bar with drop down lists and a tool bar containing several text icons.
The left window displays the parameters of the current data acquisition. It comprises a table with three columns defining respectively, the observation number (integer from 1 to n), the date of observation (real number of time unit intervals between dates) and the colour code attributed to each time period. At any time the user can add observations (i.e. new dates of data acquisition) or remove an extra observation.
The right window is a graphical display in which the coloured links are drawn using mouse clicks in respect to the defined colour coding. The general principle is to overlay on the digital image standing as a desktop picture, the interesting coloured line segments and points (i.e. the junctions between two consecutive links) describing the roots. Apices are the tips of end-links. At this location, upon date and colour changes, a left click on the mouse button sets a continuative link. At all link junctions, a right click defines the origin of a nascent root i.e. defines a branching point. The user can also insert new break-points on ancient links, i.e. a link can be divided a posteriori. This feature is particularly important in the case of Example 2 due to the root emergence lag-time. Data acquisition is made for the selected observation number and associated colour, which can be changed at any time using the “Date>” and “Date<” text icons of the tool bar. The current observation number and its associated colour are displayed in the bar (top right position). Each time a new link is drawn, the software connects it to the existing network, thus rendering coherent the entire structure of connections. Each link, but the former, is connected to the preceding link, either at a distal (continuation) or a lateral position (branching). Thus, gradually the information is captured and displayed as segmented coloured lines overlying the root image (Fig. 2), giving the user real-time control of data acquisition.
The software captures and creates two types of data file: one is concerned with the links (link file) and the other with the correspondence between observation numbers, dates and colour codes.
Using the command bar with drop down list, the user can select several features of the software. In the File menu it is possible to load (in the RAM) and save (on Disk) the data files and choose amongst different tools described hereafter. These tools are also displayed in the tool bar as text icons (Fig. 2). They are dedicated to the data acquisition process and have been implemented to facilitate data capture, from the viewpoints of ergonomics and precision. In particular, they allow the user to:
-
(Zoom+ and Zoom-) select the image view size in order to refine root tracing delineation;
-
(Date> and Date<) shift the observation number for data capture;
-
(Add, Insert and Stop) activate or deactivate the process of data acquisition;
-
(Erase) suppress unwanted links;
-
(Move) displace an existing link. In the case of Example 2 described above, roots move between successive images of a time series. Thus, in order to overlay the foregoing root system on the selected image, several links must be displaced without affecting their length.
Results
The Java language chosen to develop DART enables it to run on all systems which support the Java software. It has been tested on Microsoft Windows, Mac OS X and Linux operating systems and represents the only free software dedicated to RSA studies in the Mac users community. Table 1 presents the major features of the most important software described in the scientific literature.
Figure 3 shows an example of the graphical output that can be displayed in DART to depict the time-evolving tomato RSA taken from Example 2. In this case, early observations (not shown) after germination, revealed the development of a taproot and several laterals. From the beginning of DART measurements onwards (i.e. 18–25 days after sowing or das), it should be noticed that the main parent root (taproot, order 1) and all secondary laterals (i.e. long and short, order 2) were traced in DART but only a few long secondary laterals were analysed in detail for higher order laterals (order ≥3). Figure 3 exemplifies only the topological capacities of the software, and does not pretend to be fully comprehensive example of tomato root architecture. Since all laterals (order ≥3) were not drawn here, we did not compute the total plant root length, but rather we preferred to compare characteristics (growth rates for instance) of individual roots sampled from a population of comparable order (see Figs. 4 and 5).
Graphical outputs displayed in DART to depict the root system architecture of a tomato plant at 28 das (see Fig. 1d). Each colour is typical of a date of observation between 15 and 28 das
Although organizing the data into such interconnected links has proven useful for the acquisition process and computer filing, it is obviously not the ultimate appropriate form of the dataset required to study RSA. Such structure has been devised to easily calculate root-based information, as the individual root is the essential object commonly considered to be relevant to analyse the development of the entire root system. The software is able to make such calculation and exports the dataset as a table file in which individual roots are lines with several attributes set in the following columns:
-
identification number
-
branching order
-
date of emergence
-
parent root identification number
-
distance between the branching point to the parent root base
-
root length at each observation date
This table serves as a starting database for further statistical analyses using appropriate statistical software, such as the GNU project R language (www.r-project.org) for instance.
