Abstract
The objective of this work was to evaluate the effect of endophytic diazotrophic bacteria Herbaspirillum and Burkholderia on the Fusarium oxysporum f. sp. cubense (Foc) establishment and on the plantlets growth of the ‘Maçã’ banana (Musa spp., group ABB). Two assays were carried out in a greenhouse at the National Center of Tropical Agroindustry in Fortaleza city, Ceará State (Brazil), using randomized block designs in the factorial arrangements 4 × 2 and 8 × 2. On the first trial plantlets were inoculated with the Burkholderia sp. AB202; Herbaspirillum sp., BA227, both of strains and controls bacteria, with and without Foc and cultivated in pots filled with an autoclaved mixture of washed sand and vermiculite (ratio 3:2 v v−1), during 4 months. On a second assay, plants were subjected to following conditions: absence and presence of strains AB202, AB213 (Burkholderia spp.), BA227, BA234 (Herbaspirillum-like), AB202 plus BA227, AB213 plus BA234, the mixture of the four bacterial strain, absence and presence of the Foc; and cultivated in pots filled with autoclaved Haplic Arenosol, during 2 months. The plant association with diazotrophs and the Foc was confirmed, and factors interacted significantly on the most probable number of bacteria and the colony forming units of the pathogen on roots and plant rhizomes. The potential of the endophytes on the inhibition of Foc propagate units and on the plant growth promotion was demonstrated. The higher biomass was observed four and 2 months after plant inoculation with AB202 and BA234. Results showed that these endophytes may be used as potential biofertilizer and biocontrol agents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Banana trees are widely cultivated in tropical countries. Brazil, in 2005, produced 6,802.99 Tons on 499.23 ha (FAO 2007). It has a high consumer demand worldwide, being an important source of nutrients. Although Brazil detains a high crop production, it has a low efficiency, mainly in the Northeast region, where only 12 Tons per ha is harvested. A low technological level is adopted by farmers and diseases are frequently uncontrolled on production areas. Fusarium wilt disease is a serious constraint to many cultivars grown in different regions, caused by the soil-borne fungus Fusarium oxysporum f. sp. cubense (Foc), Smith (Cordeiro 1999).
The pathogen Foc is widely spread in different producing regions, infecting many genotypes, including the cultivar ‘Maçã’ from subgroup ‘Prata’, which represents 95% of the whole national banana plantations (Cordeiro 1999; Silva et al. 2003). In some plantations of the Brazilian Northeastern region Foc is so destructive that farmers obtain no more than one harvest, turning the area unsuitable to new crop (Weber and Freire 2003).
Substitution of the susceptible cultivar by resistant hybrids is effective against Fusarium wilt (Cordeiro 1999; Silva et al. 2003). However, consumers become displeased due to their ‘Maçã’ banana preference (Matsuura et al. 2004). Thus that kind of banana has a higher local cost when compared to the others and the farmers search for new areas to continue planting that cultivar. As chemical control presents low effectiveness (Campanioni et al. 2005; Nel et al. 2006), biological control is an attractive option, mainly during plantlet formation.
Considering the soil antagonists, such as Pseudomonas fluorescens (Saravanan et al. 2003), P. aeruginosa (Ayyadurai et al. 2006), Burkholderia cepacia (Pan et al. 1997), Trichoderma harzianum (Thangavelu et al. 2003), Trichoderma sp. (Nel et al. 2006) and Streptomyces sp. (Getha et al. 2005), which reduce symptoms of Fusarium wilt, screening of plant growth-promoting rhizobacteria (PGPR) would contribute to optimise the yield. The PGPR competitively colonize plant roots, and stimulate plant growth and/or reduce the incidence of plant diseases (Haas and Défago 2005). Pseudomonas strains 84 and 4B when introduced to banana roots of tissue-cultured plants at de-flasking stage improved plant growth and reduced infection of Foc in the rhizome under green house conditions (Smith et al. 2003). Recently, Jaizme-Vega et al. (2004) found that bacterization of banana cv. Grand Nain and ITC-1297 plantlets with Bacillus spp. significantly improved plant growth and foliar mineral contents.
