Abstract
Key message
Endogenous and exogenous GA3 responses to DoEXP and DoXTH depend on the DoGA20ox1, DoGA3ox1, DoGA2ox3, DoGA2ox4, DoGID1a, and DoDELLA1 to regulate yam tuber growth.
Abstract
Yam tuber undergoes significant alteration in morphogenesis and functions during growth, and gibberellins (GA) are considered potentially important regulators of tuber growth. However, it is little known about the regulation of GA metabolism and GA signaling components genes in tuber growth of yam. In this study, the cloning and expressions of GA3 level, GA metabolism and signaling genes, and cell wall genes in tuber growth in response to GA3 and GA biosynthesis inhibitor paclobutrazol (PP333) treatments were studied. The contents of GA3 accumulated at the tuber growth, with the highest levels in the early expansion stage. DoGA20ox1, DoGA3ox1, and four DoGA2ox genes were significantly abundant in the early expansion stage of tuber and gradually declined along with tuber growth. Three DoGID1 and three DoDELLA genes were showed different expression patterns in the early expansion stage of tuber and gradually declined along with tuber growth. Five DoEXP and three DoXTH genes expression levels were higher in the early expansion stage than in other stages. Exogenous GA3 increased endogenous GA3 levels, whereas the expression levels of DoGA20ox1, DoGA3ox1, DoGID1a, and DoDELLA1 were down-regulated in the early expansion stage of tuber by GA3 treatment, DoGA2ox3 and DoGA2ox4 were up-regulated. PP333 application exhibited opposite consequences. Thus, a mechanism of GA3 regulating yam tuber growth by DELLA-dependent pathway is established.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Yams are monocotyledonous plants belonging to the family Dioscoreaceae, and the tuber is its harvested organ. Yam tuber is an important storage source of starch, sugars, proteins, amino acids, vitamins, and small amounts of essential minerals. It is widely cultivated in tropical and subtropical regions. This biological process of yam tuberization could be divided into three phase: initiation, expansion, and maturation stage, and the expansion stage could be further divided into three periods: early expansion, middle expansion, and late expansion stage (Gong et al. 2016; Matsumoto et al. 2005). Tuber expansion was initiated, which was accompanied rapidly by a massive deposition of starch and storage proteins (Aksenova et al. 2012). It is very important to study the mechanism of tuberization for improving the yield and quality of yam.
Various endogenous and environmental factors influenced yam tuberization (Yoshida et al. 2007). It has been shown that high sucrose level, low nitrogen levels, short days, and high temperature promoted yam tuber tuberization (Feng et al. 2007; Chen et al. 2010; Agele et al. 2010; Yoshida et al. 2007). Plant hormones play an important role in tuber tuberization of yam; for example, gibberellins (GA), indole acetic acid (IAA), and abscisic acid (ABA) play a key role at the beginning of the tuber expansion stage, and trans-zeatin (tZ) and jasmonic acid (JA) are also involved in tuber growth (Chen et al. 2007; Gong et al. 2016). GA is involved in growth and development processes including seed germination, stem elongation, flower induction and development, and fruit set. Exogenous GA increased tuber weight, caused a significant tuber yield in yam (Gong et al. 2015, 2016; Yoshida et al. 2008), while inhibited tuber expansion in potato (Cheng et al. 2018; Hartmann et al. 2011). It shows that there is a different role of GA in regulating tuber growth between potato and yam, but the mechanism of GA-regulating tuber growth in yam is unknown.
The biosynthesis and signal transduction pathways of GA have been determined in arabidopsis and rice (Murase et al. 2008; Ueguchi-Tanaka et al. 2007; Sun 2011; Hedden and Phillips 2000). The biosynthesis of bioactive GAs has involved the action of two enzymes: GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox). The final level of bioactive GAs is also regulated by inactivation, which is catalyzed by the GA 2-oxidase (GA2ox) enzyme (Salazar-Cerezo et al. 2018; Hedden and Sponsel 2015; Hedden and Thomas 2012). It showed that these genes took part in the control of tuber growth by affecting endogenous GA levels. The overexpression in potato showed that StGA20ox1 participated in the biosynthesis of bioactive GAs, resulting in the high growth plants, which required a short photoperiod for tuber growth (Carrera et al. 2000). StGA3ox2 delayed tuber induction and decreased average tuber weight (Bou-Torrent et al. 2011; Roumeliotis et al. 2013), while StGA2ox1 degraded GA and promoted tuber tuberization (Kloosterman et al. 2007). The GIBBERELLIN-INSENSITIVE DWARF1 (GID1) and DELLA protein are the key components of the signal transduction pathways of GA (Murase et al. 2008; Ueguchi-Tanaka et al. 2007). DELLA protein is the key inhibitor of GA signaling pathway (Fleet and Sun 2005). GA signaling pathway is mainly realized by removing the inhibition of DELLA protein. After binding with active GA and sensing gibberellin signal, GA receptor GID1 transmits the signal to DELLA protein and induces a series of downstream reactions (Sun 2011; Li et al. 2018b; Van de Velde et al. 2017). It is necessary to study the effects of bioactive GA synthesis and signaling-related genes on tuberization in yam because of the differences of GA on tuber growth in potato and yam. Gibberellin 2-β-dioxygenase gene (Gibberellin regulatory protein gene) regulated GA3 production in yam, and in turn, affected tuber growth (Ao, 2020). GA20ox2, GID1C2, and DELLA2 were responsive to GA3 in yam tuber growth (Xing et al. 2020), and the expression of DoGID1A increased during yam bulbil sprouting (Long et al. 2019). This implies that GA metabolism and signaling components genes may regulate yam tuber growth. Until now, the mechanism of GA3 regulation of these genes expression in yam is unknown.
Expansins (EXP) and xyloglucan endotransglycosylase/hydrolase (XTH) play important roles in crop growth and development requiring cell wall extension, modification, and cell enlargement (Xu et al. 2014; Yang et al. 2020; Kushwah et al. 2020; Ratke et al. 2018). Several reports have shown that EXP and XTH genes participated in tuber growth. Overexpression of IbEXP1 suppressed the proliferation of cambium and metaxylem in the formation of sweetpotato storage roots (Noh et al. 2013). StEXP was expressed highly in potato tuber (Li et al. 2017). Furthermore, EXP and XTH genes acted as positive regulators in Chinese yam tuber growth (Zhou et al. 2020).
