Abstract
Key message
Rice MERISTEM ACTIVITYLESS (MAL), a RING-H2 finger domain (RFD)-containing gene, regulates meristem cell viability after the initiation of root primordia mediated by cytokinin signaling.
Abstract
Genes in the RING-H2 finger domain (RFD) family play various roles during plant development and in biotic/abiotic stress responses. Rice gene MERISTEM ACTIVITYLESS (MAL), being contained in the RING-H2 finger domain (RFD), is characterized by a transmembrane domain at the N-terminal and a C3H2C3 zinc finger domain at the C-terminal. To elucidate the physiological and molecular functions of MAL, we generated MAL knockdown transgenic plants by RNA interference. MAL RNA-interfered (MRi) transgenic plants exhibited a phenotype with shorter crown root length and lower crown root number, accompanied by a lower cell division rate. The low division rate was observed in the root meristem exactly where MAL was expressed. Furthermore, transcriptome data revealed that cell wall macromolecule metabolism-related genes and redox-related genes were enriched in MAL RNAi lines. Most of these differentially expressed genes (DEGs) were induced by exogenous cytokinin. Hence, we conclude that MAL, as a novel regulatory factor, plays a major role in maintaining cell viability in the meristem after the initiation of root primordial formation, mediated by cytokinin signaling and reactive oxygen species (ROS).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding the mechanisms behind plant root development is important for regulating root growth in order to improve crop performance. The root system of rice is composed of three root types that are formed during consecutive developmental stages. Among these types, crown roots (CRs) [also named shoot-borne roots or adventitious roots (ARs)], which are specific to cereals, initiate from the ground meristem located in stem nodes of coleoptile sections during the postembryonic development process (Marcon et al. 2013) and constitute the main part of the fibrous root system (Zhao et al. 2009). The elongation of crown roots primarily expands the volume of soil explored. It is now widely accepted that many aspects of crown root development, including the temporal sequences of crown root formation, the growth rate and angle, influence the ability of the root system to exploit different spatial and temporal soil niches and are key to optimizing the capture of soil resources. The formation of crown roots leads to an increased number of functional root meristems, which has a profound impact on root architecture. Therefore, it is vital to characterize the key genes which function in crown root meristem development. Over the past decades, significant progress has been made in understanding rice crown root morphogenesis. For instance, developmental stages such as initiation, emergence, and elongation have been clearly distinguished (Itoh et al. 2005; Coudert et al. 2010; Kitomi et al. 2011b), and several key genes involved in the control of crown root development have been identified and characterized (Inukai et al. 2005; Liu et al. 2005, 2009; Kitomi et al. 2011a; Zhao et al. 2009; Zhou et al. 2017). However, the molecular mechanisms of crown root development remain elusive.
As is known, the formation of proper root morphology is mainly dependent on the maintenance of the root apical meristem (RAM), which is controlled by phytohormones, transcription factors, as well as Ca2+ signaling (Lee et al. 2013; Leitao et al. 2019). It has been reported that cytokinin and auxin are the main hormones involved in RAM maintenance in Arabidopsis (Dello Ioio et al. 2007; Dello Ioio et al. 2008; Moubayidin et al. 2009; Di Mambro et al. 2017). In general, cytokinin functions to stimulate cell differentiation by suppressing auxin signaling and transport, whereas auxin plays a role in promoting cell division by inhibiting cytokinin signaling (Lee et al. 2013). Previous studies have also revealed that auxin and cytokinin signaling mutually interact with each other in the root stem cell niche in Arabidopsis. For example, PIN-FORMED (PIN) and the AUXIN RESISTANT 1 (AUX1)/LIKE AUX1 (LAX) family transporters form an auxin gradient along the RAM and an auxin maximum at the quiescent center (QC) (Blilou et al. 2005; Grieneisen et al. 2007; Ugartechea-Chirino et al. 2010). Spatial profiling of PLETHORA (PLTs) expression in RAM can be determined from the auxin distribution, which is vital for stem cell niche specification and its daughter cell proliferation (Scheres and Krizek 2018). Meanwhile, AUXIN RESPONSE FACTOR 5 (ARF5) and ARF7 regulate the expression of PLT1 and PLT2 during the development of the embryonic root (Aida et al. 2004). 26S proteasome (26SP) containing AAA-ATPase 5a (RPT5a) has been reported to be involved in adjusting the activity of TIR1/AFB-dependent auxin signaling to an appropriate level required for maintenance of RAM size, especially under high-boron conditions in Arabidopsis (Sakamoto et al. 2019). CROWN ROOTLESS 5 (CRL5), which is activated by AUXIN RESPONSE FACTOR (ARF1), promotes crown root development by repressing type A cytokinin-response factor, RESPONSE REGULATOR 1 (RR1) (Kitomi et al. 2011a). OsNAC2, a NAC transcription factor, mediates the crosstalk between auxin and cytokinin signaling pathways to regulate the initiation and elongation of rice roots (Mao et al. 2020). WUSCHEL-Related Homeobox 11 (WOX11) is also induced by auxin and cytokinin, and interacts with ETHYLENE-RESPONSIVE FACTOR 3 (ERF3) protein, which might enhance WOX11-mediated repression of RR2 or inhibit its function on RR2 activation during crown root elongation (Zhao et al. 2009, 2015). Root meristem size in rice is also found to be modulated by an AP2 transcription factor SHOEBOX mediated by GA biosynthesis, and SHOEBOX regulates the elongation and proliferation of meristem cells in a developmental stage-specific manner (Li et al. 2015). Intriguingly, nuclear Ca2+ signaling in the nucleus of Arabidopsis root cells is correlated with meristem development and auxin homeostasis to sustain primary root (PR) growth (Leitao et al. 2019). The miR156/SPL module also plays a role in Arabidopsis PR growth by altering root meristem activity, mainly via regulating cytokinin responses (Barrera-Rojas et al. 2019). Taken together, these findings suggest the important role of root apical meristem (RAM) formation and maintenance in controlling root system architecture. However, the regulatory mechanisms underlying the control of RAM activity remain largely unknown.