Endophytic diazotrophic bacteria have been isolated from banana plants (Weber et al. 1999) and some strains are plant growth-promoting (Weber et al. 2000) and also are able to inhibit Foc growth in vitro (Muniz et al. 2007). Since micropropagated plantlets are sensitive to soil pathogens (Ayyadurai et al. 2006), inoculation with selected microorganisms would be desirable. Hence, the present investigation was undertaken to evaluate the effect of endophytic diazotrophic bacteria Herbaspirillum and Burkholderia on the Foc establishment and on the plantlets growth of the ‘Maçã’ banana.
Material and methods
Plant material
The banana ‘Maçã’ plantlets were obtained from shoot tip culture. The proliferation medium was MS added with 30 g l−1 of sucrose and 2.5 mg l−1 of 6-benzylaminopurine. The shoots were elongated on same medium without growth regulators, at Embrapa Tropical Agroindustry and Biotece Tissue Culture Laboratories, both located in Fortaleza, Ceará, Brazil. The explants used were 8 to 10 cm height, with three to four leaves.
Microbial isolation and characterization
Endophytic diazotrophic bacteria were isolated from root and stem samples of pineapple (Ananas comosus L. Merril) cultivar ‘Cayenne Champac’(Champaka) (AB202) collected in Ceará, variety ‘Pérola’ (AB213) from Paraíba State, banana (Musa spp.) cultivar ‘Pacovan’ (BA227) and ‘Prata’ (BA234) from Ceará State. Following the purification steps, bacterial strains were characterized by the morphology of colonies on solid JNFb medium and by their ability of using different carbon sources in semi-solid and semi-specific media, according to Weber et al. (1999), and by sequencing and phylogenetic analysis of 16S rRNA genes. The molecular data based-method was performed at the Center of Chemical, Biological and Agricultural Research (CPQBA, Campinas State University, São Paulo State, Brazil). Briefly, total genomic DNA of the bacteria was purified according to Young and Blakesley (1991) and the fragments of the 16S rRNA genes were amplified and sequenced following Rainey et al. (1996), using an automatic system MegaBace 500 (GE Healthcare). The data obtained were compared with 16S rRNA sequence data from type strains available in the public databases GenBank (http://www.ncbi.nlm.nih.gov/BLAST/) and RDP (Ribosomal Database Project, Wisconsin, USA, http://www.cme.msu.edu/RDP/html/index.html); using nucleotide blast and RDP sequence match routines. All bacterial strains were stored at the laboratory of Soil Microbiology of Embrapa Tropical Agroindustry, Fortaleza, Ceará, at −18°C in liquid Dygs medium (Rodrigues Neto et al. 1986) with glycerol (50%). The Foc was isolated from rhizome tissue of symptomatic plants of the cultivar ‘Maçã’ obtained from local production areas. Typical structures of that pathogen were confirmed during the purification step on potato dextrose agar (PDA), and plates were maintained in a refrigerator at 5 ± 1°C before preparing the fungal inoculum.
The bacterial strains were deposited at CBMAI (Brazilian Collection of Environmental and Industrial Microorganisms, Campinas, São Paulo State), under the accession numbers CBMAI 255 (= AB202), CBMAI 256 (= AB213) and CBMAI 258 (= BA227), except for BA234, which was deposited at Embrapa Agrobiology, in Seropédica, Rio de Janeiro State.
Microbial inoculation
Bacterial strains were grown in liquid Dygs medium on a rotary shaker (125 rpm) for 24 h, at 30°C, whilst the Fusarium strain was grown in the medium with same formulation on a rotary shaker (125 rpm) for 48 h, at 30°C. A final concentration of 5 × 107 colony forming units (CFU) per ml of the bacterial suspensions was obtained as measured by optical densities (OD) of 0.7, using a Genesys 336002 Spectrophotometer (λ = 550 nm).