In our previous study, endogenous GA3 played important roles in the early expansion stage (Gong et al. 2016), and exogenous GA3 increased tuber weight and yield in Dioscorea opposita var. Guihuai 16 (Gong et al. 2015). Despite GA3 may be closely related to yam tuber growth, little is known about GA metabolism and signaling components genes that mediated the regulation of tuber growth. In this study, we analyzed all potential GA-related, EXP, and XTH genes in the GH16 tuber transcriptomics (The data in the NCBI SRA database: PRJNA533985) and found that DoGA20ox1, DoGA3ox1, DoGA2ox1, DoGA2ox2, DoGA2ox3, DoGA2ox4, DoGID1a, DoGID1b, DoGID1c, DoDELLA1, DoDELLA2, DoDELLA3, DoEXP1, DoEXP2, DoEXP3, DoEXP4, DoEXP5, DoXTH1, DoXTH2, and DoXTH3 had the complete open reading frame (ORF), and shown significant differential expression during tuber expansion stage. Our work aimed to investigate the GA3 level, potential GA-related genes, and EXP and XTH genes expression of tuber growth in response to GA3 and PP333 treatments. The results will provide useful information for the mechanism of GA-regulating tuber growth.
Materials and methods
Field experiment and sampling
GH16 (Guihuai 16) was planted at the Farm of Guangxi University in 2018–2019. Its healthy tubers' germination and planting patterns were consistent as the previous description (Gong et al. 2016). According to the life cycle of GH16 tuber, GA3 and PP333 were treated as previously describing, 200 mg/L exogenous GA3 and PP333 was sprayed on the leaves of GH16 on June 20, which was the beginning of tuber expansion of GH16 (Gong et al. 2016), and water was used as the control (CK). Each experiment consisted of three plots: each plot included 60 tuber plants and arranged in a randomized complete block design. Tubers were collected at early expansion stage (0, 1, 3, 5, 7, 15, 30 days), middle expansion stage (60 days), late expansion stage (90 days), and maturation stage (120 days) after spraying. For each experiment, each plot represented a biological replicate. Five plants were selected randomly from every repetition each time. The distal ends (5 mm long) of five fresh tubers from a plot were washed with distilled water, cut down into pieces, and mixed as a biological repetition. All samples were immediately frozen in liquid nitrogen and stored at − 80 °C. The experiments were conducted from 2018 to 2019.
Quantitation of endogenous GA3
Extraction and quantitation of endogenous hormones were carried as previously describing (Gong et al. 2016). The extracted and purified samples were subjected to UHPLC–QqQ-MS/MS before it was separated on the Agilent C18 column. The mobile phase A, consisting of 0.1% formic acid in the water, the mobile phase B, consisting of 100% methanol, were used for chromatographic separation, and the gradient changes were 65% A and 35% B for initial conditions which were maintained for 2 min, changing linearly to 0% A, 100% B for 2 min, and finally maintained at 65% A, 35% B for 3 min. The conditions of mass spectrometry were used in ESI mode.
Gene cloning and bioinformatics analysis
Genes of potential GA related, EXP, and XTH in GH16 tuber were searched from differentially expression by DEGseq and were functionally characterized by NR database in previous transcriptome database (The data in the NCBI SRA database: PRJNA533985) (Zhou et al. 2020). All gene sequences were identified by BLASTx and predicted complete ORF were submitted to NCBI (Table S1). The sequences of the primers are shown in Table S2. Total RNA was extracted from different tuber growth stages, using the MiniBEST Plant RNA Extraction Kit (TaKaRa), and cDNA was synthesized using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa). Multiple alignments of the deduced amino acid sequence were performed with the sequences from different species using Clustal W and GeneDoc software, and a phylogenetic tree was constructed using the neighbor-joining method and 1000 bootstraps in MEGA 6.0, accession numbers of protein for organisms are shown in Table S3. The basic physical, chemical properties, and conserved domains of the proteins were predicted by the Expert Protein Analysis System(http://expasy.org/proteomics) and SMART software(http://smart.embl-heidelberg).
Quantitative real-time PCR analyses
Total RNA was reverse transcribed into cDNA using the PrimeScript RT reagent Kit (TaKaRa), and specific primers were designed to generate a 100–200 bp PCR product by Primer Premier 5.0 (Table S4). Real-time RT-qPCR was performed on a CFX96 Real-Time PCR Detection System (BIO-RAD), using iTaq™ universal SYBR® Green Supermix (BIO-RAD), according to the manufacturer’s protocol. All reactions were performed in triplicate. The 2−ΔΔCt method was used to estimate relative expression level. DoActin was used as internal controls which were designed to generate a 150 bp PCR product from corresponding cDNAs by Primer Premier 5.0. All gene expressions had three biological replicates. Heat map was constructed using HemI program.
Accession numbers
Sequence data from this article can be found in the NCBI databases using following accession numbers: DoGA20ox1(MW377784), DoGA3ox1(MW377785), DoGA2ox1(MW377786), DoGA2ox2(MW377787), DoGA2ox3(MW377788), DoGA2ox4(MW377789), DoGID1a(MW377790), DoGID1b(MW377791), DoGID1c(MW377792), DoDELLA1(MW377793), DoDELLA2(MW377794), DoDELLA3(MW377795), DoEXP1(MW377796), DoEXP2(MW377797), DoEXP3(MW377798), DoEXP4(MW377799), DoEXP5(MW377800), DoXTH1(MW377801), DoXTH2(MW377802), DoXTH3(MW377803).
Yeast two-hybrid assays
Y2H assays were performed with the Matchmaker Gold Yeast two-hybrid system (Clontech). DoGID1a, DoGID1b, and DoGID1c full-length ORFs were inserted into the NdeI-EcoRI site of the pGBKT7 bait vector (GAL4 binding-domain), respectively. DoDELLA1, DoDELLA2, and DoDELLA3 were fused into the NdeI-XhoI site of the pGADT7 prey vector (GAL4 activation-domain), respectively. The sequences of the primers are shown in Table S5. Bait and prey vectors (200 ng) were transformed into Y2H Gold yeast strains, using Yeastmaker yeast transformation system 2. A positive colony was picked from QDO/X/A. One microliter of the positive yeast was spread in a 96-well plate containing medium with or without 100 μMGA3. The growth of yeast colonies was observed 3 days after incubation at 30 °C. All assays were repeated three times.
Statistical analyses
The data based on three independent biological replicates were analyzed using SPSS19.0 software. Data were presented in tables and bar graphs as the mean ± standard error (SE). Statistical significance for gene expression among treatments was determined by Student’s t test, one-way Analysis of Variance (ANOVA), and significant value set at α = 0.05.
Results
Endogenous GA3 quantities response to GA3 and PP333 application
In this study, whether gibberellin is involved in tuber growth, GA3 levels were detected in the early expansion stage (0, 1, 3, 5, 7, 15, and 30 days), middle expansion stage (60 days), late expansion stage (90 days), and maturation stage (120 days) after spraying with GA3 or PP333.