The ARABIDOPSIS TOXICOS EN LEVADURA (ATL) family is one of the plant-specific RING-H2-type ubiquitin ligases (Aguilar-Hernandez et al. 2011). ATL54 plays an essential role in secondary cell wall formation in Arabidopsis (Noda et al. 2013). Meanwhile, as a member of RING-H2, EL5 with RING-type E3 activity is involved in maintenance of root meristem viability via the effect of cytokinin on nitrogen in rice (Koiwai et al. 2007; Takai et al. 2002; Mochizuki et al. 2014). Further analysis suggests that EL5 membrane localization is necessary for its physiological function in rice root development (Koiwai et al. 2007). In this study, we identified another member of RING-H2, viz. MAL (MERISTEM ACTIVITYLESS), and found that it plays a role in CR growth by altering root meristem activity, mainly via modulating cytokinin responses, in rice. MAL exhibits a high transcription level in crown root meristem. Cytological analysis showed that MAL is involved in meristematic cell division activity. RNA-interfered plants exhibited root growth with shorter root length and fewer crown roots. MAL showed increased sensitivity to exogenous cytokinin. Transcriptome profiling of root tips revealed many cytokinin-related genes with an altered expression level in MAL RNAi plants. Taken together, these results suggest that a MAL-mediated cytokinin-triggered pathway has an effect on root meristem size in rice.
Materials and methods
Plant materials and growth conditions
Rice cultivar (Oryza sativa ssp. Japonica/Geng) Zhonghua11 (ZH11) was used in this study. Seeds were surface-sterilized and germinated on MS medium [0.8% agar and 3% (w/v) sucrose] at 28 °C (in light) and 24 °C (in dark) with a 14/10 h light/dark cycle. Full-length cDNA used in the following experiment was amplified from total cDNA of ZH11 using primers MAL-F and MAL-R (Table S3).
Vector construction and rice transformation
To construct the RNAi vector, a 304-bp cDNA fragment (Fig. S1A) of MAL was amplified from the MAL cDNA using primers MAL-RNAi F and MAL-RNAi R (Table S3) and inserted into the Kpn I and BamHi I sites (for forward insert) and the SacI and Spe I sites (for the reverse insert) of the pDS1301 vector (Zhao et al. 2009).
Agrobacterium tumefaciens (strain EHA105)-mediated transformation of ZH11 was conducted according to the method of Zhao et al. (2009).
RNA isolation and RT-qPCR
Total RNA of rice root tips was isolated using TRIzol (Invitrogen, 15596026) according to the manufacturer’s instructions. For reverse transcription, 4 µg total RNA was digested by 1 µg DNase I. Oligo dT was conjugated with poly A tail by treatment at 65 °C for 10 min and cooled down on ice for 2 min. MLV (reverse transcriptase, Invitrogen, 1906298) with RRI (RNase inhibitor, Takara, 2313A) and DTT (dithiothreitol, Invitrogen, 1906300) were added for reverse transcription at 37 °C for 1.5 h. Products were diluted by adding 140 µl ddH2O. RT-qPCR analysis was performed in a 96-well plate with an ABI StepOnePlus real-time PCR system (Applied Biosystems). The following thermal profile was used for all reactions: 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The rice ACTIN1 gene was used as internal control. The relative expression level of each gene in transgenic plants was compared with the wild type, after normalization with ACTIN1 transcript and averaged from three biological replicates. Bars represent the standard deviation (SD). The sequences of the primers used are listed in Table S3.
In situ hybridization
In situ hybridization and immunological detection were conducted as described by Zhao et al (2009). The MAL fragment was amplified from the MAL cDNA using the primer set MAL-RNAi F and MAL-RNAi R (Table S3) and inserted into pGEM-T (Promega, A3600), followed by transcription with either T7 (Roche, 10881767001) or SP6 (Roche, 10810274001) transcriptase by using the Digoxigenin RNA labeling kit (Roche, 11277073910).
