Abstract
Key message
Genome-wide identification of WD40-like genes reveals a duplication of COP1-like genes, one of the key players involved in regulation of flowering time and photomorphogenesis, with strong functional diversification in Rosaceae.
Abstract
WD40 proteins play crucial roles in a broad spectrum of developmental and physiological processes. Here, we conducted a systematic characterization of this family of genes in Rosa chinensis ‘Old Blush’ (OB), a founder genotype for modern rose domestication. We identified 187 rose WD40 genes and classified them into 5 clusters and 15 subfamilies with 11 of RcWD40s presumably generated via tandem duplication. We found RcWD40 genes were expressed differentially following stages of vegetative and reproductive development. We detected a duplication of CONSTITUTIVE PHOTOMORPHOGENIC1-like genes in rose (RcCOP1 and RcCOP1L) and other Rosaceae plants. Featuring a distinct expression pattern and a different profile of cis-regulatory-elements in the transcriptional regulatory regions, RcCOP1 seemed being evolutionarily conserved while RcCOP1L did not dimerize with RcHY5 and RcSPA4. Our data thus reveals a functional diversification of COP1-like genes in Rosacaeae plants, and provides a valuable resource to explore the potential function and evolution of WD40-like genes in Rosaceae plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
WD40 proteins are a group of transcriptional regulators containing multiple WD40 repeats (Smith et al. 1999; Neer et al. 1994). Each repeat contains a core region ~ 40 amino acids with conserved Glycine-Histidine (GH) dipeptide at the N-terminals and tryptophan-aspartate (WD) dipeptide at C-terminals that are separated by 11–24 amino acids in variable lengths (Stirnimann et al. 2010). Normally, a WD40 repeat contains a four-stranded anti-parallel β-sheet and five to seven such repeats form a bladed propeller scaffold where protein and protein interaction takes place (Andrade et al. 2001; Xu and Min 2011). WD40 proteins normally have additional domains to recruit other factors to form protein–protein complexes (van Nocker and Ludwig 2003; Jain and Pandey 2018; Dayebgadoh et al. 2019). WD40 proteins are involved in many developmental and physiological processes in plants (Wu et al. 2008; Park et al. 2019; Pazhouhandeh et al. 2011; Long et al. 2006; Xu et al. 2019). For example, the Arabidopsis Musashi1 (MSI1) interacts with LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) to inhibit H3K27 methylation (Derkacheva et al. 2013). MSI1 interacts also with CULLIN4-DAMAGED DNA BINDING PROTEIN 1B (CUL4-DDB1) complex to regulate gene imprinting during seed development and floral transition (Dumbliauskas et al. 2011; Pazhouhandeh et al. 2011).
One important representative WD40s is Arabidopsis COP1, the core factor in photomorphogenesis and light signal transduction (Yi et al. 2002; Marine 2012). Arabidopsis COP1 owns a typical seven‐bladed β‐propeller structure and has a zinc finger motif, a coiled helix region, and seven WD40 repeats at C‐terminal. All the three domains play essential roles in protein–protein interactions (Holm et al. 2001; Yi and Deng 2005; Zhu et al. 2008). COP1 promotes ubiquitination and degradation of the Elongated Hypocotyl 5 (HY5) by interacting with SUPPRESSOR OF PHYA (SPA1) and other components of E3 ubiquitin ligase complex, and regulates blue-light signaling via protein complex with Cryptochrome 1 (CRY1) and 2 (CRY2) (Lau and Deng 2012; Liang et al. 2019; Liu et al. 2016; Yang et al. 2000). Furthermore, COP1 can regulate circadian clock and flowering time via interacting with EARLY FLOWERING 3 (ELF3) and GIGANTEA (GI) (Wang et al. 2015). COP1 also interacts with its closet homolog SPA proteins in Arabidopsis to modulate the accumulation of photomorphogenesis-promoting transcription factors in nucleus to tune photomorphogenesis (Lian et al. 2011; Liu et al. 2011; Zuo et al. 2011; Sheerin et al. 2015).
Despite of its pivotal roles in developmental and physiological processes, WD40 genes are moderately conserved with their members systematically identified and characterized in human, wheat, yeast, silkworm and a few other organisms (Zou et al. 2016; Mishra et al. 2014; Salih et al. 2018; Li et al. 2014; Zhu et al. 2015; Hu et al. 2018). However, a thorough investigation of WD40 genes has not been done in horticultural and fruiting plants, for example, Rosaceae plants. Rosaceae includes many important crops, like Fragaria (strawberry), Malus domestica (apple), Prunus persica (peach), Pyrus communis (pear) and rose, which is one of the most important ornamental plants. With the development of a set of genomic resources, roses have now becoming a model woody species for understanding specific traits that not present in current model species (Raymond et al. 2018; Hibrand Saint-Oyant et al. 2018; Li et al. 2018, 2019; Bendahmane et al. 2013; Dong et al. 2017).
In this study, we systematically identified the WD40 family genes in rose (OB). We investigated their chromosome distribution, gene structure, and tissue-specific expression. We found a duplication in COP1-like genes and evaluated their protein interaction spectrum. Our work laid a foundation for further functional exploration of WD40 family in rose and other Rosacaeae plants.
