Abstract
Most environmental perturbations have a direct or indirect deleterious impact on photosynthesis, and, in consequence, the overall energy status of the cell. Despite our increased understanding of convergent energy and stress signals, the connections between photosynthesis, energy and stress signals through putative common nodes are still unclear. Here we identified an endoplasmic reticulum (ER)-localized adenine nucleotide transporter1 (ER-ANT1), whose deficiency causes seedling lethality in air but viable under high CO2, exhibiting the typical photorespiratory phenotype. Metabolic analysis suggested that depletion of ER-ANT1 resulted in circadian rhythm disorders in sucrose synthesis and induced sucrose signaling pathways, indicating that the ER is involved in the regulation of vital energy metabolism in plants. In addition, the defect of ER-ANT1 triggers ER stress and activates the unfolded protein response in plant cells, suggesting ER stress and photorespiration are closely linked. These findings provide an important evidence for a key role of ER-localized ER-ANT1 in convergent energy and stress signals in rice. Our findings support the idea that ATP is a central signal involved in the plant response to a variety of stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The endoplasmic reticulum (ER) is a highly dynamic organelle that acts as a protein-processing factory in all eukaryotic cells (Staehelin 1997). Appropriate folding of proteins in the lumen of the ER is necessary for their corresponding functions. Under conditions of nutrient depletion, cell differentiation or other perturbations, misfolded or unfolded proteins accumulate in the ER, a situation known as ER stress. ER stress activates the unfolded protein response (UPR) to restore protein-folding homeostasis to the organelle (Wu and Kaufman 2006). The UPR was first characterized in yeast, in which Ire1 (inositol-requiring enzyme 1) is the only ER stress sensor (Kozutsumi et al. 1988). In animals, the UPR has evolved three integral ER membrane proteins that constitute the cell’s ER stress surveillance system: activating transcription factor-6 (ATF6), protein kinase R-(PKR-) like ER protein kinase (PERK) and IRE1 (Eichmann and Schäfer 2012). In plants, two UPR signaling pathways, namely IRE1 and ATF6, have been found (Lu et al. 2012). The UPR allows for transcriptional induction of ER chaperone genes to increase the protein-folding capacity, reducing the load of newly translated proteins entering the ER and preventing further protein aggregation, and increase the degradation of misfolded proteins by the ER-associated degradation (ERAD) machinery and autophagic degradation, thus protecting ER function. However, severe and/or prolonged ER stress markedly impairs ER function, threatening cellular integrity, and can lead to apoptosis if homeostasis is not restored (Oyadomari et al. 2002).
ER protein-folding capacity is dependent on the abundance of chaperones, which demand a sufficient ATP supply for their activity (Braakman et al. 1992; Ostrovsky et al. 2009). In addition, activation of ATF6 can be regulated by specific ATP-dependent dissociation of BiP (IgG binding protein) (Walker and Atkin 2011). Furthermore, adenine nucleotides are able to activate IRE1 through induction of a conformational change (Papa et al. 2003; Sidrauski and Walter 1997). Based on these precedents, the initiation and maintenance of the UPR are likely to be influenced by cellular energy levels. These findings raise the question of whether depletion of ATP in the ER lumen results in ER stress and activation of the UPR.
In higher plants, the photorespiratory cycle is a highly compartmentalized process with enzymatic reactions in chloroplasts, peroxisomes and mitochondria. The metabolic pathway is well characterized and the enzymes required have been identified (Maurino and Peterhansel 2010). Recently, several transporters, such as DiT1, DiT2, PLGG1 (in chloroplasts) and BOU (in mitochondria), which catalyze the high fluxes of photorespiratory intermediates between the involved compartments have been identified (Eisenhut et al. 2013; Pick et al. 2013; Renné et al. 2003). Interestingly, a study of a maize mutant lacking glycolate oxidase showed that this cycle is vital even in this model C4 plant (Zelitch et al. 2009). Moreover, recent research has shown that photorespiratory metabolism is not confined to chloroplasts, peroxisomes and mitochondria, but also extends to the cytosol (Timm et al. 2008). More recently, a study of an Arabidopsis mutant lacking ER-ANT1 shows that ER adenine nucleotide transport is related to photorespiration (Hoffmann et al. 2013). The photorespiratory phenotype of er-ant1 mutants was caused by reduced GDC activity, which resulted from increased oxidative stress in er-ant1 plants (Hoffmann et al. 2013). The observation suggests that there is a clear metabolic connection between defects of AtER-ANT1 in the ER and ROS production. However, what connects ER-ANT1 to ROS formation is elusive.