Exogenous hormone treatment
Plants were transferred to media with or without 10−6 M NAA, 10−5 M 6-BA for the indicated hours after germination on agar medium. For kinetics assay, total RNA of root tips (3–5 mm) was extracted and analyzed by RT-qPCR as mentioned before (Zhao et al. 2015; Jiang et al. 2017).
5-Ethynyl-2′-deoxyuridine (EdU) staining
EdU staining was performed as described by Li et al. (2015). Briefly, root tips of 5 days after germination (DAG) rice seedling was labeled in 50 μM EdU (RiboBio, C10310) solution for 2 h. Root tips were fixed by 4% paraformaldehyde, then embedded in 5% agarose. These samples were sectioned for 100 μm and incubated with Apollo. Images were captured with a Leica TCS SP2 confocal laser scanning microscope.
RNA-Seq analysis
Total RNA was extracted from root tips of 5 DAG MRi and wild type (ZH11) plants using TRIzol reagent (Invitrogen, 15596026). The collected root tips were just the sections where MAL is expressed. RNA libraries were collected according to the protocol provided by Illumina. Briefly, 4 mg total RNA was used for mRNA purification and cDNA synthesis. After first- and second-strand cDNA was synthesized, indexing adapters (TruSeq ChIP Sample Preparation kit, Illumina) were added at 5′ and 3′. DNA fragments were enriched by PCR with 10 cycles. The purified DNA libraries were sequenced with the Illumina HiSeq-3000 system. High-throughput sequencing resulted in the generation of ~ 40 million raw reads for each sample. Two independent biological repeats were analyzed.
For data analysis, after removing low-quality tags with Trimmomatic (v0.32) (Bolger et al. 2014), clean tags were aligned to the rice genome (RGAP, v7.0) by TopHat (v2.1.1). Cufflinks (v2.2.1) software suite was used to assemble transcripts and find differentially expressed genes (DEGs) (fold change > 2, P < 0.05) (Trapnell et al. 2012).
To search for cytokinin-inducible genes, published data (GSE39429) were used (https://ricexpro.dna.affrc.go.jp/) (Sato et al. 2013). The cytokinin induction ability of all genomic genes was calculated. Fold change was defined as treatment value (experimental group versus mock group) divided by pretreatment value. Genes with fold changes greater than or equal to 2 times were defined as cytokinin-inducible genes. The website https://ricearray.org/ was used for gene ontology (GO) enrichment analysis.
Results
MAL is expressed in the apical meristem zone of crown roots
To study the molecular mechanism of MAL during the crown root formation process, we first analyzed MAL expression patterns in different rice tissues/organs. Total RNA was isolated from leaf (Le), stem (St), crown root tips (about 5 mm) (Rt), panicle (Pa), seeds (3 days after pollination, DAP) (Se), and callus (Ca). RT-qPCR results revealed that MAL was ubiquitously expressed in all examined tissues/organs and exhibited a high transcript level in root tips but a relatively low level in 3-DAP seeds (Fig. 1A). These results indicate that MAL might play a role in controlling root growth. To provide insight into the spatiotemporal variation of MAL during crown root formation in more detail, we further explored the expression profile of MAL at rice crown root initiation, emergence, and elongation stages by in situ hybridization. The results showed that the transcript of MAL was mainly detected in crown root tips, especially in the meristem region [Fig. 1B(a)] as well as during crown root emergence (Fig. 1B(c)). Together, these results indicate a role for MAL in establishing and maintaining the RAM in rice.
Expression patterns of MAL. A Detection of MAL transcript by RT-qPCR in leaf (Le), stem (St), crown root tip (Rt), panicle (Pa), seed (Se), and callus (Ca). ACTIN1 gene was used as internal control. Bars show mean ± SD of three technical replicates. B In situ hybridization detection of MAL transcripts in crown root tip (a, b) and crown root primordia (c, d) with an antisense or sense probe. Bar: 50 µm in (a, b) and 250 µm in (c, d)
MAL-downregulated transgenic plants displayed abnormal crown root development
To further reveal the functions of MAL during rice crown root development, a 304-bp segment downstream zinc finger domain specific to MAL was inserted in inverted repeats fashion to build a construct by RNA interference (RNAi) (Fig. S1A). The construct was transformed into a Japonica (Geng dao) variety ‘Zhonghua11’ (ZH11). More than 10 independent transgenic lines were produced, and the relative transcript levels of MAL in the root tips of transgenic plants were analyzed using specific primer pairs. Most transgenic plants showed decreased expression of the endogenous gene MAL, suggesting the effect of RNAi (Fig. S1B). Two of theRNAi transgenic lines (MRi-2, MRi-3) were selected for subsequent analysis. Seven days after germination, MRi seedlings developed fewer crown roots compared with wild type (Fig. 2a, b). In addition, the primary root length and the plant height were also decreased (Fig. 2a, b; Table 1). To further examine whether MAL modulated the initiation of crown root primordia, cross sections of the coleptilar nodal region of 3- and 5-DAG MRi and ZH11 seedlings were stained with toluidine blue, respectively. Histological observation revealed that emergence of crown roots in MRi transgenic lines was retarded compared with wild type (Fig. 2c). Furthermore, 1-month-old MRi seedlings also displayed reduced plant height and many fewer crown roots (Fig. S1C). These results indicate that MAL might be essential for crown root initiation and emergence.