Materials and methods
Plant materials and RNA isolation
OB plants were grown in a glasshouse without additional light in Kunming Botanical Garden, Kunming Institute of Botany, Chinese Academy of Sciences. Leaf materials at just open stage were collected for total RNA extraction with a RNAprep Pure Plant Kit (Tiangen, Beijing) as described by (Li et al. 2018).
RcWD40 protein identification
To identify WD40 proteins in OB, the hmmsearch program (HMMER3.0 package https://hmmer.org/) was employed against the Rose Genome Database (https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/) by using the hidden Markov model (HMM) of the WD40 domain (PF00400) as the query file with E value ≤ 10−10 (Eddy 1998). Predicted WD40 protein were manually annotated using BLASTp in NCBI (https://www.ncbi.nlm.nih.gov/). After removing redundant sequences, all candidate proteins were evaluated via Pfam (https://pfam.xfam.org) (Finn et al. 2014) and Smart (https://smart.embl-heidelberg.de/) (Ponting et al. 1999), and rectified by TBtools (Chen et al. 2018) program to eliminate redundant sequences.
Gene and protein structure and physiological characteristics for RcWD40s
The coding sequence, protein sequence, the sequence of 1.5 Kb regions upstream of transcriptional starting site (TSS) and full-length of genomic sequence were downloaded from Rose Genome Database (https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2). Exon/intron organization was predicted with Gene Structure Display Server (GSDS) (https://gsds.cbi.p-ku.edu.cn/) (Hu et al. 2015). Physicochemical properties were analyzed with EXPASY (https://www.expasy.org/tools/protparam.html) for molecular weight, predicted isoelectric point (PI), sub-cellular localization, negatively and positively charged residues, aliphatic index, grand average of hydrophobicity, and instability index (Wu et al. 2012). The conserved domain was determined by Smart (https://smart.embl-heidelberg.de/) and visualized by Illustrator for Biological Sequences (IBS) (Ren et al. 2009). The cis-regulatory-elements (CREs) for transcription factor binding sites (TFBSs) were predicted by JASPAR (https://jaspar2016.genereg.net/) (Wasserman et al. 2004) for the 1.5 kb upstream regions of TSS.
Chromosomal localization and gene duplication analysis
Physical coordinates of RcWD40s were extracted from the Rose Genome Database (https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/) and used for chromosome mapping with MapChart (Voorrips 2002). The criteria of tandem duplication and segmental duplication events were designated as the followings: 1) the physical distance for two or more genes on one chromosome was less than 100 Kb; 2) sequence identity above 80% of the full length; 3) No insertion between the duplicated genes. Genome segments longer than 1 Kb with sequence identity over 90% were considered as segmental duplication (Hanada et al. 2008; Zhao et al. 2013; Leister et al. 2004; Lynch et al. 2000).
Gene ontology (GO) enrichment assays
GO annotation was downloaded from the Rose Genome Databases (https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2). The online website OmicShare (https://www.omicshare.com/tools/Home/Soft/gogsea) was used to test the statistical enrichment of RcWD40 genes with adjusted p-value less than 0.05 considered as enriched.
Expression analysis
RNA-seq data for OB was retrieved from the database of LIPM (https://www.lipm-browsers.toulouse.inra.fr/plants/R.chinensis) (Dubois et al. 2012) and the heatmap was drawn by using TBtools for visualization (https://www.github.com/CJ-Chen/TBtools). COP1-like sequences were identified with BLASTp against strawberry, apple, peach, and pear genomes and downloaded from the Genome Database for Rosaceae GDR (https://www.rosaceae.org/). RNA-seq reads of COP1-like genes were retrieved from https://www.science.umd.edu/CBMG/faculty/Liu/lab/ using ids gene14041/FvH4_5g22570.1 and gene19736/FvH4_3g01260 for strawberry, and from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62415 with ids MDP0000245133, MDP0000241199, and MDP0000195882 for apple.
For quantitative real-time PCR (qRT-PCR) analysis, twelve rose tissues including young roots, shoot apical meristem, young shoots, old shoots, young leaves, old leaves, closed flower without sepal, petals at anthesis, stamen, axillary buds, young prickles, and young fruits were collected from four-month old cutting-propagating plants and immediately frozen in liquid nitrogen for RNA extraction. OminiPlant RNA Kit (DNase I; Transgene) was used to extract total RNA with BioPhotometerD30 (Eppendorf) used for quantification. The HiScript® II Q RT SuperMix for qPCR (+ gDNA wiper) (Vazyme, Nanjing) was used for first-strand cDNA synthesis. Gene-specific primers for RcCOP1 and RcCOP1L were designed with primer-blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast) using RcPP2A as reference (Supplementary file: Table S6). PCR was carried out using ChamQ Universal SYBR qPCR Master Mix (Vazyme) on QuantStudio 7 Flex (Applied biosystems) with 384-well format. Relative expression was determined according to Hu et al. (2014) and Klie and Debener (2011).
Phylogenetic analysis of RcWD40s and AtWD40s
For phylogenetic clustering of RcWD40s, we obtained 231 Arabidopsis WD40s from TAIR10 (https://www.arabidopsis.org/) (van Nocker and Ludwig 2003), and aligned them with Clustal X version 2.1. A maximum-likelihood (ML) tree was constructed using MEGA5 with 1000 times of bootstrap replicates (Tamura et al. 2011). A similar procedure was applied for phylogenetic analysis of COP1-like genes in Rosaceae plants.