A recent study showed that a plasma membrane-localized receptor for extracellular ATP plays a variety of roles in plant stress resistance (Choi et al. 2014), implicating ATP as an energy signal that converges onto plant stress responses. However, little is known about the role of ATP as a mediator and/or integrator in linking photosynthesis and ER function for the global regulation of stress and energy signals. In this paper, we characterized a seedling lethal rice mutant er-ant1with impaired photoautotrophic growth. Our study provides an important evidence for a key role of ER-localized ER-ANT1 in convergent energy and stress signals in rice.
Materials and methods
Plant materials and growth conditions
All rice (Oryza sativa) lines used in this study were derived from the japonica cv Zhonghua11. The er-ant1 mutant described in this paper was derived from an F4 generation plant of a cross between Ds-T-DNA homozygote (d6) and Ac-T-DNA homozygote (B5) transgenic lines. The plants were grown under natural light conditions or in a growth chamber with a 13/11 h light/darkness photoperiod at a photon flux density of 50–150 µmol m−2 s−1 and constant temperature of 28 °C, after incubating seeds in darkness for 48 h at 26 °C to ensure synchronized germination. To investigate the effect of sucrose on seedling growth, the WT and er-ant1 seeds were grown on 1/2 MS agar with or without sucrose (2 %, w/v) and coconut endosperm (10 %, v/v) for 14 days. To investigate the effect of CO2 on gene expression involved in sugar metabolism and ER stress signaling, plants were grown under 1.5 % CO2 and a light/dark cycle of 13/11 h for 1 week and subsequently shifted to ambient air with a 13 h photoperiod of 400 µmol m−2 s−1 illumination. Samples were collected 12 or 24 h of light after plants were shifted to ambient air.
Chlorophyll fluorescence analysis
Chlorophyll fluorescence images were obtained at room temperature using an IMAGING-PAM Chlorophyll Fluorometer with pulse-modulated excitation (IMAG-MAX/GS, Heinz Walz GmbH, Germany) according to the instructions provided by the manufacturer. Values of Fo (for minimum fluorescence yield) and Fm (for maximum fluorescence yield) were averaged to improve the signal-to-noise ratio. Image data acquired from each experiment were normalized to a false-color scale. The highest and lowest Fv/Fm values were represented by the purple and red extremes of the color scale, respectively.
Transmission electron microscopy
Samples of the WT and mutant leaves were prepared for transmission electron microscopy by cutting them into small pieces, fixing in 2.5 % glutaraldehyde in phosphate buffer for 4 h at 4 °C, rinsing and incubating them in 1 % OsO4 overnight at 4 °C, rinsing again in phosphate buffer, dehydrating in an ethanol series, infiltrating with a graded series of epoxy resin in epoxy propane, and then embedding them in Epon 812 resin. Thin sections were obtained using a diamond knife and a Reichert OM2 ultramicrotome, stained in 2 % uranyl acetate, pH 5.0, followed by 10 mM lead citrate, pH 12, then viewed with a transmission electron microscope (JEM-1230; JEOL).
Isolation of the Ds flanking sequences and the revertant
Genomic sequences flanking the Ds insertions were isolated by TAIL-PCR (Liu and Whittier 1995). Specific tertiary PCR fragments were directly sequenced. The primers used for TAIL-PCR are shown in Supplementary Table 2. The presence of the Ds insertion was verified by diagnostic PCR in each mutant using genomic DNA and primers specific for the Ds left border (Ds5′-1a) and gene-specific primers (OsAAC-DsL and OsAAC-DsR) that flank the Ds insertion. The er-ant1 mutation heterozygous plants with the Ac transposase gene were grown to isolate revertants. The presence of Ac in the genome was determined by PCR with primers P6 and P7. The revertant was determined by PCR with primers Dsyb, Dsyc and Ds3′-1a (Supplementary Table 2).
RT-PCR and quantitative RT-PCR
Total RNA was extracted from frozen samples with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The RNA was pre-treated with DNase I, and first-strand cDNA was generated using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). Quantitative RT-PCRs were performed using a SYBR Premix Ex Taq™ RT-PCR kit (Takara) following the manufacturer’s instructions. Investigated genes and their specific primer sets are shown in Supplementary Table 2. The rice 25 S rRNA gene was used as an internal control for all analysis. Three replicates were performed for each analysis, and average values and standard deviations are shown.