In the control, the content of endogenous GA3 in tuber increased first, and then decreased, it had the highest values on 3 days and was higher in the early expansion stage than in other stages. There were less changes among middle, late, and mature stages (Fig. 1). Compared to the control, exogenous GA3 could significantly increase the contents of endogenous GA3 from 1 to 30 days and showed one obvious peak level of 224.08 ng/g on 3 days, then it decreased significantly from 60 to 120 days. There was an opposite trend after exogenous PP333 application, it remarkably decreased the content of endogenous GA3 from 1 to 15 days, then had no change on other days. The endogenous GA3 levels were higher in tuber expansion stage, especially the early expansion stage, suggesting that the GA3 application had a significantly positive effect on increasing endogenous GA3 content.
Comparison of endogenous GA3 contents after GA3 and PP333 treatment. The data represent the mean ± standard error (SE) of three biological with three replicates each. Letters indicate significant at 5% levels, among different treatment. The analyzed tuber stage: early expansion stage (0–30 days), middle expansion stage (60 days), late expansion stage (90 days), and maturation stage (120 days). The same as below
Isolation and characterization of GA3 metabolism and signal component genes
To explain the accumulation profile of GA3 during tuber growth, one GA20ox gene (DoGA20ox1), one GA3ox gene (DoGA3ox1), four GA2ox genes (DoGA2ox1, DoGA2ox2, DoGA2ox3, DoGA2ox4), three GA receptor genes (DoGID1a, DoGID1b, and DoGID1c), and three DELLA genes (DoDELLA1, DoDELLA2, DoDELLA3) were obtained with complete open reading frame (ORF), which found in the GH16 tuber transcriptomics, and these genes have shown significant differential expression during tuber expansion stage (Table S1).
DoGA20ox1 contains an ORF of 1167 bp encoding a protein of 388 amino acids. Conserved domains analysis indicated that DoGA20ox1 belonged to 2-oxoglutarate-dependent deoxygenates family, including DIOX-N domain and 2OG-FeII-Oxy domain (Fig. S1A). Phylogenetic tree analysis indicated that DoGA20ox1 clustered into monocot group, and showed a close relationship with Oryza sativa, Zea mays, and Triticum aestivum (Fig. 2A).
Phylogenetic trees of the amino acid sequence alignment of GA metabolism and signal components genes of yam tuber. A DoGA20ox1. B DoGA3ox1. C DoGA2ox genes (DoGA2ox1, DoGA2ox2, DoGA2ox3, and DoGA2ox4). D GA receptor genes (DoGID1a, DoGID1b, and DoGID1c). E DoDELLA genes (DoDELLA1, DoDELLA2, DoDELLA3). F DoEXP genes (DoEXP1, DoEXP2, DoEXP3, DoEXP4, and DoEXP5). G DoXTH genes (DoXTH1, DoXTH2, and DoXTH3). Ortholog species information showed in Table S3
DoGA3ox1 contains an ORF of 1053 bp encoding a protein of 350 amino acids. Conserved domains analysis indicated that DoGA3ox1 belonged to 2-oxoglutarate-dependent deoxygenates family, including DIOX-N domain and 2OG-FeII-Oxy domain (Fig. S1B). Phylogenetic tree analysis indicated that DoGA3ox1 clustered into the dicotyledon group and showed a close relationship with Arabidopsis and Solanum tuberosum (Fig. 2B).
DoGA2ox1, DoGA2ox2, DoGA2ox3, and DoGA2ox4 were 525, 756, 852, and 1050 bp in ORF-encoding proteins of 174, 251, 283, and 349 amino acids, respectively. Conserved domains analysis indicated that DoGA2oxs belonged to 2-oxoglutarate-dependent dioxygenases family, including 2OG-FeII-Oxy domain (Fig. S1C). Phylogenetic tree analysis indicated that DoGA2ox1 and DoGA2ox2 clustered into same group, and DoGA2ox4 had highly homologous to the GA2ox in Zea mays, Triticum aestivum (Fig. 2C).
DoGID1a, DoGID1b, and DoGID1c were 1149, 381, and 1065 bp in ORF-encoding proteins of 382, 126, and 354 amino acids, respectively. Conserved domains analysis indicated that DoGID1s had the conserved HSL motifs HGG and GXSXG, with the amino acids related to HSL activity are S, D, and V (Fig. S1D). Phylogenetic tree analysis indicated that DoGID1a had highly homologous to the GID1 in Zea mays, Triticum aestivum, and Oryza sativa (Fig. 2D). Interestingly, DoGID1b and DoGID1c clustered into dicotyledonous group.
DoDELLA1, DoDELLA2, DoDELLA3 were 1908, 1749, and 1407 bp in ORF-encoding proteins of 635, 582, and 468 amino acids, respectively. DoDELLAs contain highly conserved domains including DELLA domain and GRAS domain (Fig. S1E). Phylogenetic tree analysis indicated that the DoDELLA1 proteins had close relatives with Zea mays, Oryza sativa, and Triticum aestivum (Fig. 2E). Interestingly, DoDELLA2 and DoDELLA3 protein clustered into same group.
Isolation and characterization of cell wall genes
DoEXP1, DoEXP2, DoEXP3, DoEXP4, and DoEXP5 were 770, 810, 555, 762, and 777 bp in ORF-encoding proteins of 256, 269, 184, 253, and 259 amino acid, respectively. Conserved domains analysis indicated that DoEXPs had the cellulose-binding-like domain (CBD), GGACG motif, and FRRV motif (Fig. S1F). Phylogenetic tree analysis indicated that DoEXPs clustered into the three groups. DoEXP1, DoEXP4, and DoEXP5 clustered into α-expansin protein, DoEXP3 clustered into β-expansin protein, and DoEXP2 clustered into α-expansin-like protein (Fig. 2F).
DoXTH1, DoXTH2, and DoXTH3 were 987, 882, and 879 bp in ORF-encoding proteins of 328, 293, and 292 amino acids, respectively. Conserved domain analysis indicated that DoXTHs had the conserved catalytic domain (DEIDFEFLG) and N-glycosylation site (Fig. S1G). Phylogenetic tree analysis indicated that DoXTH1 and DoXTH3 had highly homologous to the XTH in Arabidopsis thaliana (Fig. 2G). Interestingly, DoXTH2 was clustered into a different group with DoXTH1 and DoXTH3.
Expression profiles of genes related GA biosynthesis and signaling during tuber growth
The expressions of DoGA20ox1, DoGA3ox1, DoGA2ox1, DoGA2ox2, DoGA2ox3, DoGA2ox4, DoGID1a, DoGID1b, DoGID1c, DoDELLA1, DoDELLA2, and DoDELLA3 were assessed during tuber expansion stage. DoGA20ox1 expression levels showed higher transcript from 1 to 15 days, and gradually declined along with tuber growth, reaching low levels on 120 days (Figs. 3A and 8). The expression level of DoGA3ox1 increased first, then decreased with tuber growth, and had the highest levels on 30 days and 60 days (Figs. 3B and 8). All DoGA2ox genes were expressed in tuber growth, showed different accumulation patterns from 1 to 15 days, gradually declined from 30 to 120 days (Figs. 3C, D, E, F, and 8). The expression level of DoGID1a and DoGID1c increased from 1 to 60 days, decreased gradually at late stages of tuber growth (Figs. 4A, C and 8). The expression level of DoGID1b also decreased gradually with tuber growth (Figs. 4B and 8). The expression of DoGID1c was in general lower than that of DoGID1a and DoGID1b in tuber growth. The expression level of DoDELLA1 increased gradually in 3 days and decreased gradually at late stages of tuber growth (Figs. 4E and 8). Conversely, DoDELLA2 and DoDELLA3 steadily increased or decreased along with tuber growth, respectively (Figs. 4E, F and 8).