Cultures of different bacterial strains were used to inoculate the banana plantlets. Groups of four uniform size seedlings were transferred into tubes containing 10 ml of sterilized saline solution (1 mg K2HPO4, 0.5 mg MgSO4, 0.2 mg NaCl, 0.5 mg CaCl2.2H2O, 0.8 μg CuSO4.5H2O, 24 μg ZnSO4.7H2O, 28 μg H3BO3, 20 μg NaMoO4.2H2O, 23.5 μg MnSO4.H2O and 10 μl FeEDTA 1.64% solution), adjusted to pH 6.5. Aliquots of 4.0 ml of respective strain suspension were added to each tube. Plants immersed only in the media served as control. Plants were maintained at 28°C in growth chamber for 24 h. After bacterial suspensions discharge, each treatment tube was added with Foc spores suspension. Another part of the treatment tubes was added by saline suspensions without Foc to comparison. The plantlets were then maintained at 28°C in growth chamber for another 24 h. Plants were used for pots greenhouse experiments.
Banana pots experiment
The experiment was conducted at the greenhouse comprising two factorial assays, arranged in a randomized complete block design. The first assay occurred from February to May 2006 and had eight treatments with six replicates. Soil used in plastic pots (2.5 l) was autoclaved washed sand plus vermiculite (3:2, v v−1) with trace nutrients composition. The treatments were:
-
Four levels of bacterial inoculation: Burkholderia sp. AB202 (=CBMAI 255), Herbaspirillum sp. BA227 (=CBMAI 258), a combination of them, and uninoculated control;
-
Two levels of fungal inoculation: Foc and uninoculated control
The second assay occurred from May to July 2006 and had 16 treatments and six replicates. It was carried out in pots containing 2.5 l of an autoclaved Haplic Arenosol soil, characterized according to van Raij et al. (2001), and presenting pH in CaCl2 (0.01 M) = 7.0; Organic Matter = 8.5 mg dm−3; P (resin) = 7.0 mg dm−3; K+ (resin) = 1.9 mmolc dm−3; Ca+2 (resin) = 19.5 mmolc dm−3; Mg+2 (resin) = 3.5 mmolc dm−3; Na+ (Merlich) = 3,0 mmolc dm−3; Cu (DTPA) = 0,2 mg dm−3; Fe (DTPA) = 9 mg dm−3; Mn (DTPA) = 11,6 mg dm−3 and Zn (DTPA) = 1,1 mg dm−3. The treatments were as follows:
-
Eight levels of bacterial inoculation: Burkholderia strains AB202 (=CBMAI 255) and AB213 (=CBMAI 256), Herbaspirillum strains BA227 (=CBMAI 258) and BA234 (=BR12141), a combination of AB202 + BA227 strains, a combination of AB213 + BA234 strains, a combination of all of them, uninoculated control;
-
Two levels of fungal inoculation: Foc and uninoculated control
The soil was maintained humid in the pots by a nebulisation system, which was daily activated for four or five times for every 5 min. Fortnightly, each vase received 50 ml of Hoagland solution (Hoagland and Arnon 1950), modified by N content (4.2 mg NH4NO3).
Plant surviving and diazotrophic bacteria and Foc colonization
Sampling occurred 120 days after cultivation for the first assay and after 75 days for the second assay. Plants were divided into roots (main and lateral roots) and aerial (basal stem and leaves) pieces. Samples for bacterial density determination were thoroughly washed with water, cut into small pieces from 1 g, and transferred to sterile tubes containing 10 ml of 1% T chloramine for 5 min. The samples were then washed with distilled water, macerated with mortar and pestle, and the homogenate was serially diluted (until 10−7) and inoculated on vials containing JNFb medium (Döbereiner et al. 1995), using three replicates and incubated at 30°C. Dilutions were also plated on PDA medium and incubated at 30°C. After 72 h of incubation, vials were evaluated to determine the most probable number (MPN) of diazotrophic bactéria. Plates were counted for CFU Foc determination. Remaning plant portions were dried for biomass measuring using the formula: [100 (X−Y) Y−1], where X corresponded to inoculated plant weight and Y, control plant weight.