Protein interaction assays with yeast-two-hybrid (Y2H)
Known and potential protein interaction partners for AtCOP1 were predicted using STRING (https://string-db.org/) (Szklarczyk et al. 2019). The sequences of potential orthologs for these partners were identified with BLASTp using best match in the rose genome. cDNA clones of these rose orthologs were isolated from leaf RNA pools and sequenced using gene specific primers (Supplementary file: Table S6). The GAL4 activation domain (AD) of pGADT7 and the GAL4 DNA-binding domain (BD) of pGBKT7 were digested by Ndel, Xhol and EcoR1, BamH1, respectively. RcCOP1 and RcCOP1L were fused to AD and the potential interacting proteins were fused to BD using Clone ExpressIIOne Step Cloning Kit (Vazyme, Nanjing, China). Protein pairs were then co-transfected into Y2H gold yeast strains, and selected according to procedures described in manual of Matchmaker Gold Yeast Two-Hybrid system (ClonTech, Beijing, China). The positive interaction between AtAGL16 and AtSVP was used as positive control (Hu et al. 2014). Final interaction pattern was photographed after incubation at 28 °C for 3 days.
Bimolecular fluorescent complimentary assay (BiFC)
BiFC analysis was carried out according to Hu et al. (2014) using pFGC-cYFP and pFGC-nYFP vectors (Kim et al. 2008). Full-length CDSs of RcCOP1 and RcCOP1L were inserted into pFGC-nYFP vectors, while RcHY5 and RcSPA4 CDSs were cloned into pFGC-cYFP vectors. See Supplementary Table S6 for primers information. All plasmids were introduced into Agrobacterium tumefaciens strain GV3101 and later used to test the pairwise interactions in Nicotiana benthamiana (tobacco) leaves following protocols described in Hu et al. (2014). YFP fluorescence signals were observed and documented under a confocal laser scanning microscope (Olympus Fluoview Ver. 2.0c Viewer).
Results and discussion
Rose genome has 187 WD40 proteins featuring different properties
We identified 187 WD40 proteins in rose genome mainly based on sequence similarity for WD40-domain, and named them as RcWD40-1 to RcWD40-187 following their positions on rose chromosomes for convenience (Fig. 1; Supplementary file: Table S1) (Raymond et al. 2018).
Uneven distribution of RcWD40s on rose chromosomes. a RcWD40s distribution on seven chromosomes (indicated above). Ticks on each chromosome marked the RcWD40s. Tandem duplicated genes were marked in red and the two RcCOP1-like genes were labelled by blue dots. Scale bar was in Megabase (Mb). Note the uneven distribution of RcWD40s on all chromosomes. b Statistics of RcWD40s on rose chromosomes. Y-axis on the left showed the number of RcWD40s on each chromosome, while the Y-axis on the right side gave the number of RcWD40s per Mb of each chromosome
RcWD40 proteins varied dramatically in their WD40 domain, length, size and other physicochemical properties. Their length ranged from 84 to 1756 amino acids with the Isoelectric Point (IP) values being between 4.39–9.48 and Molecular Weights (MW) being between 9,735 Da-364,711 Da. The number of WD40 domains varied between 1–13. RcWD40 proteins had numerous atypical WD40 domains, while 98 WD40 proteins were stable and the remaining 89 proteins being unstable (Wu et al. 2012). Other physical and chemical information about RcWD40 proteins were included in Supplementary file: Table S1.
Uneven distribution on rose chromosomes and gene duplication
RcWD40 genes located extensively and unevenly on the seven rose chromosomes (Fig. 1a; Supplementary file: Table S1). Chr2 harbored 35 RcWD40s with about 0.40 gene per Mb genome sequence, and Chr6 and Chr7 had 33 (0.47 gene per Mb) and 32 (0.46 gene per Mb) genes, respectively. In contrast, Chr1 featured only 18 genes, the least abundant chromosome with about 0.26 gene per Mb genome sequence (Fig. 1b).
We further observed an uneven distribution of RcWD40 genes on different chromosome arms (Fig. 1a). Except Chr3, on which RcWD40 genes were more likely present in the center, all the other six chromosomes seemed have RcWD40 genes at either upper (Chr5) or lower (Chr1, Chr4, and Chr6) or both (Chr2 and Chr7) chromosome arms. The distribution of RcWD40 genes appeared anti-collocating with the distribution of transposable elements on chromosomes (Raymond et al. 2018; Hibrand Saint-Oyant et al. 2018), but was consistent with previous observation in other species (Zou et al. 2016; Mishra et al. 2014; Salih et al. 2018; Li et al. 2014; Zhu et al. 2015; Hu et al. 2018; Ouyang et al. 2012).
We next examined whether tandem duplication and segmental duplication contributed to amplification of RcWD40 genes. We identified 11 members (5.9%) on Chr1, 5, 6 and 7 showing a signature of tandem duplication event (Fig. 1a). No RcWD40 gene was duplicated via potential segmental genome duplication (Zhao et al. 2013; Leister et al. 2004; Lynch et al. 2000). These results implied that, in addition to random duplication, tandem duplication played a role in amplification of RcWD40 genes in rose.
RcWD40 proteins feature different structure and function
RcWD40 genes varied significantly in their exon and intron organization (Supplementary Fig. S1; Supplementary file: Table S1). The gene featuring maximum number of introns was RcWD40-181, which had 38 introns, while 24 RcWD40 genes had no intron. The introns number of other genes ranged between 2 and 14.