To assess whether the increased GA3 levels after application GA3 resulted from transcriptional regulation, the expression levels of GA biosynthesis and signaling genes after GA3 or PP333 treatment were measured during tuber growth. It showed that DoGA20ox1, DoGA3ox1, DoGA2ox3, DoGA2ox4, DoGID1a, and DoDELLA1 expression displayed in response to GA3. At the early expansion stage (1, 3, 5, 7, 15, and 30 days), GA3 treatment decreased DoGA20ox1, DoGA3ox1, DoGID1a, and DoDELLA1 expression levels, while PP333 treatment enhanced their expression levels. In the meanwhile, GA3 treatment increased DoGA2ox3 and DoGA2ox4 expression levels, while PP333 treatment decreased their expression levels, compared to the controls (Figs. 3, 4, and 8).
Properties of DoGID1-DoDELLA interaction in Y2H assays
To further characterize the biochemical properties of DoGID1s and DoDELLAs, Y2H assays were performed. Y2H assays showed that DoGID1a, DoGID1b, and DoGID1c interacted with all DoDELLAs, except DoGID1a and DoDELLA3 (Fig. 5). The binding results confirmed the essential GA-induced assembly of stable GA-DoGID1-DoDELLA complex in yeast (Fig. 5). DoDELLA1 and DoDELLA2 were effectively able to interact with DoGID1a, DoGID1b, and DoGID1c in the presence of GA3 (growth in GA3 medium and darker color), except for DoGID1b-DoDELLA3 and DoGID1c-DoDELLA3 (growth in GA3 medium). Conversely, DoGID1a did not bind to DoDELLA3, even in the presence of GA3. Nevertheless, this may be different in the case of DoGID1s and DoDELLAs, where sequence and conformational differences may confer some levels of specificity in DoGID1s and DoDELLAs. These data showed DoGID1s interacted with DoDELLA1 and DoDELLA2 in a GA-mediated manner in Y2H assay.
Expression profiles of cell wall genes during tuber growth
To understand the EXP and XTH genes response to tuber growth, the expression levels of five DoEXP and three DoXTH genes of the cell wall were detected in tuber growth. The expression levels of DoEXP1, DoEXP2, and DoEXP5 genes were higher in the early expansion stage than in other stages except for DoEXP3 and decreased gradually in late stages. DoEXP4 was increased in the tuber growth stage (Figs. 6 and 8). The expression level of DoXTH1, DoXTH2, and DoXTH3 increased on 30 days and decreased gradually in late stages (Figs. 7 and 8). Compared to the control, GA3 treatment increased significantly the expression levels of five DoEXP and three DoXTH genes from 1 to 30 days, while that of were reduced by PP333 treatment from 1 to 30 days (Figs. 6, 7 and 8). Interestingly, GA3 treatment increased significantly increased DoEXP4 and DoEXP5 expression during tuber growth. According to the expression level and pattern, DoEXP4 and DoEXP5 have more roles in tuber cell expansion than other genes.
Discussion
Yam is one of the most commercial tuber crops. Yam tuber growth and development are an attractive theoretical model for studying the development of underground organs. In recent years, GA has been used to explore the physiological factors affecting the growth of yam tubers. While endogenous GA has been detected in yam, GA3 was higher in early expansion stage than other stages (Gong et al. 2016), and the application of GA3 to yam produced new tubers and promoted tuber expansion with an increase in tuber weight and yield (Yoshida et al. 2008; Gong et al. 2015). The concentration of bioactive GAs is determined by the balance between gene expression of biosynthesis and deactivation, in which GA20ox, GA3ox, and GA2ox genes encode key enzymes of biosynthesis and inactivation of GA, and GA binds its receptor, GID1, to form a complex pathway that mediates the degradation of DELLA proteins to regulate plant growth (Hedden and Sponsel 2015; Middleton et al. 2012; Hedden and Thomas 2012). However, the regulating mechanism of GA3 on yam tuber growth by bioactive GAs remains elusive.
Endogenous GA3 level in tuber growth is controlled by GA biosynthetic genes
GA3 and GA4 were detected in yam tuber (Gong et al. 2016). High endogenous GA3 is responsible for the beginning of tuber dormancy and growth (Ao et al. 2020; Zhu and Hou 2011). Furthermore, GA20ox regulated GA3 production in yam tuber (Ao et al. 2020). GA20ox2, GID1C2, and DELLA2 were responsive to GA3 in yam tuber growth (Xing et al. 2020). It is suggested that GA3 may be closely related to tuber growth, and GA-related genes are responses to GA3 regulation in yam. In rice, GA4 could be rapidly inactivated and degraded in GA3-treated cells by GA-inactivating enzymes; however, GA3 was not easily inactivated by inactivating enzymes and remained active after GA3 treatment, which promoted GID1–SLR interaction (Ueguchi-Tanaka et al. 2007). Hence, it is necessary to investigate the GA3 level and clarify the molecular mechanism in tuber growth. In this study, GA3 had the highest level from 1 to 30 days in the early expansion stage, and then decreased in middle, late, and mature stages, which shows that the increment of GA3 may induce cell expansion, and improve tuber growth. The result is similar to previous reports in yam (Gong et al. 2016) and other species. During carrot root growth, the highest GA levels were observed early enlarge stage (42 days), this level subsequently decreased (Wang et al. 2015). GA levels are critical for early tissue or organ development which is consistent with the role of GA during early plum fruit formation (El-Sharkawy et al. 2014).
However, little is known the correlation between GA levels and the gene expressions of GA metabolism components in yam tuber. In this study, the accumulation of GA3 was consistent with the expression of DoGA20ox1 during the early expansion stage of yam tuber, contrary to the expression of DoGA2ox3 and DoGA2ox4. With the tuber growth, the content of GA3 gradually decreased, and the expression of these genes also gradually decreased. Taken together, the decrease in endogenous GA3 in yam tuber growth is explained by GA biosynthetic gene expressions. The down-regulation of DoGA20ox1 transcript and the apparent accumulation of DoGA2ox3 and DoGA2ox4 reflect an important role for the genes related to GA3 synthesis during tuber growth. This scenario is similar to the results observed in other species during plant development. GA accumulation in anthesis coincided with transient up-regulation of VvGA20ox1, VvGA20ox3, and VvGA3ox2 in grapevine (Giacomelli et al. 2013). Hence, endogenous GA3 accumulates in the early expansion stage by DoGA20ox1-active synthesis and DoGA2ox3- and DoGA2ox4-active inactivation. However, the DoGA3ox1, DoGA2ox1, and DoGA2ox2 expression patterns were incompletely consistent with the GA3 levels possibly because of the feedback mechanism, or other hormones may play vital roles in tuber growth (Gong et al. 2016), which indicating a complex mechanism in endogenous GA3 accumulation.