Statistical analysis
Bacteria MPN data were transformed to Bacteria MPN and Foc CFU data, to log x + 1, prior statistical analysis. Data were subjected to ANOVA at the significant level and means were compared by Duncan’s multiple range test (P < 0.05).
Results
Characterization of diazotrophs
The bacteria tested were able to use malate, succinate, mannitol and sorbitol as sole carbon source in semi-solid N-free media. Phenotypic data of carbon source utilization (Table 1) combined with 16S rRNA gene analysis (Fig. 1) allowed the identification of the strains AB202 and AB213 as Burkholderia sp. and Burkholderia anthina, respectively. Strain AB202 presented high similarity values with B. cepacia and B. vietnamiensis (99%) in the BLASTn search, and was recovered in a tight cluster with Burkholderia cepacia in the phylogenetic tree (Fig. 1). However, this grouping was not supported by high bootstrap value, indicating that this closer relationship is not reliable. In that sense, identification of strain AB202 was not conclusive at the species level. Both strains presented medium or good growth on the sugar substrates d-raffinose, sucrose, d-fructose, d-glucose, l-arabinose and N-acetyl-glucosamine. The two other strains, BA227 and BA234, exhibited none or poor growth with d-raffinose, sucrose and d-fructose semi-solid N-free media. Strain BA234 exhibited blue centered colonies on solid NFb medium, forming veil-like membranes near to the surface of semi-solid N-free JNFb medium, typical characteristics of Herbaspirillum genera, as described by Döbereiner et al. (1995). 16S rRNA sequence analysis indicated that strain BA227 was phylogenetically related to different species of Herbaspirillum (Fig. 1); however a conclusive identification was not achieved.
Phylogenetic analysis based on partial 16S rRNA gene sequences of the endophytic diazotrophic bacteria and related microorganisms using the Kimura 2p evolutionary model and the neighbor joining method for tree reconstruction. Bootstrap values (1000 replicate runs) greater than 70% are listed. GenBank accession numbers are listed after species names. Thiobacillus denitrificans NCIMB 9548T was used as outgroup
Greenhouse experiment
Colonization of banana plants was successfully accomplished by Burkholderia spp. and by Herbaspirillum sp., as well as by Foc (Tables 2 and 4). Bacteria isolated from the inoculated plants showed the same physiological characteristics as those we observed prior inoculation (Table 1).
First assay ANOVA showed a significant factor interaction on bacteria MPN and on Foc CFU from roots and rhizomes of the inoculated plants, except for bacteria MPN from root plants (P < 0.05) (Table 2). Bacterial population within those root plants was compared aiming to verify the Burkholderia sp. (AB202) inoculation benefit on the plants in the absence of the pathogen. Rhizome plants did not show variation between bacteria population in the absence of Foc, except for Herbaspirillum sp. (BA227) and the combination of AB202+BA227.
In the second assay, where the substrate was a Haplic Arenosol, largest diazotrophic bacteria populations were detected on roots in the presence of AB213, BA227, BA234, plus the Foc, when compared to the control samples (Table 4). Increments on the bacteria populations were detected after inoculations with AB202, BA227, the combination of all of them, in the absence of Foc. In the presence of Foc, larger bacteria populations were found out at the rhizomes of banana plants inoculated with AB202, BA234, AB213+BA234, AB202+BA227+AB213+BA234, when compared to control samples. These results indicate the preference of the bacteria to colonize specific plant tissues.