Figure 4 shows an example of the time course change of individual tomato root growth (elongation) curves computed from a database and analysed using this methodology. At each date of observation, the figure highlights the existence of a large variability in absolute growth rate of roots of similar order (i.e. order 2 in this example), indicating clearly that growth rate is age dependent. Individual growth rate is low for young roots but increases with root age until reaching a maximum value (near 6–7 cm d-1). The concomitant drop in root growth rate observed 26 das was due to an abrupt change in the fairly stable climatic time series, where cloudy weather at 25 das decreased daily PPFR (photosynthetic photon fluence rate, mol m−2 over the waveband 400–700 nm) by 76%.
Figure 5 shows the distribution of inter-branch distances between short laterals held by their parent roots within the entire root system of a tomato plant grown in hydroponics (i.e. example 2). The figure indicates that most roots branch at a very short distance (mode = 1 mm) from their neighbours. Such a dataset allows us to describe the root system characteristics and in this example, it shows clearly that the population (n > 2000 roots) is not distributed symmetrically but is positively skewed, inferring that it may be important to consider more parameters than the simple mean to compare root growth characteristics between plants or treatments.
Discussion
DART has been developed to analyse complex root system architectures (RSA) in developmental process and root filiations studies. The information compiled in Table 1 shows that this software differs from those designed (and mainly used) to analyse minirhizotron pictures (RMS from Ingram and Leers 2001; MR-RIPL 2.0 & 3.0 http://rootimage.msu.edu/; MSU ROOTs Tracer http://roots.rsgis.msu.edu/; Rootfly http://www.ces.clemson.edu/~stb/rootfly/; RooTracker www.biology.duke.edu/rootracker/; RootView http://www.mv.helsinki.fi/aphalo/RootView.html). DART converts the information captured by images into a structured dataset suitable to study root development and analyse the RSA. Compared to other packages (Armengaud et al. 2009; Kimura et al. 1999; WinRHIZO™ www.regentinstruments.com; WR-RIPL 2.0 http://rootimage.msu.edu/; Zeng et al. 2008) this software does not use automated procedures to compute global variables. For fine roots, automatic measurements are highly dependent upon image quality and resolution, leading sometimes to deceptive results (Ryser 2006) particularly when real objects (i.e. roots) cannot be extracted from the background noise. Because DART relies on manual measurements, it cannot generate errors per se. DART exploits the expertise of human vision and provides technical help from the computer to visualize, organize and file the information carried by the image. According to root system complexity, it may take several hours to produce with DART a database with a confident information derived from the analysis. This adds up to the time required by the activity of root observations.
In DART, all markers are set manually. Their placement is adapted to the precision implemented throughout the entire process of image acquisition (time intervals, image quality and resolution), thereby minimizing the risks of errors during this phase. Hence, a zooming feature allows accurate placement of marker points. When roots are tortuous, the user may increase the number of links to refine the tracing of curvilinear shapes using a series of line segments. However, increasing the number of links increases also the time of data acquisition and eventually generates larger data files. Consequently, the user must find a balance between the constraints of accuracy, time, file storage capacity and dataset processing.
DART is particularly fitted for the examples treated in this paper, which are typical of experiments conducted on root systems. Obviously, the main use of DART should be the architectural analyses of intact and complex root systems in developmental studies. Nonetheless, it can analyse discontinuous root systems provided that each entity is treated as a single dataset. This constraint may prevent using DART for images holding numerous fractions of root systems, as is the case in minirhizotron studies. In contrast, other systems allow constructing images of intact root systems from multiple small sized pictures joined together. In this case, DART appears particularly effective to interpret time-based changes in the large, branched root system (see Fig. 3). Thus, in order to cope with root movements between successive frames in a time-series, DART gives the possibility of displacing a connecting point from its original position to a new one. In doing so, the father link changes orientation not its length while all descending links are moved accordingly. Thus the entire shape of the root system can be rearranged rapidly without altering its morphological characteristics.
DART is also functional to study complex RSA characterized at a single date as long as the analysis is concerned with topology and general descriptors such as length or branching densities. In this case however, no dynamic information will be gained. DART becomes appropriate to handle intricate systems bearing approximately more than twenty branched roots. For simpler root systems, picking other image analysis packages should prove adequate.