We classified the RcWD40 proteins into 15 subfamilies based on their domain structure. There were 127 proteins only containing WD40 domain and were grouped as subfamily A. The remaining 60 RcWD40 proteins had additional domains and were classified into subfamily B to O, respectively (Fig. 2). Gene ontology (GO) annotation revealed that RcWD40 genes were involved in many aspects of biological processes like metabolism and development as well as responses to stimulus and rhythmic regulation (Supplementary Fig. S2; Supplementary file: Table S2). Interestingly, RcWD40 genes were significantly enriched for GO terms related to DNA repair or integrity, root or shoot development, and responses to shade and others stimulus (Fig. 3; Supplementary file: Table S3).
Diversified protein domain structure of RcWD40s. The WD40 repeats were shown in blue, while the other domains were marked in different colors. Texts in red above each cartoon indicated the types of domains. For each subfamily (from A to O), numbers in brackets gave the protein numbers for that subfamily, which was indicated by a representative protein shown below
Rose WD40 proteins experienced significant expansion and contraction along specific phylogenetic lineages
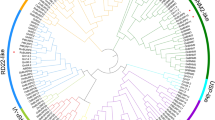
To better understand the evolution of rose WD40 proteins, we conducted a phylogenetic analysis with the 187 RcWD40 proteins and 231 Arabidopsis proteins (Fig. 4). Rose and Arabidopsis WD40 proteins could be grouped into five clusters (I-V), which contained 15, 45, 33, 40 and 54 rose and 39, 60, 34, 28 and 70 Arabidopsis proteins, respectively. In clade I, rose experienced a significant contraction (Chi-square test, p = 0.018) of WD40 genes which were involved in peptidyl-serine dephosphorylation, phagophore assembly, autophagy of nucleus and mitochondria, and trichome differentiation (Supplementary Fig. S3a). Clade IV showed a significant expansion (Chi-square test, p = 0.031) of rose WD40 genes that were involved in histone acetyltransferase activity (Supplementary Fig. S3b). These data suggested that rose had undergone a significant WD40 genes expansion and contraction along specific phylogenetic lineages.
Phylogenetic clustering of WD40s from rose and Arabidopsis revealed lineage-specific gene expansion and contraction with important function in rose. A maximum-likelihood tree clustered the 187 RcWD40s (marked by red circles) and 231 AtWD40s into five clusters (Cluster I-V), which were labelled with different colors. Note that cluster I and IV experienced a lineage-specific contraction and expansion in rose, respectively. Light blue triangles indicated the duplicated RcCOP1-like genes, while numbers on branches showed the bootstrap support values in percentage
RcWD40 genes featured spatial and temporal expression profiles
We explored the spatial and temporal expression profile for RcWD40 genes using the expression data for 11 rose tissues (stamen, dormant axillary buds, active axillary buds, floral buds, white young roots, young leaves and stems, early floral organs, closed flower, open flower, senescent flower and fruit) (Supplementary file: Table S4). Figure 5 showed the heatmap of 186 RcWD40 genes in these tissues. Accordingly, we classified the RcWD40 genes into four main groups (I-IV).
RcWD40s featured spatial and temporal expression profiles. A heatmap showed the Log2 values for reads per million kilobases (RPKM) in 11 rose tissues indicated below. RcWD40s were grouped into four main clusters, which were indicated by I-IV, respectively. Cluster IV was further grouped into three subgroups that were labelled with IV1-3. Note that cluster I and III showed the lowest and highest expression levels, respectively. Expression profiles for the duplicated RcCOP1-like genes were boxed in red (see also Supplementary Fig. S4)
The group I (32 genes) was the group with lowest expression in most of the tissues analyzed. Group II (44 genes) showed a relatively higher expression than group I. Group III (20 genes) had the highest expression, while group IV (90 genes) displayed a moderate but highly variable expression. Group IV was further divided into three subgroups: IV1, IV2 and IV3, which included 27, 23 and 40 genes, respectively. Those genes were more detected in special tissues, indicating that they might participate in specific physiological and biological processes under certain conditions.
Rose had two COP1-like genes with diversified expression patterns
The E3 ubiquitin ligase COP1 contained seven WD40 repeats and played essential roles in photomorphogenesis and many other biological processes (Yi et al. 2002; Marine 2012; Liu et al. 2016; Yang et al. 2000). In our phylogenetic analysis, we identified two copies of COP1-like genes showing distinct expression pattern in rose (Figs. 4, 5; Supplementary Fig. 4), in contrary to previous notion that angiosperms normally have one copy of COP1 (Artz et al. 2019). We renamed RcWD40-176 as RcCOP1 and RcWD40-96 as RcCOP1-like (RcCOP1L) based on their sequence similarity and phylogenetic relationship with AtCOP1 (Fig. 4). RcCOP1 featured the lowest expression in white young roots and dormant axillary buds and the higher expression in stamen, active axillary buds, and fruit, while RcCOP1L expression was detected in stamen at the lowest level and in the senescence flower at the highest level, indicating that these two genes had temporal and spatial expression (Fig. 5 and Supplementary Fig. 4). A further quantitative real-time PCR (qRT-PCR) assay using 12 different tissues also revealed a similar pattern (Fig. 6a). Thus, RcCOP1 and RcCOP1L might have undergone functional diversification post their duplication.