Immunoblot analysis
Total proteins from plant extracts were extracted as described previously (Zheng et al. 2013). Specific anti-ER-ANT1 peptide antibodies were raised in rabbits and affinity purified against the peptides SAAAGDGKGKRRLV (ER-ANT1, amino acids 3–16; Hangzhou Huaan Biotechnology). ER-ANT1 was detected with 1:1000 anti-ER-ANT1 polyclonal antibodies, respectively, and HEAT SHOCK PROTEIN82 (HSP82) was detected with a mouse anti-HSP82 monoclonal antibody (1:1000, AbM51099-31-PU; Beijing Protein Innovation). Proteins were visualized by incubating specimens with goat anti-rabbit secondary antibodies conjugated to alkaline phosphatase (1:5000, CW0111 or CW0102; Beijing CoWin Biotechology) in the presence of 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium as substrates.
Dithiothreitol (DTT) treatment
For ER-stress treatments, 15-day-old WT seedlings were incubated in liquid MS medium containing 2 mM DTT for 3 h. Equal volumes of H2O were used as negative controls. Total RNA samples were extracted from WT and mutant rice plants at 12 DAG (two-leaf stage) and 15 DAG (three-leaf stage).
GUS staining
For promoter analysis, approximately 673 bp of the OsER-ANT1 5′ region were amplified with primers AACP-1301F and AACP-1301R (Supplementary Table 2) and inserted into the HindIII and NcoI sites of the pCAMBIA1305.1 vector. The resulting plasmid was transformed into rice and GUS staining was performed as described previously (Jefferson 1989). Various tissues or hand-cut sections of transgenic Pro er−ant1 :GUS plants were incubated in a solution containing 50 mM NaPO4 buffer (pH 7.0), 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 0.1 % Triton X-100 and 1 mM X-Gluc at 37 °C.
Subcellular localization
To determine localization of the ER-ANT1 protein in plant cells, a 990-bp coding sequence of ER-ANT1 was amplified with the primers RTL-AAC-15F and RTL-AAC-15R (Supplementary Table 2) and the PCR product was inserted into the BglII/XbaI sites of the pRTL2-mGFP vector and fused downstream of GFP, resulting in the GFP-ER-ANT1 fusion protein-expressing plasmid pRTL2-mGFP-ER-ANT1, then transformed into Arabidopsis protoplasts with ER CD3-960-mCherry marker (Nelson et al. 2007) for co-localization studies, as reported previously (Yoo et al. 2007).
HPLC analysis of soluble carbohydrates from leaf tissue
The second and/or third leaves from WT and er-ant1-mutant plants were collected on days 10, 13 and 16 after germination at 7:00 AM and 7:00 PM. The samples, frozen in liquid nitrogen, were homogenized and then extracted in boiling water for 10 min and subsequently incubated at 70 °C for 20 min. After centrifugation (10 min, 4000 rpm), the supernatant was removed and the pellet was re-extracted twice at 70 °C for 30 min. The glucose, fructose and sucrose contents were measured from a 2-ml sample using a Dionex DX-500 (HPAE-PAD) liquid chromatograph equipped with a 4 × 250-mm Dionex 35,391 CarboPac PA1 analytical column and a 4 × 50-mm Dionex 43,096 CarboPac PA1 guard column. The column was eluted with 100 mM NaOH at a flow rate of 1 mL/min.
Accession numbers
Sequence data from this article can be found in the Rice Annotation Project Database (RAP-DB) databases under the following accession numbers: BiP (Os02g0115900, Os05g0428600, Os06g0212900); bZIP39 (Os05g0411300); bZIP50 (Os06g0622700); bZIP60 (Os07g0644100); CNX (Os04g0402100); CRT (Os07g0246200); DAD1 (Os04g0397000); FBPase (Os05g0438600); Fd-GOGAT (Os07g0658400); OsIRE1 (Os07g0471000); ER-ANT1 (Os11g0661300); GRP94 (Os06g0716700); CIPK15 (Os11g0113700); PDI (Os09g0451500); Ribophorin I (Os05g0301500); Ribophorin II (Os01g0911200); SEC61 (Os09g0347700); SnRK1 (Os02g0178000); SnRK2 (Os04g0629300); SPS1 (Os01g0919400); STT3 (Os04g0675500); SUS1 (Os03g0401300); UGPase (Os09g0553200).