The DART dataset structure (i.e. a population of links) is highly flexible, allowing for subsequent calculation of other parameters than those described here. Branching angle for instance is an intrinsic trait of Cartesian coordinate measurements not exemplified here. The same apply to topological indicators discussed by Fitter (1987). The functional importance of other root descriptors, such as root diameter (Boukcim et al. 2006; Lecompte and Pagès 2007; Pagès 1995; Zobel et al. 2007) or colour has been emphasized in the literature. The current version of DART does not feature the measurement of root diameter. This is because in large images (ca.150 × 55 cm in Fig. 1 b–d) pixel size is too close to fine root diameter to allow accurate characterization (this point is explained in the paper of Zobel 2008). Nonetheless, there is no impediment to technical progress and this feature may deserve future implementation. Other packages (Rootfly http://www.ces.clemson.edu/~stb/rootfly/; RooTracker www.biology.duke.edu/rootracker/; WinRHIZO™ www.regentinstruments.com) propose also to keep track of root colour (set by user to white, light brown, dark brown or black in Rootfly) or to perform root classification according to their colour (WinRHIZO™ www.regentinstruments.com). However, root colour in architectural analyses has been only scarcely examined in the literature (McCrady and Comerford 1998; Smit and Zuin 1996) but has not emerged as a major root discriminant. Furthermore, users subjectively define colour coding and local substrate environment influences colour characteristics. Thus, for the time being, DART ignores this information.
DART is specialized in the 2D analysis of root systems, but displays similarities with software developed for 3D scanning of plant shoots (Godin 2000). For the time being the structure of the dataset has been kept simple, due to the straightforward nature of the branched root architecture, but it could evolve in the future to account for new root attributes, capable of describing changes of other root parameters, such as colour or diameter, for instance.
Readers can download DART from: (http://www.avignon.inra.fr/psh/outils/dart).
References
Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A (2009) EZ-RHIZO: integrated software for the fast and accurate measurement of root system architecture. Plant J 57:945–956. doi:10.1111/j.1365-313X.2008.03739.x
Berntson GM (1997) Topological scaling and plant root system architecture: developmental and functional hierarchies. New Phytol 135:621–634. doi:10.1046/j.1469-8137.1997.00687.x
Boukcim H, Pagès L, Mousain D (2006) Local NO -3 or NH +4 supply modifies the root system architecture of Cedrus atlantica seedlings grown in a split-root device. J Plant Physiol 163:1293–1304. doi:10.1016/j.jplph.2005.08.011
Bouma TJ, Nielsen KL, Koutstaal B (2000) Sample preparation and scanning protocol for computerised analysis of root length and diameter. Plant Soil 218:185–196. doi:10.1023/A:1014905104017
Cahn MD, Zobel RW, Bouldin DR (1989) Relationship between root elongation rate and diameter and duration of growth of lateral roots of maize. Plant Soil 119:271–279. doi:10.1007/BF02370419
Cheng W, Coleman DC, Box JE Jr (1991) Measuring root turnover using the minirhizotron technique. Agric Ecosyst Environ 34:261–267. doi:10.1016/0167-8809(91)90113-C
Colin-Belgrand M, Joannes H, Dreyer E, Pagès L, suppl (1989) A new data processing system for root growth and ramification analysis: description of methods. Ann Sci For 46:305s–309s. doi:10.1051/forest:19890570
Costa C, Dwyer LM, Hamel C, Muamba DF, Wang XL, Nantais L, Smith DL (2001) Root contrast enhancement for measurement with optical scanner-based image analysis. Can J Botany-Revue Canadienne Botanique 79:23–29. doi:10.1139/cjb-79-1-23
Danjon F, Reubens B (2008) Assessing and analyzing 3D architecture of woody root systems, a review of methods and applications in tree and soil stability, resource acquisition and allocation. Plant Soil 303:1–34. doi:10.1007/s11104-007-9470-7
Devienne-Baret F, Richard-Molard C, Chelle M, Maury O, Ney B (2006) Ara-rhizotron: An effective culture system to study simultaneously root and shoot development of Arabidopsis. Plant Soil 280:253–266. doi:10.1007/s11104-005-3224-1
Fitter AH (1986) The topology and geometry of plant-root systems - influence of watering rate on root-system topology in Trifolium pratense. Ann Bot (Lond) 58:91–101
Fitter AH (1987) An architectural approach to the comparative ecology of plant root systems. New Phytol 106:61–77
Fitter AH, Stickland TR, Harvey ML, Wilson GW (1991) Architectural analysis of plant-root systems. 1. Architectural correlates of exploitation efficiency. New Phytol 118:375–382. doi:10.1111/j.1469-8137.1991.tb00018.x