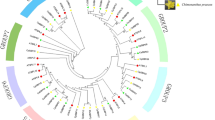
The duplicated COP1-like genes in rose and other Rosaceae plants featured distinct expression pattern. a Quantitative RT-PCR analysis revealed a differential expression of RcCOP1 (red bars) and RcCOP1L (blue bars) in twelve tissues of OB. RcPP2A was used as reference. Mean relative expression with standard deviation was shown. Significant variation was estimated with Student’s t-test, *, p < 0.05; **, p < 0.01; *** p < 0.001. b Phylogenetic clustering of COP1-like genes in Arabidopsis and five Rosaceae plants. COP1s and COP1Ls were indicated in red and blue colors, respectively. c A simplified model showing the duplication of COP1-like genes occurred prior to the split of Rosaceae plants from their common ancestors (pointed by an arrow). Numbers after each species indicated the copy numbers of COP1-like genes. d Diverged expression of strawberry COP1 (FvCOP1, red bars) and COP1L (FvCOP1L, blue bars) in 13 tissues. e Expression of apple COP1s in lateral and central seeds. Note that MdCOP1L1 showed almost no expression in the tissues examined. f Predicted cis-regulatory-elements (CREs) in the 1.5 Kb promoter regions of RcCOP1. g Predicted cis-regulatory-elements (CREs) in the 1.5 Kb promoter regions of RcCOP1L. Only CREs specific for RcCOP1 (red) and RcCOP1L (blue) were shown with names given above each tick in f and g. For d and e, RPKM values for each tissue were given
We next asked whether the duplication of rose COP1-like genes occurred also in other Rosaceae plants. We identified the COP1-like genes from strawberry, apple, peach and pear and performed a phylogenetic reconstruction (Fig. 6b). Interestingly, all these Rosaceae species had two or three copies of COP1-like genes, and accordingly we named them as COP1s and COP1Ls (Fig. 6b). Therefore, duplication of COP1-like genes seemed occurred prior to the split of Rosaceae plants from their common ancestor (Fig. 6c). More importantly, post the duplication, strawberry and apple COP1-like genes have also experienced expression diversification in different tissues (Fig. 6d, e).
RcCOP1s had different cis-regulatory-element (CRE) profiles
We continued to ask whether the diverged expression of rose COP1-like genes correlated with the divergence in CRE profiles by identification of known transcription factor binding sites (TFBSs) in the 1.5 kb regions upstream of transcriptional starting site (TSS). We detected 20 TFBSs shared between RcCOP1 and RcCOP1L, while found 13 TFBSs only present in RcCOP1 and 7 TFBSs only present in RcCOP1L (Fig. 6f, 6g; Supplementary file: Table S5). Notably, the RcCOP1 specific CREs were likely bound by different WRKY and NAC transcription factors, while the RcCOP1L specific CREs were related to SQUAMOSA-LIKEs (SPL4/SPL5), MYB, GARP (KAN1) and AP2/B3 (RAV1) families of transcription factors (Fig. 6f, 6g), suggesting a different regulatory potential of COP1-like genes post duplication.
RcCOP1 proteins differed in their protein partners
As one of the key factors controlling photomorphogenesis and many other developmental and physiological processes, AtCOP1 was known to form protein complexes with a set of partners like HY5, CRY1, CRY2, FUSCA 6/9 (FUS6/9), SUPPRESSOR OF PHYA-105 1/4 (SPA1/4), UVB-RESISTANCE 8 (UVR8), and DDB1B (Kim et al. 2017). Furthermore, COP1 could form homodimers (Torii et al. 1998; Stacey et al. 2000; McNEllis et al. 1996; Xie et al. 2015). To investigate further the functional potential of RcCOP1s post duplication, we first examined the protein sequence divergence between RcCOP1s and AtCOP1 (Fig. 7). Like AtCOP1, both RcCOP1 and RcCOP1L had seven WD40 repeats and one RING-domain at the N-terminal and one coiled-coil region between RING and WD40 repeats. All these domains seemed relatively conserved than rest segments of the three proteins. However, RcCOP1L differed in many positions including the WD40 repeats, a domain where COP1 hetero-dimerize with HY5 (Holm et al. 2001; Osterlund et al. 2000), and the coiled-coil domain, where SPA4 interacts with COP1 (Laubinger and Hoecker 2003; Yi and Deng, 2005), thus suggesting a different interacting spectrum from that of RcCOP1 (Fig. 7).