DELLA participate in yam tuber growth
It is known that GID1 acts as GA receptors, whereas DELLA protein is a negative regulator of GA signaling (Murase et al. 2008; Ueguchi-Tanaka et al. 2007). Hence, the expression levels of DoGID1 and DoDELLA genes were assessed during various tuber developmental stages. DoGID1a and DoDELLA1 genes were abundantly expressed in the early expansion stage. GA3 was involved in tuber growth, particularly the early expansion stage. Similarly, GA is needed to organize the abundant cell expansion, division during tissue development. The abundance of the DoGID1a and DoDELLA1 genes during early tuber expansion stage suggests a dominant task of DoGID1a and DoDELLA1 genes in regulating GA response during this stage. Previous reports have indicated that GID1 and DELLA proteins play a key role in hypocotyl growth, stem and root elongation, bud dormancy, plant height, and fruit development in GA response (Griffiths et al. 2006; Lv et al. 2018; Li et al. 2018a). These results suggested that DoGID1a and DoDELLA1 genes are active components of the GA signal network that regulate tuber growth.
Effect of exogenous GA3 treatment on gene expression
In this study, GA3 treatment increased endogenous GA3 levels and PP333 treatment decreased endogenous GA3 levels in tuber growth, which is consistent with the previous results. In pear, GA4+7 increased unpollinated ovaries GA4+7 levels to induce parthenocarpy (Liu et al. 2018). GA3 application resulted in a significant increase in GA3 levels along with a slight acceleration in fruit size and weight in plum fruit development (El-Sharkawy et al. 2014). It can be assumed that exogenous GA3 applications increase relatively higher endogenous GA3 levels in the tuber and improve yam tuber growth.
GA homeostasis in plants is maintained by feedback regulation. When GA levels were too high in plants, GA20ox and GA3ox were subject to negative feedback by decreasing their expression, GA2ox increased expression by positive feed-forward regulation (Fukazawa et al. 2017). In Cucumber, CsGA20ox1, CsGA20ox2, and CsGA3ox1 were strongly repressed by GA3 treatment, when CsGA2ox1, CsGA2ox4, and CsGA2ox6 were simultaneously induced by GA3 treatment (Sun et al. 2018). Exogenous GA application in tomato fruit led to down-regulation of SIGA20ox, SIGA3ox and up-regulation of SIGA2ox (Chen et al. 2016).
Taken together, we propose that the expression profiles of DoGA20ox1, DoGA3ox1, and DoGA2ox were regulated by GA availability in yam tuber, especially in the early expansion stage. The down-regulation of DoGA20ox1 and DoGA3ox1, and up-regulation of DoGA2ox3 and DoGA2ox4 following by GA3 application show a feedback mechanism in response to endogenous GA3. On the other hand, PP333 application exhibited the adverse consequences. It suggests the existence of a negative feedback mechanism to regulate the expression of DoGA20ox1 and DoGA3ox1, and positive feedback regulation of DoGA2ox3 and DoGA2ox4 in tuber growth. The feedback regulation may keep suitable concentration of active GA3 after exogenous GA3 application, to improve growth and prevent overgrowth simultaneously in tuber. In addition, no significant changes were observed for the expression of DoGA2ox1 and DoGA2ox2 in tuber growth of GA3 and PP333 treatment, suggesting that individual members of the DoGA2ox gene family may play different physiological roles to response GA3 treatment, or depend on tissue types and organs.
Also, we found the expression pattern of DoGID1a and DoDELLA1 in response to GA3 or PP333 treatment in the early expansion stage. GA3 application down-regulated DoGID1a and DoDELLA1 expressions, while PP333 treatment up-regulated expressions in the early expansion stage. It is consistent with the previous results in grape and pear. The application of GA3 increased endogenous GA3 level, down-regulated VvGID1 transcripts, and resulted in the degradation of VvDELLA protein in grape and pear (Acheampong et al. 2017, 2015; Liu et al. 2018). In addition, DoGID1s interacted with DoDELLA1 and DoDELLA2 in a GA-mediated manner in Y2H assay. The potential explanation for DoGID1a and DoDELLA1 in response to GA3 is that exogenous GA3 application increase endogenous active GA3 concentration, and high-level GA3 rapidly induced DoDELLA1 degradation, resulted in the DoGID1a expression decrease.
However, the expression patterns of DoGA2ox1, DoGA2ox2 and DoGID1b, DoGID1c, DoDELLA2, and DoDELLA3 were incompletely consistent with the GA3 levels by GA3 and PP333 application, and it possibly results from the feedback mechanism of GA3 (Fukazawa et al. 2017) or their different roles in the control of active GA homeostasis. In the meanwhile, other hormones may also play vital roles in active GA biosynthesis and metabolism, indicating a complex mechanism of GA biosynthesis and catabolism (Okabe et al. 2019; Cong et al. 2019).
Previous studies in pear had demonstrated that exogenous GA4+7 barely changed the expression levels of encoded biosynthetic GA genes, since GA content was already high to support pear fruit development, while mainly influenced GA response genes, such as cell cycle and cell expansion genes (Liu et al. 2018). Some EXP and XTH genes associated with cell walls were found in the yam tuber expansion stage by transcript profiling of Guihuai 16 (Zhou et al. 2020). Plant organ growth depends on cell differentiation and enlargement through GA regulation. Overall, all DoEXP and DoXTH genes expression levels were high in the early expansion stage, except DoEXP4 was high in the late expansion stage. In the meanwhile, GA3 had a higher level in the early expansion stage, which showed that the increment of GA3 may regulate all DoEXP and DoXTH genes expression, induce cell expansion, and improve tuber growth. Exogenous GA3 treatment in maize leaf could affect cell expansion and gives rise to a dramatic change in direction for cell expansion in growing cells (Nelissen et al. 2012). Cellular elongation required cell wall remodeling enzymes, including EXPs and XTHs, to rearrange cell wall matrix polymers for cell wall loosening (Hervieux et al. 2016; Cosgrove 2000). GA3 could up-regulate EXPs and XTHs transcripts to promote cell elongation or cell wall modification in Arabidopsis, Cucumber, and Persimmon (Sanchez-Montesino et al. 2019; Sun et al. 2017; Han et al. 2016). GA3 application up-regulated all DoEXP and DoXTH genes in the expansion stage of the tuber, while the PP333 treatment showed the opposite pattern. DoEXP4, DoEXP5, and DoXTH1 genes performed higher responses to GA3 in tuber growth. DELLA protein could regulate cell wall properties by repressing the EXP8, EXP10, and XTH28 expression either directly or indirectly in Arabidopsis (Oh et al. 2009; Dello Ioio et al. 2008). These results suggest that the accumulation of GA3 by the activities DoGA20ox1, DoGA3ox1, DoGA2ox3, DoGA2ox4, and the degradation of DoDELLA1 proteins due to increase the formation of GA-DoGID1a-DoDELLA1, increase GA response genes expression, such as DoEXP1, DoEXP2, DoEXP3, DoEXP4, DoEXP5, DoXTH1, DoXTH2, and DoXTH3, and then enhance tuber growth by inducing cell expansion.