Results
er-ant1 is a seedling-lethal mutant with impaired photoautotrophic growth
A rice mutant with pale-green leaves was obtained from a two-element Activator/Dissociation (Ac/Ds) transposon-tagging population in the japonica rice variety Zhonghua 11 (Liu et al. 2007) and was named ER-ANT1 after the gene was cloned from the mutant (see below). Under paddy field conditions, er-ant1 plants were almost indistinguishable from wild type (WT) plants until the three-leaf stage (approximately 15 days after germination, DAG) when the er-ant1 mutant displayed a clearly pale-green phenotype and retarded growth (Fig. 1a). Thereafter, the resulting chlorotic phenotype was exacerbated and these plants withered and died at approximately 25 DAG (Fig. 1a), indicating that the mutant phenotype is expressed in a growth stage-dependent manner. The three-leaf stage in rice plants is a transition phase from heterotrophic to autotrophic growth, in which the grain reserves are assumed to have been dissimilated by the seedling. Therefore, the characteristic of the seedling lethal phenotype after the three-leaf stage suggests that the er-ant1 mutant has impaired autotrophic growth. Moreover, we found that the retarded growth phenotype was relieved in normal air at low irradiance (~50 µmol m−2 s−1), allowing survival for more than 50 days with five leaves (Fig. 1b). Thus, the er-ant1 mutant phenotype was sensitive to light modulation. Genetic analysis demonstrated that the mutant phenotype was controlled by a single recessive gene (Supplementary Table 1).
Phenotypes of the er-ant1 mutant. Phenotypic characterizations of the er-ant1 mutant grown under natural field light conditions for 12, 15 or 25 days after germination (DAG) (a) or weak light conditions (~50 µmol m−2 s−1) for 12, 23, 36 or 52 DAG (b). Bars 2 cm. WT, wild type. c Comparison of chloroplast ultrastructure in mesophyll cells between the WT and er-ant1 mutant. The seedlings were grown in a growth chamber (25 °C, ~50 µmol m−2 s−1) for 15, 30 or 36 DAG. All transmission electron microscopy samples were obtained from the second leaf blade of both WT and er-ant1 mutant seedlings. Bars 1 µm. d Chlorophyll fluorescence images of WT and er-ant1 mutant seedlings grown in a growth chamber (25 °C, ~50 µmol m−2 s−1) for 12, 30 or 36 DAG. Fluorescence was visualized using a pseudocolor index as indicated on the left
To investigate the effects of the ER-ANT1 on chloroplast development, we examined the ultrastructure of chloroplasts in wild type and er-ant1mutant plants at high and low light intensities. Under paddy field conditions (high light intensities), the er-ant1 plants were close to the plasma membrane and regularly arranged, and similar to the control at 12 DAG while the outer chloroplast membrane in er-ant1 mutant became partly obscure at 15 DAG, accompanied with gradual disintegration of basal granules, and then the chloroplast became loose, deformed and having no thylakoids (Supplementary Fig. 1). Under low light (growth chamber, ~50 µmol m−2 s−1), the chloroplasts in er-ant1 plants also displayed well-developed lamellar structures equipped with normally stacked grana and thylakoid membranes at the two-leaf stage (approximately 15 DAG), which is comparable to those of wild type plants (Fig. 1c). However, the thylakoid membrane organization in the chloroplasts of er-ant1 mutants at the three-leaf stage (approximately 30 DAG) was disturbed, and their thylakoid membranes were much less abundant at 36 DAG (Fig. 1c). In contrast, the thylakoid membrane organization and abundance in wild type plants had well-structured thylakoid membranes, composed of grana connected by stroma lamellae. In addition, we investigated the effects of ER-ANT1 on PSII (photosystem II) functionality (Fig. 1d) under low light. The er-ant1 mutant exhibited PSII activity similar to the wild type plants grown in the growth chamber at the two-leaf stage (15 DAG), whereas PSII activity of er-ant1 leaves were substantially lower than those of the wild type counterparts at 30 DAG, consistent with the chloroplast development of the er-ant1 mutant. These results indicate that the er-ant1 mutant phenotype is not an indirect effect of chloroplast development and PSII functionality in rice.