RcCOP1 and RcCOP1L had significant sequence variation in different domains. Identical amino acids between AtCOP1, RcCOP1 and RcCOP1L were shaded in dark. Grey shadings represented positions with similar amino acids. The RING-finger domain, the coiled-coil region, and the seven WD repeats were underlined in dark lines. The different amino acids between RcCOP1 and RcCOP1L were marked with red boxes, and the amino acids variation between AtCOP1 and RcCOP1/RcCOP1L were marked with blue boxes
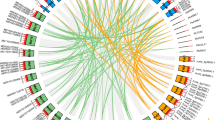
To further substantiate this, we tested the interaction spectrum of RcCOP1s with yeast-two-hybrid (Y2H) approaches. We cloned the corresponding sequences of ten rose homologs to Arabidopsis partners (Fig. 8a) and tested their interaction potential with RcCOP1 and RcCOP1L (Fig. 8b). Our assays confirmed the positive interaction between AGL16 and SVP of Arabidopsis (Hu et al. 2014) and the negative interactions between AD-RcCOP1/RcCOP1L and BD-empty vector (Supplementary Fig. S5). The same to AtCOP1, both RcCOP1 and RcCOP1L could form homodimers, while at the same time they could also interact with each other to form heterodimers (RcCOP1-RcCOP1L) (Torii et al. 1998; Stacey et al. 2000; McNEllis et al. 1996; Xie et al. 2015). Both proteins formed complexes with CRY1, CRY2, FUS6, FUS9, and SPA1, indicating that both proteins could participate in the same biological processes. Corroborating with its sequence variation in the WD40-repeats and coiled-coild domain (Fig. 7), RcCOP1 could interact physically with HY5 and SPA4 proteins of rose, while RcCOP1L did not, suggesting a functional diversification between these two proteins. A further bimolecular fluorescent complimentary (BiFC) assay further substantiated this pattern (Fig. 8c). Both proteins could not form complex with UVR8 or DDB1B, a contrasting pattern to that of AtCOP1 (Fig. 8b). Taken these together, the two RcCOP1-like proteins shared most of their partners while functional divergence could be happened.
RcCOP1 and RcCOP1L featured diversified interactomes. a A protein interaction network for Arabidopsis COP1. b Interaction spectrum of RcCOP1/RcCOP1L tested with yeast-two-hybrid assays. The interactions for Arabidopsis AGL16-SVP and empty vector on BD with AD-RcCOP1s, were used as positive and negative controls, respectively. Note that RcHY5 and RcSPA4 did not interact with RcCOP1L in comparison to RcCOP1. See methods part and supplementary Fig S5 for detailed information about negative controls. c A BiFC assay confirmed the interactome variation between RcCOP1/RcCOP1L with RcHY5/RcSPA4. Empty vectors with one-side fusion proteins were used as negative controls
Conclusion
WD40 proteins were famous for their pivotal roles in plant development and physiological processes. In this study, we systematically identified and characterized 187 WD40s from rose and detected a significant structural variation among these proteins (Figs. 1 and 2). Rose had likely undergone a lineage-specific WD40 genes expansion and contraction (Fig. 4), a similar pattern to that of MADS-box family transcription factors (Liu et al. 2018).
More importantly, we found that rose had duplicated its COP1-like genes with a strong expression and functional divergence. RcCOP1 differed significantly from RcCOP1L with their distinct expression in stamen and several other tissues/organs, a similar pattern also observed in other Rosaceae plants like strawberry and apple (Figs. 5 and 6; Supplementary Fig. S4). Interestingly, the duplication and expression diversification seemed occurred prior to the split of Rosaceae plants from their common ancestors (Fig. 6). Corroborating with the expression divergence between the two rose COP1-like genes, CREs for both genes differed obviously (Fig. 6). Furthermore, the two rose COP1-like proteins had different partners in forming protein complexes, indicating again the strong functional diversification (Fig. 8).
Taken these together, our analyses showcased a rose-specific evolution pattern for WD40 family with COP1-like genes as an example. Our efforts should pave the way to further understand the molecular regulatory mechanisms of biodiversity in roses as well as in other Rosaceae species (like apple and strawberry), which is one of the most important plant families for human being.
References
Andrade MA, Perez-Iratxeta C, Ponting CP (2001) Protein repeats: structures, functions and evolution. J Struct Biol 134(2–3):117–131
Artz O, Dickopf S, Ranjan A, Kreiss M, Abraham ET, Boll V, Rensing SA, Hoecker U (2019) Characterization of spa mutants in the moss physcomitrella provides evidence for functional divergence of SPA genes during the evolution of land plants. New phytol 224(4):1613–1626
Bendahmane M, Dubois A, Raymond O, Bris ML (2013) Genetics and genomics of flower initiation and development in roses. J Exp Bot 64(4):847–857
Chen C, Xia R, Chen H, He Y (2018) TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 289660
Dayebgadoh G, Sardiu ME, Florens L, Washburn MP (2019) Biochemical Reduction of the Topology of the Diverse WDR76 Protein Interactome. J Proteome Res 18(9):3479–3491
Derkacheva M, Steinbach Y, Wildhaber T, Mozgová I, Mahrez W, Nanni P, Bischof S, Gruissem W, Hennig L (2013) Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J 32(14):2073–2085
Dong X, Jiang X, Kuang G, Wang Q, Zhong M, Jin D, Hu J (2017) Genetic control of flowering time in woody plants: Roses as an emerging model. Plant Divers 39(2):104–110