Godin C (2000) Representing and encoding plant architecture: a review. Ann Sci For 57:413–438. doi:10.1051/forest:2000132
Himmelbauer ML, Loiskandl W, Kastanek F (2004) Estimating length, average diameter and surface area of roots using two different Image analyses systems. Plant Soil 260:111–120. doi:10.1023/B:PLSO.0000030171.28821.55
Iijima M, Oribe Y, Horibe Y, Kono Y (1998) Time lapse analysis of root elongation rates of rice and sorghum during day and night. Ann Bot (Lond) 81:603–607. doi:10.1006/anbo.1998.0611
Ingram KT, Leers GA (2001) Software for Measuring Root Characters from Digital Images. Agron J 93:918–922
Johnson MG, Tingey DT, Phillips DL, Storm MJ (2001) Advancing fine root research with minirhizotrons. Environ Exp Bot 45:263–289. doi:10.1016/S0098-8472(01)00077-6
Kimura K, Kikuchi S, Yamasaki S (1999) Accurate root length measurement by image analysis. Plant Soil 216:117–127. doi:10.1023/A:1004778925316
Lecompte F, Pagès L (2007) Apical diameter and branching density affect lateral root elongation rates in banana. Environ Exp Bot 59:243–251. doi:10.1016/j.envexpbot.2006.01.002
McCrady RL, Comerford NB (1998) Morphological and anatomical relationships of loblolly pine fine roots. Trees-Structure Funct 12:431–437
Ortiz-Ribbing LM, Eastburn DM (2003) Evaluation of digital image acquisition methods for determining soybean root characteristics. Crop Management, 1-9
Pagès L (1995) Growth-patterns of the lateral roots of young oak (Quercus robur) tree seedlings - Relationship with apical diameter. New Phytol 130:503–509. doi:10.1111/j.1469-8137.1995.tb04327.x
Pagès L, Bengough AG (1997) Modelling minirhizotron observations to test experimental procedures. Plant Soil 189:81–89. doi:10.1023/A:1004288430467
Pagès L, Vercambre G, Drouet JL, Lecompte F, Collet C, Le Bot J (2004) Root Typ: a generic model to depict and analyse the root system architecture. Plant Soil 258:103–119. doi:10.1023/B:PLSO.0000016540.47134.03
Russ JC (1999) The image processing handbook. CRC Press, IEEE Press, Boca Raton, Florida, USA. pp. 771
Ryser P (2006) The mysterious root length. Plant Soil 286:1–6. doi:10.1007/s11104-006-9096-1
Shabala SN, Newman IA (1997) Root nutation modelled by two ion flux-linked growth waves around the root. Physiol Plant 101:770–776. doi:10.1111/j.1399-3054.1997.tb01062.x
Smit AL, Zuin A (1996) Root growth dynamics of Brussels sprouts (Brassica olearacea var gemmifera) and leeks (Allium porrum L) as reflected by root length, root colour and UV fluorescence. Plant Soil 185:271–280. doi:10.1007/BF02257533
Smit AL, Bengough AG, Engels C, van Noordwijk M, Pellerin S, van de Geijn SC (2000) Root methods: a handbook. Springer-Verlag, Berlin, Heidelberg, New York, London, Paris, Tokyo, Hong Kong, p 587
Taub DR, Goldberg D (1996) Root system topology of plants from habitats differing in soil resource availability. Funct Ecol 10:258–264. doi:10.2307/2389851
Vamerali T, Ganis A, Bona S, Mosca G (1999) An approach to minirhizotron root image analysis. Plant Soil 217:183–193. doi:10.1023/A:1004616217070
Vamerali T, Guarise M, Ganis A, Bona S, Mosca G (2003) Analysis of root images from auger sampling with a fast procedure: a case of application to sugar beet. Plant Soil 255:387–397. doi:10.1023/A:1026147607879
Walter A, Spies H, Terjung S, Küsters R, Kirchgebner N, Schurr U (2002) Spatio-temporal dynamics of expansion growth in roots: automatic quantification of diurnal course and temperature response by digital image sequence processing. J Exp Bot 53:689–698. doi:10.1093/jexbot/53.369.689
Zeng G, Birchfield ST, Wells CE (2008) Automatic discrimination of fine roots in minirhizotron images. New Phytol 177:549–557
Zobel RW (2003) Sensitivity analysis of computer-based diameter measurement from digital images. Crop Sci 43:583–591
Zobel RW (2008) Hardware and software efficacy in assessment of fine root diameter distributions. Comput Electron Agric 60:178–189
Zobel RW, Kinraide TB, Baligar VC (2007) Fine root diameters can change in response to changes in nutrient concentrations. Plant Soil 297:243–254
Acknowledgements
We are grateful to Dr. D. J. Pilbeam (University of Leeds, UK) and Dr. C. Jourdan (CIRAD, France) for their critical reviews and suggestions to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Jonathan P. Lynch.
Rights and permissions
About this article
Cite this article
Le Bot, J., Serra, V., Fabre, J. et al. DART: a software to analyse root system architecture and development from captured images. Plant Soil 326, 261–273 (2010). https://doi.org/10.1007/s11104-009-0005-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-009-0005-2