Knockout of OsER-ANT1 obstructs rice photorespiration
To determine the Ds insertion site in the er-ant1 mutant, we isolated genomic flanking sequences of the Ds insertion by thermal asymmetric interlaced (TAIL)-PCR (Liu and Whittier 1995). Analysis of both Ds flanking sequences revealed that the Ds element is inserted into the first intron of the ER-ANT1 (LOC_Os11g43960) gene on rice chromosome 11, resulting in a duplication of an 8-bp target site as expected for a Ds insertion (Fig. 2a, b). Co-segregation analysis of the Ds insertion with the pale-green phenotype of the er-ant1 mutant showed that the Ds insertion was responsible for the mutant phenotype (Supplementary Table 1). Co-segregation of the Ds and the mutant phenotype was further confirmed by a revertant with normal growth similar to the WT plant (Fig. 2c). In the revertant, a 6-bp duplication was kept as a Ds excision footprint (Fig. 2b). Expression analysis showed that ER-ANT1 was knocked out in the er-ant1 mutant and recovered in the revertant (Fig. 2d, e). Taken together, the above analyses suggest that the Ds insertion is responsible for the er-ant1 phenotypes.
Identification of the er-ant1 mutant gene and the revertant of the er-ant1 mutant. a Mapping of the er-ant1 mutant gene. The Ds insertion was mapped to rice genomic BAC clone OSJNBa0010P23 on chromosome 11. The Ds was inserted in the 652 bp upstream region of the second exon of the ER-ANT1 gene. The untranslated regions are shown in gray rectangles, the exons in black rectangles and the introns in thick black lines. Ds 5′ and Ds 3′, the 5′ and 3′ end of the Ds transposon, respectively; Bar, the BASTA resistant gene. b Identification of Ds excision footprint in the revertant of the er-ant1 mutant. The 8 base-pairs (bp) sequence, GTTGTGGT (in red), is the target site for Ds duplication. The 6-bp footprint, TGTGGT, in the revertant is marked in green. c Phenotype of the revertant of the er-ant1 mutant. Seedlings of the WT, er-ant1 mutant and revertant were gown in field light conditions for 28 days. Bar 4 cm. RT-PCR analysis of ER-ANT1 transcript levels (d) and immunoblot analysis of endogenic ER-ANT1 (e) in the WT, er-ant1 mutant and revertant. RT-PCR of 25 S rRNA is shown below as an internal control (d). Anti-HSP82 is shown at the bottom to indicate approximately equal loading (e). Seedlings were grown as in (c)
Comparison of deduced amino acid sequences suggested that ER-ANT1 exhibited a high degree of sequence identity with mitochondrial ADP/ATP carriers (AACs), including Arabidopsis AtER-ANT1 (Supplementary Fig. 2). AtER-ANT1 plays an important role in photorespiratory metabolism (Hoffmann et al. 2013). These observations suggest that rice ER-ANT1 may also be involved in photorespiration. To confirm this, we compared the growth of the er-ant1 mutant and wild-type plants under both ambient air and high-CO2 conditions (1.5 %). As shown in Fig. 3a, er-ant1 is sensitive to ambient air as demonstrated by chlorotic leaves and impaired growth. These effects are almost fully compensated when the mutant is grown at 1.5 % CO2 (Fig. 3b). However, 2 weeks after transfer of the plants to ambient air (~420 ppm), er-ant1 mutant plants were dead (date no shown). This demonstrates that er-ant1 is a classic photorespiratory mutant and ER-ANT1 is an irreplaceable member of the photorespiratory pathway in rice.
Photorespiratory phenotype of the er-ant1 mutant. Seedlings of the wild type (WT) and the er-ant1 mutant were grown in ambient air (0.04 % CO2) for 18 days and then kept under the same conditions (a), or transferred to high-CO2 conditions (1.5 % CO2) (b) for a further 7 days. Day/night 13/11 h; Temperature, 28 °C; Light intensity, 100 µmol m−2 s−1; Bars 10 cm
Expression pattern and subcellular localization of ER-ANT1
To gain a better understanding of ER-ANT1 function, we examined its spatial and temporal expression patterns using a GUS reporter system. Analysis of transgenic plants harboring the PRO ER−ANT1:GUS construct showed that ER-ANT1 was expressed constitutively across tissue types, suggesting that it performs essential housekeeping functions, but with preferential expression in actively dividing zones (Fig. 4a). In the root, the ER-ANT1 promoter was active in root tips, while the signal became weaker as roots grew longer and was barely detectable in old roots (Fig. 4a). We also noticed that the GUS signal was strong in the rachis, branches and the dividing zones of florets. Further quantitative RT-PCR (qRT-PCR) analysis indicated that ER-ANT1 was highly expressed in leaves; low-level expression was detected in roots (Fig. 4b).