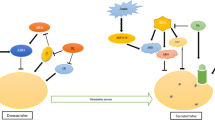
In brief, endogenous GA3 biosynthesis is catalyzed by DoGA20ox1 and DoGA3ox1, and its deactivation is catalyzed by DoGA2ox3 and DoGA2ox4 in yam tuber growth. High GA3 promotes tuber growth by stimulating the degradation of the growth repressing DoDELLA1 proteins. In GA3 treatment, the down-regulation of DoGA20ox1 and DoGA3ox1, and up-regulation of DoGA2ox3 and DoGA2ox4 transcript accumulation are in response to a high endogenous GA3 level as a feedback mechanism. A model is presented in Fig. 9. Exogenous GA3 results in a significant increase in endogenous GA3 levels along with a feedback mechanism by downregulating DoGA20ox1 and DoGA3ox1 expression levels and upregulating the expression of DoGA2ox3 and DoGA2ox4 to maintain GA homeostasis. A high GA3 level can rapidly bind DoGID1a and allows the DoDELLA1 to be targeted for degradation, leading to increase DoEXP1, DoEXP2, DoEXP3, DoEXP4, DoEXP5, DoXTH1, DoXTH2, and DoXTH3 expression, cell division and cell expansion, and improve tuber growth. This work helps increase our understanding of the cross-talk between endogenous and exogenous GA3 in yam tuber.
Probable mechanism of endogenous GA3 regulates tuber growth in yam. The accumulation of active GA3 is controlled by the activities of DoGA20ox1, DoGA3ox1, DoGA2ox3, and DoGA2ox4, and the degradation of DoDELLA1 proteins due to the formation of GA-DoGID1-DoDELLA. Active GA3 improves GA response genes expression, such as DoEXP1, DoEXP2, DoEXP3, DoEXP4, DoEXP5, DoXTH1, DoXTH2, and DoXTH3, and enhance tuber growth
Conclusion
The profiles of GA3 accumulation with tuber growth may be the result of the combined action of DoGA20ox1, DoGA3ox1, DoGA2ox3, and DoGA2ox4 expression and DoDELLA1 protein degradation, and the formation of GA-DoDID1a-DoDELLA1, while promoting the expression of GA response genes DoEXP1, DoEXP2, DoEXP3, DoEXP4, DoEXP5, DoXTH1, DoXTH2, and DoXTH3.
References
Acheampong AK, Hu JH, Rotman A, Zheng CL, Halaly T, Takebayashi Y, Jikumaru Y, Kamiya Y, Lichter A, Sun TP, Or E (2015) Functional characterization and developmental expression profiling of gibberellin signalling components in Vitis vinifera. J Exp Bot 66(5):1463–1476. https://doi.org/10.1093/jxb/eru504
Acheampong AK, Zheng C, Halaly T, Giacomelli L, Takebayashi Y, Jikumaru Y, Kamiya Y, Lichter A, Or E (2017) Abnormal endogenous repression of GA signaling in a seedless table grape cultivar with high berry growth response to GA application. Front Plant Sci 8:850. https://doi.org/10.3389/fpls.2017.00850
Agele SO, Ayankanmi TG, Kikuno H (2010) Effects of synthetic hormone substitutes and genotypes on rooting and mini tuber production of vines cuttings obtained from white yam (Dioscorea rotundata, Poir). Afr J Biotechnol 9(30):4714–4724
Aksenova NP, Konstantinova TN, Golyanovskaya SA, Sergeeva LI, Romanov GA (2012) Hormonal regulation of tuber formation in potato plants. Russ J Plant Physiol 59(4):451–466. https://doi.org/10.1134/S1021443712040024
Ao LJY, Ji X, Shao Y, (2020) Effect of endogenous hormones and expressions of relevant genes on tuber growth of Bikeqi Yam. Fujian J Agric Sci 35(09):964–973
Bou-Torrent J, Martinez-Garcia JF, Garcia-Martinez JL, Prat S (2011) Gibberellin A1 metabolism contributes to the control of photoperiod-mediated tuberization in potato. PLoS ONE 6(9):e24458. https://doi.org/10.1371/journal.pone.0024458
Carrera E, Bou J, Garcia-Martinez JL, Prat S (2000) Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J 22(3):247–256. https://doi.org/10.1046/j.1365-313x.2000.00736.x
Chen FQ, Fu Y, Wang DL, Gao X, Wang L (2007) The effect of plant growth regulators and sucrose on the micropropagation and microtuberization of Dioscorea nipponica Makino. J Plant Growth Regul 26(1):38–45. https://doi.org/10.1007/s00344-005-0147-2
Chen SW, Shiwachi H, Sanada A, Toyohara H (2010) Theobroxide and day-length effects on the growth of yam (Dioscorea spp.). JISSAAS 16(1):22–30
Chen S, Wang X, Zhang L, Lin S, Liu D, Wang Q, Cai S, El-Tanbouly R, Gan L, Wu H, Li Y (2016) Identification and characterization of tomato gibberellin 2-oxidases (GA2oxs) and effects of fruit-specific SlGA2ox1 overexpression on fruit and seed growth and development. Hortic Res 3:16059. https://doi.org/10.1038/hortres.2016.59
Cheng L, Wang Y, Liu Y, Zhang Q, Gao H, Zhang F (2018) Comparative proteomics illustrates the molecular mechanism of potato (Solanum tuberosum L.) tuberization inhibited by exogenous gibberellins in vitro. Physiol Plant 163(1):103–123. https://doi.org/10.1111/ppl.12670
Cong L, Yue R, Wang H, Liu J, Zhai R, Yang J, Wu M, Si M, Zhang H, Yang C, Xu L, Wang Z (2019) 2,4-D-induced parthenocarpy in pear is mediated by enhancement of GA4 biosynthesis. Physiol Plant 166(3):812–820. https://doi.org/10.1111/ppl.12835
Cosgrove DJ (2000) Expansive growth of plant cell wall. Plant Physiol Biochem 38(1/2):109–124
Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322(5906):1380–1384. https://doi.org/10.1126/science.1164147
El-Sharkawy I, Sherif S, El Kayal W, Mahboob A, Abubaker K, Ravindran P, Jyothi-Prakash PA, Kumar PP, Jayasankar S (2014) Characterization of gibberellin-signalling elements during plum fruit ontogeny defines the essentiality of gibberellin in fruit development. Plant Mol Biol 84(4–5):399–413. https://doi.org/10.1007/s11103-013-0139-8
Feng F, ChunHai YE, YingZhi LI, WeiFei XU (2007) Effects of growth regulators, carbon sources and photoperiod on in vitro formation and growth and development of microtubers of Dioscoreafordii Prain et Burk. Plant Physiol Commun 43(6):1045–1049