Bacteria inoculation also influenced Foc propagules. In the first assay, a reduction of the pathogen propagules was detected on roots of plants bacterized with AB202 and the combination of AB202 plus BA227 strains, when compared to control plants (Table 2). This reduction was also observed in rhizomes of plants bacterized with BA227 and the combination of all the strains. Also, survival rate was higher in the presence of AB202 strain and for the combination of all of them (Table 3), indicating the Foc infection process was blocked. The Foc detection on control plants in the second assay (Table 4) is probably due to the short distance between the pots inside the greenhouse (20 to 40 cm). Even so, roots presented a lowest Foc CFU, when BA227 strain was introduced. There was a reduction of the Foc CFU in plantlets inoculated with BA234 strain and the combination of AB202 plus BA227, when compared to the control. Surviving plant rates were maximal when AB202, AB213 and the combination of AB213 plus BA234 strains were introduced (Table 4). These inhibition and survival values may be due to Herbaspirillum-like bacteria action against Foc infection.
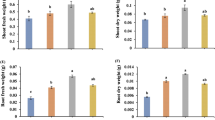
Plantlets also showed a better growth, with largest amount of dry root biomass after inoculation of AB202 in the first assay (Table 3). In the presence of that bacterial strain a relative increment of 35.6% in the root biomass and 23.1% in the total plant biomass were observed. In the second assay (Table 5), an increase on the dry root biomass was observed with BA234 strain inoculation, but those medium values didn’t differ of AB213, BA227 and combination of all strains inoculated. Contrary to the first assay, AB202 strain was less effective on root growth induction, may be due to soil substrate and shorter cultivation period. Accumulated total biomass didn’t vary significantly between the strains, although a relative increment of 26.7% was observed in the presence of BA234 strain and 18.3% with AB213 strain.
Discussion
Previous studies of endophytic bacteria have relied on phenotypic and genotypic characterizations for their identification (Mao et al. 2006; Lodewyckx et al. 2002). Comparative sequence analysis of 16S rRNA genes provides a reliable approximation to species level and cultivation studies help to enhance tentative allocation. Patterns of carbon assimilation in Herbaspirillum sp. were similar to the observations made by Weber et al. (1999). Affiliation to this genus was confirmed by BA227 16S rRNA sequences. Carbon assimilation patterns for strains BA227 and BA234 strains differed from species H. seropedicae (Baldani et al. 1986), H. rubrisubalbicans (Baldani et al. 1996) and H. frisingense (Kirchhof et al. 2001) and as such they may be included in a new diazotrophic species.
Bacterial colonization of plants was effective, with some variations in populations. These bacteria MPN variations in both assays show a trend of the colonization site within plants and the strain AB202 seems to be greatly adapted to roots in plants grown using poor fertile soil. B. vietnamiensis has been reported from the rhizosphere of rice plants (Gillis et al. 1995) and bacteria associated with B. cepacia have been found in larger numbers on banana roots (Weber et al. 1999). According to these authors, growth of Burkholderia-like bacteria prefers substrates with higher sugar concentrations, in comparison to other endophytic bacteria, making them more competitive for colonization sites.
In this work, diazotrophic bacteria were also detected in roots and rhizomes of control banana plants (Tables 2 and 4), in accordance with Weber et al. (2000) and Weber et al. (2003), who isolated diazotrophic bacteria from roots and rhizomes of micropropagated banana plantlets cv. ‘Prata Anã’ and ‘Caipira’, and from pineapple, respectively. Considering that micropropagated banana plantlets are sensitive to soil pathogens (Ayyadurai et al. 2006), we suggest the inoculation of plantlets with selected diazotrophic bacteria.