Dumbliauskas E, Lechner E, Jaciubek M, Berr A, Pazhouhandeh M, Alioua M, Cognat V, Brukhin V, Koncz C, Grossniklaus U, Molinier J, Genschik P (2011) The Arabidopsis CUL4-DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. EMBO J 30(4):731–743
Dubois A, Carrere S, Raymond O, Pouvreau B, Cottret L, Roccia A, Onesto JP, Sakr S, Atanassova R, Baudino S, Foucher F, Le Bris M, Gouzy J, Bendahmane M (2012) Transcriptome database resource and gene expression atlas for rose. BMC Genomics 13:638
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14(9):755–763
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42(Database issue):D222–D230
Hibrand Saint-Oyant L, Ruttink T, Hamama L, Kirov I, Lakhwani D, Zhou NN, Bourke PM, Daccord N, Leus L, Schulz D, Van de Gesst H, Hesselink T, Van Laere K, Debray K, Balzergue S, Thouroude T, Chastellier A, Jeauffre J, Voisine L, Gaillard S, Borm TJA, Arens P, Voorrips RE, Maliepaard C, Neu E, Linde M, Le Paslier MC, Bérard A, Bounon R, Clotault J, Choisne N, Quesneville H, Kawamura K, Aubourg S, Smulders MJM, Schijlen E, Bucher E, Debener T, De Riek J, Foucher F (2018) A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat Plants 4(7):473–484
Holm M, Hardtke CS, Gaudet R, Deng XW (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20(1–2):118–127
Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148(2):993–1003
Hu JY, Zhou Y, He F, Dong X, Liu LY, Coupland G, Turck F, de Meaux J (2014) miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell 26(5):2024–2037
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297
Hu R, Xiao J, Gu T, Yu X, Zhang Y, Chang J, Yang G, He G (2018) Genome-wide identification and analysis of WD40 proteins in wheat (Triticum aestivum L.). BMC Genomics 19(1):803
Jain BP, Pandey S (2018) WD40 repeat proteins: signalling scaffold with diverse functions. Protein J 37(5):391–406
Kim JY, Song JT, Seo HS (2017) COP1 regulates plant growth and development in response to light at the post-translational level. J Exp Bot 68(17):4737–4748
Klie M, Debener T (2011) Identification of superior reference genes for data normalization of expression studies via quantitative PCR in hybrid roses (Rosa hybrida). BMC Res Notes 4:518
Kim KC, Lai Z, Fan B, Chen Z (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20(9):2357–2371
Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17(10):584–593
Laubinger S, Hoecker U (2003) The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J 35(3):373–385
Leister D (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20(3):116–122
Li Q, Zhao P, Li J, Zhang C, Wang L, Ren Z (2014) Genome-wide analysis of the WD-repeat protein family in cucumber and Arabidopsis. Mol Genet Genomics 289(1):103–124
Li S, Zhong M, Dong X, Jiang X, Xu Y, Sun Y, Cheng F, Li DZ, Tang K, Wang S, Dai S, Hu JY (2018) Comparative transcriptomics identifies patterns of selection in roses. BMC Plant Biol 18(1):371
Li S, Yang G, Yang S, Just J, Yan H, Zhou N, Jian H, Wang Q, Chen M, Qiu X, Zhang H, Dong X, Jiang X, Sun Y, Zhong M, Bendahmane M, Ning G, Ge H, Hu JY, Tang K (2019) The development of a high-density genetic map significantly improves the quality of reference genome assemblies for rose. Sci Rep 9(1):5985
Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25(10):1023–1028
Liang T, YangY LH (2019) Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol 221(3):1247–1252
Liu B, Yang Z, Gomez A, Liu B, Lin C, Oka Y (2016) Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana. J Plant Res 129(2):137–148
Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25(10):1029–1034
Liu J, Fu X, Dong Y, Lu J, Ren M, Zhou N, Wang C (2018) MIKCC-type MADS-box genes in Rosa chinensis: the remarkable expansion of ABCDE model genes and their roles in floral organogenesis. Hortic Res 5:25
Long JA, Ohno C, Smith ZR, Meyerowitz EM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312(5779):1520–1523
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155
Marine JC (2012) Spotlight on the role of COP1 in tumorigenesis. Nat Rev Cancer 12(7):455–464
McNellis TW, Torii KU, Deng XW (1996) Expression of an N-Terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis. Plant Cell 8(9):1491–1503
Mishra AK, Muthamilarasan M, Khan Y, Parida SK, Prasad M (2014) Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setaria italica L.). PLoS ONE 9(1):e86852
Neer EJ, Schmidt CJ, Nambudripad R, Smith TF (1994) The ancient regulatory-protein family of WD-repeat proteins. Nature 371(6495):297–300
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405(6785):462–466
Ouyang Y, Huang X, Lu Z, Yao J (2012) Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genomics 13:100
Ponting CP, Schultz J, Milpetz F, Bork P (1999) SMART: Identification and annotation of domains from signaling and extracellular protein sequences. Nucleic Acids Res 27(1):229–232
Park HJ, Baek D, Cha JY, Liao X, Kang SH, McClung CR, Lee SY, Yun DJ, Kim WY (2019) HOS15 Interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell 31(1):37–51
Pazhouhandeh M, Molinier J, Berr A, Genschik P (2011) MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc Natl Acad Sci USA 108(8):3430–3435