Transgenic plants with downregulation of MAL (MRi) displayed severe crown root development effects. a Root phenotypes of MAL RNAi (MRi) and ZH11 plants at 7 DAG (days after germination). ZH11 represents wild type, MRi-2 and MRi-3 represents two independent transgenic lines. Bar = 3 cm. b Statistical analysis of primary root length, crown root length, and crown root number for each genotype. Data represent the mean ± SD of 30 independent biological samples. c Crown root primordia numbers in MAL RNAi and wild-type (ZH11) seedlings at 3 DAG (left) and 5 DAG (right) as indicated. Arrowheads indicate emerging crown root primordia. Bar = 100 μm. Significance determined by t-test: Asterisks indicate significant difference between wild type and MRi lines at *P < 0.05 and **P < 0.01 level
MAL participates in regulating meristem size and cell division activity
Continuous root growth and development are sustained by RAM, a reservoir of undifferentiated cells that gives rise to mature root architecture. To determine whether MAL regulates root meristem size, longitudinal sections of root tip of wild-type and MRi transgenic lines were stained with toluidine blue. The root meristem size of MRi plants was shorter than that of wild type (MRi-2, 379.67 ± 34.62**; MRi-3, 355.00 ± 58.41**; wild type, 496.88 ± 62.53; data obtained from three lines of transgenic genotype with n > 15 for each line) (Fig. 3a, b). Root meristem size is correlated with cell number and cell longitudinal length in the meristem region, so we examined both of them in root-tip longitudinal sections of transgenic and wild-type plants. The results showed that the root meristem of MRi plants contained fewer cells (MRi-2, 56.33 ± 6.56**; MRi-3, 52.50 ± 8.19**) than wild type (81.00 ± 2.55) (Fig. 3b), while no clear difference in cell longitudinal length in root meristem was observed between MRi and wild-type plants (Fig. 3b). These data suggest that MAL knockdown impaired cell proliferation of root meristem in rice. Consistently, EdU staining indicated that MRi noticeably decreased cell proliferation in root meristem (Fig. 3c, d). All of these data suggest that MAL is involved in promoting cell division of root meristem in rice.
MRi plant roots show decreased root meristem size and activity of meristem. a Median longitudinal sections through crown root tips of 5-DAG ZH11 and MAL RNAi (MRi) seedlings. Red arrowheads indicate the proximal end of the root meristem. Black box marks region selected for enlargement in meristem region. b Quantification analysis done at the fourth cortical layer in the whole meristem region. c Median longitudinal view of EdU staining in root meristem of ZH11 (left) and MRi (right) seedlings. d Quantification analysis of EdU-stained cell number and signal intensity (using ImageJ software) at the fourth cortical layer in the whole meristem region. Bar = 150 µm in (a, b). Error bars represent SD (n = 10). *P < 0.05, **P < 0.01, t-test
MAL is involved in response to exogenous cytokinin and in cytokinin-related pathway
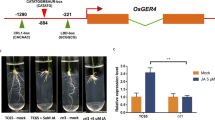
Cytokinin and auxin have central, but opposite, regulatory functions in root development. To ascertain whether MAL expression is responsive to exogenous cytokinin and auxin treatments, analysis of the promoter sequence of MAL gene was performed by using the PLACE program (https://www.dna.affrc.go.jp/PLACE/?action=newplace). The results showed that MAL promoter included 36 cytokinin response element NGATT (Ross et al. 2004; Sakai et al. 2000), 4 auxin response element TGTCTC (Ulmasov et al. 1999) or GAGAC (Maruyama-Nakashita et al. 2005), and 4 ABA response element MACGYGB (Kaplan et al. 2006) (Fig. S2A). Furthermore, we examined the expression levels of MAL in 7-DAG seedling root tips treated with 10−5 M 6-benzylaminopurine (6-BA) and 10−6 M 1-naphthaleneacetic acid (NAA) during different time courses by RT-qPCR, respectively. These results showed that, apart from NAA treatment, the transcript of MAL was greatly induced by 6-BA 0.5 h post treatment, compared with control (Fig. 4a; Fig. S2B). Additionally, in situ hybridization results also revealed that the transcript level of MAL in cytokinin-treated crown roots was higher than in control [Fig. 4B(a, b)]. Collectively, these results indicate that MAL might be involved in cytokinin-mediated crown root development.