Expression pattern of ER-ANT1 and subcellular localization of ER-ANT1. a ER-ANT1 expression analysis using ER-ANT1 promoter–GUS lines. GUS staining is found in various organs, including the germinating seed, root, leaf, sheath, flower, developing seed and panicle. b Quantitative RT-PCR (qRT-PCR) analysis of ER-ANT1 expression in various organs, including root, shoot and leaf blade of young seedlings at the two-leaf stage; root, leaf blade, leaf sheath and stem in the rooting stage and young panicle of 8 cm. Each column shows the mean relative expression of three repeats of ER-ANT1. Error bars indicate SD. c Co-localization of ER-ANT1 proteins with endoplasmic reticulum markers in Arabidopsis protoplast cells. Protoplasts were transformed with a control, cytoplasmatic expressed, GFP construct or with a ER-ANT1:GFP fusion construct. ER-ANT1-mGFP, green fluorescence from ER-ANT1–GFP fusion protein; ER-mCherry, ER CD3-960-mCherry marker; Bright field, bright-field image under transmitted light; Merged, Merged image of ER-ANT1-mGFP, ER-mCherry and bright field. Bar 25 μm
No signal peptide was identified in ER-ANT1 by either the TargetP or the SignalP prediction servers (Emanuelsson et al. 2000). However, ER-ANT1 exhibits a high degree of sequence identity with AtER-ANT1 (Supplementary Fig. 2), an ER-localized ATP/ADP transporter from Arabidopsis (Leroch et al. 2008), suggesting that they might share the same subcellular localization. To ascertain its subcellular location, we fused GFP to the N-terminus of ER-ANT1 and co-expressed this fusion protein with red-fluorescent-protein (RFP) -tagged ER marker (Nelson et al. 2007) in Arabidopsis protoplast cells. The ER-ANT1 fusion was identical to the RFP signals of the ER marker (Fig. 4c), suggesting an ER-specific localization for the ER-ANT1 transporter.
Absence of ER-ANT1 causes a deficiency in sucrose synthesis
Because the er-ant1 mutant exhibits a seedling-lethal phenotype after the three-leaf stage (Fig. 1a), we wondered if the ER-ANT1 mutation has a deleterious effect on carbohydrate metabolites. Metabolic analysis revealed that the concentrations of glucose and fructose in the er-ant1 mutant were lower than in WT, but its diurnal change (low at AM to high at PM) is consistent with the WT (Fig. 5a). Leaves from mutant plants had much lower concentrations of sucrose (<4.5 mg g−1 FW) than the WT. In contrast to the diurnal change of sucrose concentration in the WT, the diurnal change of sucrose metabolism in the er-ant1 mutant was almost nonexistent, suggesting that the pattern of sucrose metabolism is altered in the absence of ER-ANT1. These results indicate that ER-ANT1 deficiency has a greater effect on sucrose than on glucose or fructose.
Effects of ER-ANT1 mutation on carbohydrate metabolism. a Assays for carbohydrate metabolism of the wild type (WT) and er-ant1 mutant. The tested leaf tissues were collected from seedlings 10, 13 or 16 days after germination (DAG) at both 7:00 AM and 7:00 PM. Mean and SD of three biological replicates are shown. FW, fresh weight. b Restoration of external sucrose supplies in the er-ant1 mutant. Seedlings were grown on MS medium with no sugar or 2 % sucrose for 14 days or with 10 % coconut endosperm for 30 days. Bars 4 cm. c The effect of ER-ANT1 mutation on the expression of sucrose metabolism-related genes. Total RNAs were extracted from the wild-type (WT) and er-ant1 mutant seedlings at the two-leaf stage (12 days after germination, DAG) and at the three-leaf stage (15 DAG), respectively
Given the distinctively reduced concentration of sucrose in the er-ant1 mutant, we hypothesized that a supplemental supply of sucrose should improve seedling growth and complement, at least partially, the mutant phenotype. As expected, in the presence of sucrose, the growth of the er-ant1 mutant was significantly improved, though still reduced compared with the WT (Fig. 5b). This suggested that sucrose was partially able to rescue the er-ant1 phenotype, but did not restore it to the WT. Addition of coconut endosperm, which supplements nutrition, also only partially complements seedling growth, indicating that the lack of photosynthetic products is not the only reason for the mutant phenotype (Fig. 5b).