Fleet CM, Sun TP (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8(1):77–85. https://doi.org/10.1016/j.pbi.2004.11.015
Fukazawa J, Mori M, Watanabe S, Miyamoto C, Ito T, Takahashi Y (2017) DELLA-GAF1 complex Is a main component in gibberellin feedback regulation of GA20 oxidase 2. Plant Physiol 175(3):1395–1406. https://doi.org/10.1104/pp.17.00282
Giacomelli L, Rota-Stabelli O, Masuero D, Acheampong AK, Moretto M, Caputi L, Vrhovsek U, Moser C (2013) Gibberellin metabolism in Vitis vinifera L. during bloom and fruit-set: functional characterization and evolution of grapevine gibberellin oxidases. J Exp Bot 64(14):4403–4419. https://doi.org/10.1093/jxb/ert251
Gong M, Luo H, Yuan H, Wei S, Yang X, He L (2015) Effects of exogenous gibberellin and paclobutrazol on tuber expansion and bulbil formation of Chinese yam. Acta Hortic Sin 42(6):1175–1184
Gong M, Luo H, Wang A, Zhou Y, Huang W, Zhu P, He L (2016) Phytohormone profiling during tuber development of Chinese yam by ultra-high performance liquid chromatography–triple quadrupole tandem mass spectrometry. J Plant Growth Regul 36(2):362–373. https://doi.org/10.1007/s00344-016-9644-8
Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, Thomas SG (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 (12):3399–3414. https://doi.org/10.1105/tpc.106.047415
Han Y, Ban Q, Hou Y, Meng K, Suo J, Rao J (2016) Isolation and characterization of two persimmon xyloglucan endotransglycosylase/hydrolase (XTH) genes that have divergent functions in cell wall modification and fruit postharvest softening. Front Plant Sci 7:624. https://doi.org/10.3389/fpls.2016.00624
Hartmann A, Senning M, Hedden P, Sonnewald U, Sonnewald S (2011) Reactivation of meristem activity and sprout growth in potato tubers require both cytokinin and gibberellin. Plant Physiol 155(2):776–796. https://doi.org/10.1104/pp.110.168252
Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5(12):523–530
Hedden P, Sponsel V (2015) A century of gibberellin research. J Plant Growth Regul 34(4):740–760. https://doi.org/10.1007/s00344-015-9546-1
Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444(1):11–25. https://doi.org/10.1042/BJ20120245
Hervieux N, Dumond M, Sapala A, Routier-Kierzkowska AL, Kierzkowski D, Roeder AH, Smith RS, Boudaoud A, Hamant O (2016) A mechanical feedback restricts sepal growth and shape in Arabidopsis. Curr Biol 26(8):1019–1028. https://doi.org/10.1016/j.cub.2016.03.004
Kloosterman B, Navarro C, Bijsterbosch G, Lange T, Prat S, Visser RGF, Bachem CWB (2007) StGA2ox1 is induced prior to stolon swelling and controls GA levels during potato tuber development. Plant J 52(2):362–373. https://doi.org/10.1111/j.1365-313X.2007.03245.x
Kushwah S, Banasiak A, Nishikubo N, Derba-Maceluch M, Majda M, Endo S, Kumar V, Gomez L, Gorzsas A, McQueen-Mason S, Braam J, Sundberg B, Mellerowicz EJ (2020) Arabidopsis XTH4 and XTH9 contribute to wood cell expansion and secondary wall formation. Plant Physiol 182(4):1946–1965. https://doi.org/10.1104/pp.19.01529
Li X, Dai D, Yang T, Hao X (2017) Genome-wide identification and expression analysis of the expansin gene family in potato. Acta Agriculturae Boreali-Sinica 32(5):37–44
Li WJ, Zhang JX, Sun HY, Wang SM, Chen KQ, Liu YX, Li H, Ma Y, Zhang ZH (2018a) FveRGA1, encoding a DELLA protein, negatively regulates runner production in Fragaria vesca. Planta 247(4):941–951. https://doi.org/10.1007/s00425-017-2839-9
Li X, Liu W, Li B, Liu G, Wei Y, He C, Shi H (2018b) Identification and functional analysis of cassava DELLA proteins in plant disease resistance against cassava bacterial blight. Plant Physiol Biochem 124:70–76. https://doi.org/10.1016/j.plaphy.2017.12.022
Liu L, Wang Z, Liu J, Liu F, Zhai R, Zhu C, Wang H, Ma F, Xu L (2018) Histological, hormonal and transcriptomic reveal the changes upon gibberellin-induced parthenocarpy in pear fruit. Hortic Res 5:1. https://doi.org/10.1038/s41438-017-0012-z
Long W, Meng J, Xu S, Zhang X, Duan Y, Yang R (2019) Cloning and expression analysis of gibberellin receptor gene DoGID1A in Dioscorea opposita. J Agric Biotechnol 27(11):1933–1941
Lv L, Huo X, Wen L, Gao Z, Khalil-Ur-Rehman M (2018) Isolation and role of PmRGL2 in GA-mediated floral bud dormancy release in Japanese Apricot (Prunus mume Siebold et Zucc.). Front Plant Sci 9:27. https://doi.org/10.3389/fpls.2018.00027
Matsumoto R, Kikuno H, Shiwachi H, Toyohara H, Takebayashi Y, Jikumaru Y, Kamiya Y (2005) Growth of vine cuttings and fluctuations of concentrations of endogenous plant hormones in water yam (Dioscorea alata L.). Trop Agric Dev 57(1):23–30
Middleton AM, Ubeda-Tomas S, Griffiths J, Holman T, Hedden P, Thomas SG, Phillips AL, Holdsworth MJ, Bennett MJ, King JR, Owen MR (2012) Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. Proc Natl Acad Sci USA 109(19):7571–7576. https://doi.org/10.1073/pnas.1113666109
Murase K, Hirano Y, Sun TP, Hakoshima T (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456(7221):459–463. https://doi.org/10.1038/nature07519
Nelissen H, Rymen B, Jikumaru Y, Demuynck K, Van Lijsebettens M, Kamiya Y, Inze D, Beemster GT (2012) A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr Biol 22(13):1183–1187. https://doi.org/10.1016/j.cub.2012.04.065
Noh SA, Lee H-S, Kim Y-S, Paek K-H, Shin JS, Bae JM (2013) Down-regulation of the IbEXP1 gene enhanced storage root development in sweetpotato. J Exp Bot 64(1):129–142. https://doi.org/10.1093/jxb/ers236
Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G (2009) Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21(2):403–419. https://doi.org/10.1105/tpc.108.064691