MAL is responsive to exogenous cytokinin treatment. A Kinetics of induction of MAL in response to 6-BA. The transcript level of MAL in roots of 7-DAG light-grown wild-type seedlings treated with 6-BA (black) or water (white) for the indicated durations.PCR signals normalized with those of ACTIN1 transcripts. Bars are mean ± SD of three technical replicates. B In situ hybridization detection of MAL transcripts in crown root tip treated with 6-BA (a) and mock (b) with antisense (a and b) or sense probe (c). Bar = 250 µm
Knockdown of MAL affects expression of cytokinin-related genes
To reveal the physiological function of MAL, the transcript changes in crown root tips of two different MRi transgenic lines and wild type (ZH11) were investigated. The sequence data displayed high reproducibility (coefficient of association, R > 0.99) (Table S1). Above 97% of the reads were of high quality, of which about 95% aligned well to the rice genome sequence. Above 80% of the aligned reads were unique (Table S1). The uniquely aligned reads were used to calculate the relative abundance of transcripts. A total of 1179 differentially expressed genes (DEGs) (405 upregulated, 774 downregulated) exhibited greater than twofold change (P < 0.05) in MRi plants versus wild type (Fig. 5a). Most of the DEGs fell into the posttranslation, proteolysis, metabolism, stress, and cell components categories (Fig. 5b), while about 35% of the DEGs had no annotated function. More interestingly, we found that a noticeable number of the down- (103) and upregulated (17) genes could be inducible during cytokinin treatment (https://ricexpro.dna.affrc.go.jp/; Fig. 5c), including cytokinin dehydrogenase precursor (LOC_Os01g56810) and cis-zeatin O-glucosyltransferase (LOC_Os04g47720) (Table S2). We selected 12 DEGs to verify the accuracy of the RNA-Seq results and found a high correlation coefficient between the two datasets (Fig. 6a). To further determine whether these genes involved in cytokinin metabolism and signaling were regulated by MAL, we examined the relative expression levels of not only these DEGs, but also other cytokinin responsive regulators (RRs) and cytokinin oxidases/dehydrogenases in wild type (ZH11) and MRi root tips. We found that all these genes were highly activated in MRi transgenic plants compared with wild type (Fig. 6b, Fig. S3). This result suggests that MAL regulates rice root development by mediating cytokinin homeostasis/signaling.
Knockdown of MAL affects the transcripts of a large number of cytokinin-regulated genes. a Differentially expressed gene numbers of MRi compared with wild type. b Functional categorization of 1,179 genes with altered expression in MRi crown root by gene ontology analysis. c Relative transcript level of identified differentially expressed genes (17/103 up/down) that are responsive to cytokinin
Discussion
Root growth is mainly dependent on cell proliferation in the RAM zone (Takatsuka and Umeda 2014; Dello Ioio et al. 2012). The root cell proliferation speed and root meristem size are determined by the cell division rate in the root meristem zone. In rice, crown root growth is modulated by many factors, such as phytohormones (Lin and Sauter 2019; Sun et al. 2015; Li et al. 2015), transcription factors (Inukai et al. 2005; Liu et al. 2005; Zhao et al. 2009; Li et al. 2015), microRNAs (miRNAs) (Cho and Paszkowski 2017; Bian et al. 2012), and the interplay between them. In this study, we identified MAL as a novel regulatory gene involved in the development of rice crown roots by controlling meristem size. Our results suggest that MAL has a high expression level in root meristem (Fig. 1). Knockdown of MAL significantly decreased the number and length of crown roots (Fig. 2, Fig. S1). Histological analysis further showed that MRi plants displayed retardation of crown root primordia initiation and impairment of cell mitosis activity in RAM (Fig. 2b). These results suggest that MAL preferentially acts in root development after initiation of root primordia by maintaining the viability of meristematic cells. The expression profile and histological phenotype of MAL were similar to those of WOX11 (Zhou et al. 2017), and the transcript level of MAL was also reduced in wox11 (Fig. S3A). However, no WOX11 binding site was found in the MAL promoter and gene body. These results demonstrate that the expression of MAL might be regulated indirectly by WOX11. In addition, expression of WOX11 was mainly detected in emerging crown root primordia and later growth stage (Zhao et al. 2009), whereas MAL was expressed during the initiation of crown root primordia [Fig. 1B(c)]. These results suggest that MAL and WOX11 might be involved in early development stages of crown roots. To investigate whether MAL and WOX11 regulated crown root development by a similar pathway, the expression levels of downregulated genes in wox11 (Jiang et al. 2017) were also detected in MRi plants (Fig. S3B). However, these gene transcripts were not significantly different in MRi lines compared with wild type, indicating that MAL controlled crown root development via a different pathway than WOX11. It has been reported that CRL5 regulates crown root development through activation of OsRR1 (Kitomi et al. 2011a). OsRR1 and OsRR2 regulate OsCKX4 by directly binding to its promoter (Gao et al. 2014). Interestingly, in MRi crown root tips, the expression levels of CRL5, OsRR1/2, and OsCKX4 were clearly decreased (Fig. S3A; Fig. S6B). These results demonstrate that MAL might act upstream of CRL5 and be involved in crown root development through cytokinin signaling.