Bacterial–fungal pathogen interactions have been extensively reported, including studies with Burkholderia sp. strains against Fusarium sp. (Whipps 2001). In this work we observed a reduction of Foc unit propagules, suggesting that endophytic colonization reduces Fusarium wilt severity in the cultivar ‘Maçã’ plantlets. Since the first experimental evidence for the involvement of bacteria-mediated induced resistance against Fusarium wilt (Van Peer et al. 1991), several studies have suggested that treatments with selected endophytic bacteria could induce plants to defend themselves against pathogen attack (Kloepper 1993). Induced resistance has also been associated to bacterial presence in roots (Loon et al. 1998). Few plantlets showed internal symptoms of Fusarium wilt in this study, although this characteristic could not be differentiated between treatments. Determination of propagules in roots and rhizomes is therefore a practical measure and can be employed as a technique for banana genotypes to obtain a degree of sensibility to Foc colonization. This antagonism was also detected via an in vitro assay, and Muniz et al. (2007) observed antagonism of AB202, AB213, BA234 and BA227 strains against Foc with inhibition values corresponding to 78, 78, 74 and 21 cm2 of culture radial growth, respectively. Previous data obtained from Herbaspirillum species (Baldani et al. 1986, 1996; Kirchhof et al. 2001) and other endophytic diazotrophic bacteria from grasses in Brazil (Baldani and Baldani 2005) did not show antagonism against fungi, indicating a possibility of strains BA227 and BA234 representing different bacterial species.
Gillis et al. (1995) observed production of siderophores by B. vietanamiensis TW75T and antagonist effect against Fusarium and other soil fungi (Bevivino et al. 1994). In addition, Weber and Freire (2003), using the cultivar ‘Maçã’ plantlets colonized by AB202, observed reduction of Fusarium wilt in field banana. Increments in biomass may be also related to endophytic bacterization. Larger biomass increments were detected by Weber et al. (2000) working with cv. ‘Prata Anã’ and ‘Caipira’ after inoculation of diazotrophic bacteria isolated from fruit plants.
Based upon the results obtained we suggest the inoculation of endophytic diazotrophic bacteria as biofertilizers and antagonists to Foc on ‘Maçã’ banana plantlets. This should be considered as an additional option in the integrated management of Fusarium wilt, considering that B. cepacia (Pan et al. 1997), P. fluorescens (Saravanan et al. 2003), P. aeruginosa (Ayyadurai et al. 2006), T. harzianum (Thangavelu et al. 2003), Trichoderma sp. (Nel et al. 2006), non-pathogenic F. oxysporum (Forsyth et al. 2006) and Streptomyces sp. (Getha et al. 2005) have been indicated for biocontrol of Fusarium wilt in banana.
Conclusions
-
1.
The diazotrophic endophytic bacteria Burkholderia and Herbaspirillum characterized in this study showed the potential to reduce Foc attack and to promote the growth of banana ‘Maçã’ plants.
-
2.
Foc propagules can be determined directly from banana plant roots and rhizomes, aiming at the establishment of the resistance degree to Foc.
-
3.
Strains of Burkholderia sp. (AB202) and Herbaspirillum-like (BA234) induce increments on banana plant biomass production, after four and 2 months of cultivation, respectively.