Okabe Y, Yamaoka T, Ariizumi T, Ushijima K, Kojima M, Takebayashi Y, Sakakibara H, Kusano M, Shinozaki Y, Pulungan SI, Kubo Y, Nakano R, Ezura H (2019) Aberrant stamen development is associated with parthenocarpic fruit set through up-regulation of gibberellin biosynthesis in Tomato. Plant Cell Physiol 60(1):38–51. https://doi.org/10.1093/pcp/pcy184
Ratke C, Terebieniec BK, Winestrand S, Derba-Maceluch M, Grahn T, Schiffthaler B, Ulvcrona T, Ozparpucu M, Ruggeberg M, Lundqvist S-O, Street NR, Jonsson LJ, Mellerowicz EJ (2018) Downregulating aspen xylan biosynthetic GT43 genes in developing wood stimulates growth via reprograming of the transcriptome. New Phytol 219(1):230–245. https://doi.org/10.1111/nph.15160
Roumeliotis E, Kloosterman B, Oortwijn M, Lange T, Visser RGF, Bachem CWB (2013) Down regulation of StGA3ox genes in potato results in altered GA content and affect plant and tuber growth characteristics. J Plant Physiol 170(14):1228–1234. https://doi.org/10.1016/j.jplph.2013.04.003
Salazar-Cerezo S, Martinez-Montiel N, Garcia-Sanchez J, Perez YTR, Martinez-Contreras RD (2018) Gibberellin biosynthesis and metabolism: A convergent route for plants, fungi and bacteria. Microbiol Res 208:85–98. https://doi.org/10.1016/j.micres.2018.01.010
Sanchez-Montesino R, Bouza-Morcillo L, Marquez J, Ghita M, Duran-Nebreda S, Gomez L, Holdsworth MJ, Bassel G, Onate-Sanchez L (2019) A regulatory module controlling GA-mediated endosperm cell expansion Is critical for seed germination in Arabidopsis. Mol Plant 12(1):71–85. https://doi.org/10.1016/j.molp.2018.10.009
Sun T-p (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21(9):R338–R345. https://doi.org/10.1016/j.cub.2011.02.036
Sun Y, Luo W, Li Z, Li X (2017) Endogenous hormones levels and csexpansin 10 gene expression in the fruit set and early development of Cucumber. J Chem Soc Pak 39(1):59–64
Sun H, Pang B, Yan J, Wang T, Wang L, Chen C, Li Q, Ren Z (2018) Comprehensive analysis of cucumber gibberellin oxidase family genes and functional characterization of CsGA20ox1 in root development in Arabidopsis. Int J Mol Sci 19(10):3135. https://doi.org/10.3390/ijms19103135
Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, Matsuoka M (2007) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19(7):2140–2155. https://doi.org/10.1105/tpc.106.043729
Van de Velde K, Chandler PM, Van Der Straeten D, Rohde A (2017) Differential coupling of gibberellin responses by Rht-B1c suppressor alleles and Rht-B1b in wheat highlights a unique role for the DELLA N-terminus in dormancy. J Exp Bot 68(3):443–455. https://doi.org/10.1093/jxb/erw471
Wang GL, Xiong F, Que F, Xu ZS, Wang F, Xiong AS (2015) Morphological characteristics, anatomical structure, and gene expression: novel insights into gibberellin biosynthesis and perception during carrot growth and development. Hortic Res 2:15028. https://doi.org/10.1038/hortres.2015.28
Xing D, Huang Y, Zhao H, Li Y, Liu M, Xia W (2020) Cloning and expression analysis of genes related to gibberellin pathway in Dioscorea alala. Molecular Plant Breeding 18(6):1755–1761
Xu P, Cai X-T, Wang Y, Xing L, Chen Q, Xiang C-B (2014) HDG11 upregulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis. J Exp Bot 65(15):4285–4295. https://doi.org/10.1093/jxb/eru202
Yang J, Zhang G, An J, Li Q, Chen Y, Zhao X, Wu J, Wang Y, Hao Q, Wang W, Wang W (2020) Expansin gene TaEXPA2 positively regulates drought tolerance in transgenic wheat (Triticum aestivum L.). Plant Sci: Int J Exp Plant Biol 298:110596–110596. https://doi.org/10.1016/j.plantsci.2020.110596
Yoshida Y, Takahashi H, Kanda H, Kanahamaz K (2007) Effect of seed tuber weights on the development of tubers and flowering spikes in Japanese yams (Dioscorea japonica) grown under different photoperiods and with plant growth regulators. J Jpn Soc Hortic Sci 76(3):230–236. https://doi.org/10.2503/jjshs.76.230
Yoshida Y, Ichikawa A, Takahashi M, Takahashi H, Kanda H, Kanahama K (2008) The effects of gibberellin concentration, treatment interval and commencement timing on the development of tubers and inflorescences in Chinese yam (Dioscorea oppositifolia L.) 'Ichoimo' plant. In: Proceedings of the international symposium on endogenous and exogenous plant bioregulators (774):269. https://doi.org/10.17660/ActaHortic.2008.774.35
Zhou Y, Luo S, Hameed S, Xiao D, Zhan J, Wang A, He L (2020) Integrated mRNA and miRNA transcriptome analysis reveals a regulatory network for tuber expansion in Chinese yam (Dioscorea opposita). BMC Genom 21(1):117. https://doi.org/10.1186/s12864-020-6492-5
Zhu H, Huo X (2011) Chinese Yam (Dioscorea opposite Thunb.) dormancy-related activity and the dynamic changes of endogenous hormone. Acta Agriculturae Boreali-Sinica 26(2):198–202
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81860670), the Guangxi Natural Science Foundation of China (Grant No.2018GXNSFDA281029), and the Guangxi Innovation Team Project of Tubers of Modern Agricultural Industrial Technology System of China (nycytxgxcxtd-11-01).
Author information
Authors and Affiliations
Contributions
Y.Y.Z. performed the experiments and wrote the manuscript. Y.T.L. and M.X.G performed the experiments. F.Q., J.Z., and D.X. provided technical assistance and research guidance. A.Q.W. and L.F.H. designed the research and revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, Y., Li, Y., Gong, M. et al. Regulatory mechanism of GA3 on tuber growth by DELLA-dependent pathway in yam (Dioscorea opposita). Plant Mol Biol 106, 433–448 (2021). https://doi.org/10.1007/s11103-021-01163-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01163-7