Cytokinin plays a crucial role in determining root meristem size by controlling the switch from cell proliferation to cell differentiation (Takatsuka and Umeda 2014), and inhibiting root elongation by reducing the number of dividing cells and the size of root meristem (Hwang et al. 2012; Moubayidin et al. 2009; Schaller et al. 2015; Vanstraelen and Benkova 2012; Werner et al. 2003). In Arabidopsis, dysregulation of cytokinin-related genes could affect root meristem activity, resulting in abnormal root growth and development (Miyawaki et al. 2006; Sakamoto et al. 2006; Hirose et al. 2007; Cheng et al. 2010), and an increase in the level of endogenous cytokinin further leads to inhibition of primary root growth (Sakamoto et al. 2006). Disruption of eight of the ten type-A ARR genes affects root development by altering the size of the apical meristem (Zhang et al. 2011). Mutation of a maize type-A RR gene, ABPHL1 (ABPH1), induced an increased root meristem size (Giulini et al. 2004). In rice, overexpression of OsRR3, OsRR5, and OsRR6 affects crown root development (Hirose et al. 2007; Cheng et al. 2010). In this study, exogenous cytokinin greatly induced MAL expression in crown root tips (Fig. 4a, b). In the root tips of MAL RNAi transgenic plants, the expression levels of many genes encoding cytokinin synthesis, cytokinin response regulators (RRs), and oxidase/dehydrogenase were obviously changed (Fig. S3C). These results suggest that MAL plays a partial role in crown root development by mediating cytokinin signals, which is necessary for induction of cell division and elongation as well as maintenance of meristem identity. Similarly, EL5, another ATL member, was reported to maintain the viability of root meristem through the effect of cytokinin on nitrogen in rice (Mochizuki et al. 2014). However, in our study, the transcript level of EL5 was not affected in MAL RNAi plants roots (Fig. S3A). These results indicate that MAL and EL5 might play different roles in rice crown root development mediated by cytokinin signaling. The molecular mechanism by which MAL affects root meristem cell activity through cytokinin-triggered pathway remains to be further elucidated.
Our transcriptome data show that the DEGs in MAL RNAi plants were enriched in cell wall macromolecule metabolism and oxidation–reduction (Fig. S4), and many of them were cytokinin-induced genes, such as LOC_Os01g54620, LOC_Os01g61160, and LOC_Os01g61940 (Figs. 5c, 6a; Table S2). It was recently reported that changes in cell size were essential for the initial steps of cell differentiation, which were related to cytokinin activation of expansion (Pacifici et al. 2018), thus triggering the differentiation through the mechanical control of cell walls. The cell wall loosening consequently induced cell elongation, driving cell differentiation in the root meristem. We found that some DEGs related to cell wall formation were also responsive to exogenous cytokinin (Fig. 5c). These results indicate that MAL might be involved in cell wall metabolism by mediating cytokinin-regulated cell differentiation and growth. Higher H2O2 level in vtc-1 mutant RAM led to increased QC cell number and periclinal division (Kka et al. 2018). This result reveals that H2O2 level and cell division exhibit a certain correlation during RAM size maintenance. Accumulating evidence demonstrates that the cell redox state has an impact on its proliferation/differentiation program; For instance, in Arabidopsis embryonic roots, cells in the oxidized state were accelerated during divisions of the G1 phase, whereas cells in the reduced state were slowed down (de Simone et al. 2017). These results suggest that regulating cell redox is a key sign of cell activity in root apical meristem. Meanwhile, overexpression of IPT increased cytokinin synthesis, which alleviated inhibition of root growth in response to drought through ROS scavenging system activation in Agrostis stolonifera (Xu et al. 2016). These results demonstrate the interplay between cytokinin and oxidative state to maintain better root growth. In MRi plants, oxidation reduction-related genes were enriched (Fig. S4), and importantly, expression levels of these DEGs were also induced by cytokinin (Fig. 5c). These results demonstrate that MAL might play a role in the crosstalk between ROS and cytokinin (Fig. 7). At present, although there is limited information about the relationship between MAL, ROS, and cytokinin in regulation of root meristem size responsible for crown root growth, our results provide a theoretical basis for further research into the interaction.
Accession numbers
Sequence data from this article can be found on the Rice Genome Annotation Project website (https://rice.plantbiology.msu.edu/) under the following accession numbers: MAL, LOC_Os06g19680; EL5, LOC_Os02g57490; CKX1, LOC_Os05g31040; CKX2, LOC_Os01g10110; CKX3, LOC_Os01g09260; CKX4, LOC_Os01g71310; CKX5, LOC_Os06g37500; CKX6, LOC_Os06g37500; CKX7, LOC_Os02g12770; CKX8, LOC_Os01g56810; CKX9, LOC_Os08g35860; RR1, LOC_Os04g36070; RR2, LOC_Os02g35180; RR3, LOC_Os02g58350; RR4, LOC_Os01g72330; RR5, LOC_Os04g44280; RR6, LOC_Os04g57720; RR7, LOC_Os07g26720; RR8, LOC_Os08g28900. The RNA-Seq described in this paper have been deposited in the National Center for Biotechnology Information databases (GSE141854).