References
Ayyadurai N, Rayindran N, Rao MS, Kumar RS, Samrat SK, Manohar M, Sakthivel N (2006) Isolation and characterization of a novel banana rhizosphere bacterium as fungal antagonist and microbial adjuvant in micropropagation of banana. J Appl Microbiol 100:926–937
Baldani JI, Baldani VLD (2005) History on the biological nitrogen fixation research in gramineous plants: special emphasis on the Brazilian experience. An Acad Bras Ciênc 77:549–579
Baldani JI, Baldani VLD, Seldin L, Döbereiner J (1986) Characterization of Herbaspirillum seropedicae gen. nov., a root associated nitrogen-fixing bacterium. Int J Sys Bacteriol 36:86–93
Baldani JI, Pot TB, Kirchhof G, Falsen E, Baldani VLD, Olivares FL, Hoste B, Kersters K, Hartmann A, Gillis M, Döbereiner J (1996) Emended description of Herbaspirillum; a mild plant pathogen, as Herbaspirillum rubrisubalbicans comb. nov.; and classification of a group of clinical isolates (EF group 1) as Herbaspirillum species 3. Int J Sys Bacteriol 46:802–810
Bevivino A, Tabacchioni S, Chiarini L, Carusi MV, Del Gallo M, Visca P (1994) Phenotypic rhizosphere and clinical isolates of Burkholderia cepacia. Microbiology 140:1069–1077
Campanioni B, Mora N, Diaz L, Pérez A, Arzola M, Espinosa P, Ernández M, Ventura J, Pérez MC, Santos R, Lorenzo CJ (2005) Identification of discriminant factors after treatment of resistant and susceptible banana leaves with Fusarium oxysporum f. sp. cubense culture filtrates. Plant Breed 124:79–85
Cordeiro ZJM (1999) Doenças. In: Alves EJ (org.) A cultura da banana: aspectos técnicos, socioeconômicos e agroindustriais, 2nd edn. Embrapa-SPI, Brasília, pp 353–407
Döbereiner J, Baldani VLD, Baldani JI (1995) Como isolar e identificar bactérias diazotróficas de plantas não-leguminosas. Embrapa-SPI, Brasília, p 60
FAO (Food and Agriculture Organization of the United Nations) (2007) FAOSTAT, core production data. Available at http://faostat.fao.org/site/340/DesktopDefault.aspx?PageID=340. Accessed 4 Apr 2007
Forsyth LM, Smith LJ, Aitken AB (2006) Identification and characterization of non-pathogenic Fusarium oxysporum capable of increasing and decreasing Fusarium wilt severity. Mycol Res 110:929–935
Getha K, Vikineswary S, Wong WH, Seki T, Ward A, Goodfellow M (2005) Evaluation of Streptomyces sp. strain g10 for suppression of Fusarium wilt and rhizosphere colonization in pot-grown banana plantlets. J Ind Microbiol Biotechnol 32:24–32
Gillis M, Trân Van V, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez MP (1995) Polyphasic taxonomy in the genus Burkholderia leading to an amended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol 45:274–289
Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3:307–319
Hoagland DR, Arnon DT (1950) The water culture method for growing plants without soil. California Agriculture Experiment Station, University of California, Berkeley, CA, p 32
Jaizme-Vega MDC, Romero ASR, Guerr, MSP (2004) Potential use of rhizobacteria from the Bacillus genus to stimulate the plant growth of micropropagated bananas. Fruits 59:83–90
Kirchhof G, Eckert B, Stoffels M, Baldani JI, Reis VM, Hartmann A (2001) Herbaspirillum frisingense sp. nov., a new nitrogen-fixing bacterial species that occurs in C4-fibre plants. Int J Syst Evol Microbiol 51:157–168
Kloepper JW (1993) Plant growth-promoting rhizobacteria as biological control agents. In: Metting FB Jr (ed.) Soil microbial ecology: applications in agricultural and environmental management. Marcel Dekker, New York, NY, pp 255–274
Lodewyckx CJ, Vangronsveld F, Porteous Moore ERB, Moore S, Taghavi D, van der Lelie (2002) Endophytic bacteria and their potential applications. Crit Rev Plant Sci 21:583–606
Loon LC, Bakker PA, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483