Raymond O, Gouzy J, Just J, Badouin H, Verdenaud M, Lemainque A, Vergne P, Moja S, Choisne N, Pont C, Carrère S, Caissard JC, Couloux A, Cottret L, Aury JM, Szécsi J, Latrasse D, Madoui MA, Francois L, Fu X, Yang SH, Dubois A, Piola F, Larrieu A, Perez M, Labadie K, Perrier L, Govetto B, Labrousse Y, Villand P, Bardoux C, Boltz V, Lopez-Roques C, Heitzler P, Vernoux T, Vandenbussche M, Quesneville H, Boualem A, Bendahmane A, Liu C, Le Bris M, Salse J, Baudino S, Benhamed M, Wincker P, Bendahmane M (2018) The Rosa genome provides new insights into the domestication of modern roses. Nat Genet 50(6):772–777
Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X (2009) DOG 1.0: illustrator of protein domain structures. Cell Res 19(2):271–273
Salih H, Gong W, Mkulama M, Du X (2018) Genome-wide characterization, identification, and expression analysis of the WD40 protein family in cotton. Genome 61(7):539–547
Sheerin DJ, Menon C, Zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27(1):189–201
Smith TF, Gaitatzes C, Saxena K, Neer EJ (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 24(5):181–185
Stacey MG, Kopp OR, Kim TH, von Arnim AG (2000) Modular domain structure of Arabidopsis COP1. Reconstitution of activity by fragment complementation and mutational analysis of a nuclear localization signal in planta. Plant Physiol 124(3):979–989
Stirnimann CU, Petsalaki E, Russell RB, Müller CW (2010) WD40 proteins propel cellular networks. Trends Biochem Sci 35(10):565–574
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–613
Torii KU, McNellis TW, Deng XW (1998) Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J 17(19):5577–5587
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
van Nocker S, Ludwig P (2003) The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4(1):50
Wang CQ, Sarmast MK, Jiang J, Dehesh K (2015) The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell 27(4):1128–1139
Wu JF, Wang Y, Wu SH (2008) Two new clock proteins, LWD1 and LWD2, regulate arabidopsis photoperiodic flowering. Plant Physiol 148(2):948–959
Wu Y, Li X, Xiang W, Zhu C, Lin Z, Wu Y, Li J, Pandravada S, Ridder DD, Bai G, Wang ML, Tick HN, Bean SR, Tuinsra MR, Tesso TT, Yu J (2012) Presence of tannins in sorghum grains is conditioned by different natural alleles of Tannin1. Proc Natl Acad Sci USA 109(26):10281–10286
Wasserman WW, Sandelin A (2004) Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 5(4):276–287
Xie L, Lang-Mladek C, Richter J, Nigam N, Hauser MT (2015) UV-B induction of E3 ligase ARIADNE12 depends on CONSTITUTIVELY PHOTOMORPHOGENIC 1. Plant Physiol Biochem 93:18–28
Xu C, Min J (2011) Structure and function of WD40 domain proteins. Protein Cell 2(3):202–214
Xu X, Wan W, Jiang G, Xi Y, Huang H, Cai J, Chang Y, Duan CG, Mangrauthia SK, Peng X, Zhu JK, Zhu G (2019) Nucleocytoplasmic trafficking of the Arabidopsis WD40 repeat protein XIW1 regulates ABI5 stability and abscisic acid responses. Mol Plant 12(12):1598–1611
Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103(5):815–827
Yi C, Deng XW (2005) COP1—from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15(11):618–625
Yi C, Wang H, Wei N, Deng XW (2002) An initial biochemical and cell biological characterization of the mammalian homologue of a central plant developmental switch, COP1. BMC Cell Biol 3:30
Zhao Q, Zhu Z, Kasahara M, Morishita S, Zhang Z (2013) Segmental duplications in the silkworm genome. BMC Genomics 14:521
Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW (2008) Biochemical Characterization of Arabidopsis Complexes Containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA Proteins in Light Control of Plant Development. Plant Cell 20(9):2307–2323
Zhu Y, Huang S, Miao M, Tang X, Yue J, Wang W, Liu Y (2015) Genome-wide identification, sequence characterization, and protein-protein interaction properties of DDB1 (damaged DNA binding protein-1) binding WD40-repeat family members in Solanum lycopersicum. Planta 241(6):1337–1350
Zou XD, Hu XJ, Ma J, Li T, Ye ZQ, Wu YD (2016) Genome-wide analysis of WD40 protein family in human. Sci Rep 6:39262
Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue Light-Dependent Interaction of CRY2 with SPA1 Regulates COP1 activity and Floral Initiation in Arabidopsis. Curr Biol 21(10):841–847
Acknowledgements
We appreciate Dr. Shubin Li, Dr. Yanxia Jia, Ms. Shulan Chen, Mr. Hongyuan Yu and Yuanlin Lv for the help in cultivating rose plants and microscopy experiments. This work is supported by: the Strategic Priority Research Program of the Chinese Academy of Sciences to J-Y H (XDB31000000); the CAS Pioneer Hundred Talents Program to J-Y H (292015312D11035); Yunnan Applied Basic Research Projects to X D (2016FB040); CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, and Yunnan Recruitment Program of Experts in Science to J-Y H. This work is partially facilitated by the Germplasm Bank of Wild Species of China and Kunming Botanical Garden, Kunming Institute of Botany, Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
J-Y H and Y-B S conceptualized and designed the research; Y-B S performed most experiments and analyzed the data together with the help from X-J Z; X-J Z, M-C Z, X D, D-M Y, DW, X-D J, W-H C and J-H C generated part of the data; J-Y H and Y-B S wrote the manuscript with the help from X-J Z and all other authors; all authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests:
The authors declare no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, YB., Zhang, XJ., Zhong, MC. et al. Genome-wide identification of WD40 genes reveals a functional diversification of COP1-like genes in Rosaceae. Plant Mol Biol 104, 81–95 (2020). https://doi.org/10.1007/s11103-020-01026-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01026-7