References
Aguilar-Hernandez V, Aguilar-Henonin L, Guzman P (2011) Diversity in the architecture of ATLs, a family of plant ubiquitin-ligases, leads to recognition and targeting of substrates in different cellular environments. PLoS ONE 6(8):23934
Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119(1):109–120
Barrera-Rojas CH, Rocha GHB, Polverari L, Brito DAP, Batista DS, Notini MM, da Cruz ACF, Morea EGO, Sabatini S, Otoni WC, Nogueira FTS (2019) miR156-targeted SPL10 controls root meristem activity and root-derived de novo shoot regeneration via cytokinin responses. J Exp Bot 71(3):934–950
Bian H, Xie Y, Guo F, Han N, Ma S, Zeng Z, Wang J, Yang Y, Zhu M (2012) Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol 196(1):149–161
Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433(7021):39–44
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Cheng X, Jiang H, Zhang J, Qian Y, Zhu S, Cheng B (2010) Overexpression of type-A rice response regulators, OsRR3 and OsRR5, results in lower sensitivity to cytokinins. Genet Mol Res 9(1):348–359
Cho J, Paszkowski J (2017) Regulation of rice root development by a retrotransposon acting as a microRNA sponge. eLife 6:30038
Coudert Y, Perin C, Courtois B, Khong NG, Gantet P (2010) Genetic control of root development in rice, the model cereal. Trends Plant Sci 15(4):219–226
de Simone A, Hubbard R, de la Torre NV, Velappan Y, Wilson M, Considine MJ, Soppe WJJ, Foyer CH (2017) Redox changes during the cell cycle in the embryonic root meristem of Arabidopsis thaliana. Antioxid Redox Signal 27(18):1505–1519
Dello Ioio R, Galinha C, Fletcher AG, Grigg SP, Molnar A, Willemsen V, Scheres B, Sabatini S, Baulcombe D, Maini PK, Tsiantis M (2012) A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr Biol 22(18):1699–1704
Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17(8):678–682
Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322(5906):1380–1384
Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novak O, Ljung K, Di Paola L, Maree AFM, Costantino P, Grieneisen VA, Sabatini S (2017) Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci USA 114(36):7641–7649
Gao S, Fang J, Xu F, Wang W, Sun X, Chu J, Cai B, Feng Y, Chu C (2014) Cytokinin oxidase/dehydrogenase 4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol 165(3):1035–1046
Giulini A, Wang J, Jackson D (2004) Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430(7003):1031–1034
Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449(7165):1008–1013
Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48(3):523–539
Hwang I, Sheen J, Muller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63:353–380
Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17(5):1387–1396
Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46(1):23–47
Jiang W, Zhou S, Zhang Q, Song H, Zhou DX, Zhao Y (2017) Transcriptional regulatory network of WOX11 is involved in the control of crown root development, cytokinin signals, and redox in rice. J Exp Bot 68(11):2787–2798
Kaplan B, Davydov O, Knight H, Galon Y, Knight MR, Fluhr R, Fromm H (2006) Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18(10):2733–2748
Kitomi Y, Ito H, Hobo T, Aya K, Kitano H, Inukai Y (2011a) The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J 67(3):472–484
Kitomi Y, Kitano H, Inukai Y (2011b) Molecular mechanism of crown root initiation and the different mechanisms between crown root and radicle in rice. Plant Signal Behav 6(9):1270–1278
Kka N, Rookes J, Cahill D (2018) The influence of ascorbic acid on root growth and the root apical meristem in Arabidopsis thaliana. Plant Signal Behav 129:323–330
Koiwai H, Tagiri A, Katoh S, Katoh E, Ichikawa H, Minami E, Nishizawa Y (2007) RING-H2 type ubiquitin ligase EL5 is involved in root development through the maintenance of cell viability in rice. Plant J 51(1):92–104
Lee Y, Lee WS, Kim SH (2013) Hormonal regulation of stem cell maintenance in roots. J Exp Bot 64(5):1153–1165
Leitao N, Dangeville P, Carter R, Charpentier M (2019) Nuclear calcium signatures are associated with root development. Nat Commun 10(1):4865
Li J, Zhao Y, Chu H, Wang L, Fu Y, Liu P, Upadhyaya N, Chen C, Mou T, Feng Y, Kumar P, Xu J (2015) SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation. PLoS Genet 11(8):1005464
Lin C, Sauter M (2019) Polar auxin transport determines adventitious root emergence and growth in rice. Front Plant Sci 10:444
Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P (2005) ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J 43(1):47–56
Liu S, Wang J, Wang L, Wang X, Xue Y, Wu P, Shou H (2009) Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res 19(9):1110–1119
Mao C, He J, Liu L, Deng Q, Yao X, Liu C, Qiao Y, Li P, Ming F (2020) OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development. Plant Biotechnol J 18(2):429–442
Marcon C, Paschold A, Hochholdinger F (2013) Genetic control of root organogenesis in cereals. Methods Mol Biol 959:69–81
Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H (2005) Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J 42(3):305–314
Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, Tabata S, Sandberg G, Kakimoto T (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103(44):16598–16603
Mochizuki S, Jikumaru Y, Nakamura H, Koiwai H, Sasaki K, Kamiya Y, Ichikawa H, Minami E, Nishizawa Y (2014) Ubiquitin ligase EL5 maintains the viability of root meristems by influencing cytokinin-mediated nitrogen effects in rice. J Exp Bot 65(9):2307–2318
Moubayidin L, Di Mambro R, Sabatini S (2009) Cytokinin-auxin crosstalk. Trends Plant Sci 14(10):557–562
Noda S, Takahashi Y, Tsurumaki Y, Yamamura M, Nishikubo N, Yamaguchi M, Sakurai N, Hattori T, Suzuki H, Demura T (2013) ATL54, a RING-H2 domain protein selected by a gene co-expression network analysis, is associated with secondary cell wall formation in Arabidopsis. Plant Biotech 30(2):169–177
Pacifici E, Di Mambro R, Dello Ioio R, Costantino P, Sabatini S (2018) Acidic cell elongation drives cell differentiation in the Arabidopsis root. EMBO J 37(16):99134
Ross EJ, Stone JM, Elowsky CG, Arredondo-Peter R, Klucas RV, Sarath G (2004) Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J Exp Bot 55(403):1721–1731
Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24(6):703–711
Sakamoto T, Sakakibara H, Kojima M, Yamamoto Y, Nagasaki H, Inukai Y, Sato Y, Matsuoka M (2006) Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol 142(1):54–62
Sakamoto T, Sotta N, Suzuki T, Fujiwara T, Matsunaga S (2019) The 26S proteasome is required for the maintenance of root apical meristem by modulating auxin and cytokinin responses under high-boron stress. Front Plant Sci 10:590
Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y (2013) RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res 41:1206–1213
Schaller GE, Bishopp A, Kieber JJ (2015) The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27(1):44–63
Scheres B, Krizek BA (2018) Coordination of growth in root and shoot apices by AIL/PLT transcription factors. Curr Opin Plant Biol 41:95–101
Sun H, Tao J, Hou M, Huang S, Chen S, Liang Z, Xie T, Wei Y, Xie X, Yoneyama K, Xu G, Zhang Y (2015) A strigolactone signal is required for adventitious root formation in rice. Ann Bot 115(7):1155–1162
Takai R, Matsuda N, Nakano A, Hasegawa K, Akimoto C, Shibuya N, Minami E (2002) EL5, a rice N-acetylchitooligosaccharide elicitor-responsive RING-H2 finger protein, is a ubiquitin ligase which functions in vitro in co-operation with an elicitor-responsive ubiquitin-conjugating enzyme, OsUBC5b. Plant J 30(4):447–455
Takatsuka H, Umeda M (2014) Hormonal control of cell division and elongation along differentiation trajectories in roots. J Exp Bot 65(10):2633–2643
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578
Ugartechea-Chirino Y, Swarup R, Swarup K, Peret B, Whitworth M, Bennett M, Bougourd S (2010) The AUX1 LAX family of auxin influx carriers is required for the establishment of embryonic root cell organization in Arabidopsis thaliana. Ann Bot 105(2):277–289
Ulmasov T, Hagen G, Guilfoyle TJ (1999) Dimerization and DNA binding of auxin response factors. Plant J 19(3):309–319
Vanstraelen M, Benkova E (2012) Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol 28:463–487
Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15(11):2532–2550
Xu Y, Burgess P, Zhang X, Huang B (2016) Enhancing cytokinin synthesis by overexpressing IPT alleviated drought inhibition of root growth through activating ROS-scavenging systems in Agrostis stolonifera. J Exp Bot 67(6):1979–1992
Zhang W, To JP, Cheng CY, Schaller GE, Kieber JJ (2011) Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. Plant J 68(1):1–10
Zhao Y, Cheng S, Song Y, Huang Y, Zhou S, Liu X, Zhou DX (2015) The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 27(9):2469–2483
Zhao Y, Hu Y, Dai M, Huang L, Zhou DX (2009) The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21(3):736–748
Zhou S, Jiang W, Long F, Cheng S, Yang W, Zhao Y, Zhou DX (2017) Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell 29(5):1088–1104
Acknowledgements
We thank Qinglu Zhang, and Xianghua Li for help in field experiments and management and Qinghua Zhang for RNA-Seq sequencing. This work was supported by National Key Research and Development Program of China (2016YFD0100301-006-1 and 2016YFD0100903-3), National Natural Science Foundation of China (31671516, 31730049 and 31970806) and the Fundamental Research Funds for the Central Universities (2662018JC017).
Author information
Authors and Affiliations
Contributions
W.J. and Y.Z. designed the experiment. W.J. S.Z. H.H., and H.S. performed the experiments. Q.Z. managed the fields. W.J., S.Z., and Y.Z. analyzed the data. J.W. and Y.Z. wrote the article.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, W., Zhou, S., Huang, H. et al. MERISTEM ACTIVITYLESS (MAL) is involved in root development through maintenance of meristem size in rice. Plant Mol Biol 104, 499–511 (2020). https://doi.org/10.1007/s11103-020-01053-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01053-4