Mao S, Lee SJ, Hwangbo H, Kim YW, Park KH, Cha GS, Park RD, Kim KY (2006) Isolation and characterization of antifungal substances from Burkholderia sp. culture broth. Curr Microbiol 53:358–364
Matsuura FCAU, Costa JP, Polegatti MIS (2004) Marketing de banana: preferências do consumidor quanto aos atributos de qualidade dos frutos. Rev Bras Frutic 26(1):48–52
Muniz CR, Weber OB, Freire FCO (2007) Behaviour of Fusarium oxysporum f.sp. cubense (E.F. Smith) in the presence of endophytic bacteria in vitro. Br J Microbiol (in press)
Nel B, Steinberg C, Labuschagne N, Viljoen A (2006) The potential of nonpathogenic Fusarium oxysporum and other biological control organisms for suppressing Fusarium wilt of banana. Plant Pathol 55:217–223
Pan MJ, Rademan K, Hastings JW (1997) Ultrastructural studies on the colonization of banana tissue and Fusarium oxysporum f. sp. cubense race 4 by the endophytic bacterium Burkholderia cepacia. J Phytopathol 145:479–486
Rainey FA, Ward-Reiney N, Kroppenstedt RM, Stackebrandt E (1996) The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int J Syst Bacteriol 46:1088–1092
Rodrigues Neto J, Malavolta VA Jr, Victor O (1986) Meio simples para isolamento e cultivo de Xanthomonas campestris pv. citri tipo B. Sum Phytol 2:16
Saravanan T, Muthusamy M, Marimuthu T (2003) Development of integrated approach to manage the fusarial wild of banana. Crop Prot 22:1117–1123
Silva SO, Gasparotto L, Matos AP, Cordeiro ZJM, Ferreira CF, Ramos MM, Jesus ON (2003) Programa de melhoramento de bananeira no Brasil – resultados recentes. Embrapa Mandioca e Fruticultura, Cruz das Almas, p 36
Smith L, Keefe DO, Smith M, Hamill S (2003) The benefits of applying rhizobacteria to tissue cultured bananas. Banana Topics Newsletter 33:1–4
Thangavelu R, Palaniswami A, Doraiswamy S, Velazhan R (2003) The effect of Pseudomonas fluorescens and Fusarium oxysporum f. sp. cubense on induction of defense enzymes and phenolics in banana. Biol Plant 46:107–112
Van Peer R, Niemann GJ, Schippers B (1991) Induced resistance and phytoalexin accumulation in biological control of Fusarium wilt of carnation by Pseudomonas sp. strain WCS417r. Phytopathology 81:28–734
van Raij B, Andrade JC, Cantarella H, Quaggio JA (2001) Análise química para avaliação da fertilidade de solos tropicais. Instituto Agronômico de Campinas, Campinas, Brazil, p 285
Weber OB, Freire FCO (2003) Contribuição de bactérias diazotrópicas na cultura da bananeira: perspectivas de utilização na produção integrada. Embrapa Meio Ambiente, Jaguariúna, p 29
Weber OB, Baldani VLD, Teixeira KRS, Kirchhof G, Baldani JI, Döbereiner J (1999) Isolation and characterization of diazotrophic bacteria from banana and pineapple plants. Plant Soil 210:103–113
Weber OB, Baldani JI, Döbereiner J (2000) Bactérias diazotróficas em mudas de bananeira. Pesq Agropec Bras 35:2277–2285
Weber OB, Correia D, Rocha MW, Alvez GC, Oliveira EM, Sá EG (2003) Resposta de plantas micropropagadas de abacaxizeiro à inoculação de bactérias diazotróficas em casa de vegetação. Pesq Agropec Bras 38:1419–1426
Whipps JM (2001) Microbial interactions and biocontrol in the rhizosphere. J Exp Bot 52:487–511
Young A, Blakesley R (1991) Sequencing plasmids from single colonies with the dsDNA cycle sequencing system. Focus 13:13
Acknowledgements
We are grateful to the Brazilian Council for Scientific and Technological Development (CNPq) for the financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Euan K. James.
Rights and permissions
About this article
Cite this article
Weber, O.B., Muniz, C.R., Vitor, A.O. et al. Interaction of endophytic diazotrophic bacteria and Fusarium oxysporum f. sp. cubense on plantlets of banana ‘Maça’. Plant Soil 298, 47–56 (2007). https://doi.org/10.1007/s11104-007-9335-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-007-9335-0