Expression analysis showed that the transcript levels of sucrose biosynthesis-related genes, such as UDP-glucose pyrophosphorylase (UGPase, Os09g0553200), fructose-1, 6-bisphosphatase (FBPase, Os05g0438600) and sucrose phosphate synthase1 (SPS1, Os01g0919400), were significantly altered in er-ant1 seedlings (Fig. 5c). However, enzymatic analysis showed that the functional deficiency of ER-ANT1 has no direct effect on cytosolic FBPase (cyFBPase) and SPS, two key enzymes for sucrose biosynthesis (Supplementary Fig. 3).
Sucrose non-fermenting-1 (SNF1)-related protein kinases (SnRKs) are global regulators of carbon metabolism in plants and can be subdivided into three sub-families: SnRK1, SnRK2 and SnRK3 also known as CIPK, Calcineurin B-like Interacting Protein Kinase (Coello et al. 2011; Luan 2009). OsCIPK15 (Os11g0113700) is a key regulator of sugar metabolism in rice, induced under sucrose deprivation (Kudahettige et al. 2011; Lee et al. 2009). As expected, the transcript level of OsCIPK15 was over 20-fold higher than the control at the two-leaf stage and remained significantly elevated at the three-leaf stage (Fig. 5c). A similar trend was observed for SnRK1 (Os02g0178000) and SnRK2 (Os04g0629300) transcripts in er-ant1 seedlings.
Taken together, our findings suggest that the ER-ANT1 mutation caused a deficiency in sucrose and, thus, induced sucrose signaling pathways in the er-ant1 mutant.
Defect of ER-ANT1 triggers unfolding protein response
To determine the effect of ER-ANT1 on cellular energy, we measured ATP contents of leaves in the er-ant1 and wild-type plants grown under natural full sunlight at 9, 12 and 15 DAG. The results showed that ATP concentration in er-ant1 leaves was significantly lower compared to wild-type plants (Supplementary Fig. 4), suggesting that ER-ANT1 is involved in the modulation of cellular energy metabolism in rice. Because many metabolic reactions in the ER including appropriate folding of protein require a sufficient ATP supply, we expected that inactivation of ER-ANT1 would result in ER stress and activation of unfolding protein response (UPR). To confirm the link between the absence of ER-ANT1 and ER-stress responses, we measured the expressions of several ER stress markers by qRT-PCR in WT and mutant. As shown in Fig. 6a, the ER stress-related genes BiP (IgG binding protein, Os02g0115900, Os05g0428600, Os06g0212900), GRP94 (glucose-regulated protein 94, Os06g0716700) and PDI (protein disulfide isomerase,Os09g0451500) were up-regulated at 12 DAG in the er-ant1 mutant. It was reported that defects in protein N-glycosylation are closely associated with the UPR (Liu and Howell 2010). Higher levels of induction for protein glycosylation-related genes in the er-ant1 mutant, such as SEC61 (Os09g0347700), calnexin (CNX, Os04g0402100), calreticulin (CRT, Os07g0246200), STT3 (Os04g0675500), Ribophorin I (Os05g0301500) and Ribophorin II (Os01g0911200), were also observed. These results indicate that the deficiency in ER-ANT1 resulted in a bona fide ER stress response in rice. In contrast to the induction at 12 DAG, the abundances of the transcripts for the genes mentioned above were reduced by 15 DAG, except for defender against death 1 (DAD1, Os04g0397000). This reduction was especially remarked for GRP94, SEC61 and STT3 (Fig. 6a), suggesting the occurrence of severe and/or prolonged ER stress. Taken together, these results demonstrate that inactivation of ER-ANT1 is sufficient to trigger UPR activation.
Expression levels of ER stress-related genes in the wild type and er-ant1 mutant. Total RNAs were extracted from the wild-type (WT) and er-ant1 mutant seedlings at the two-leaf stage (12 days after germination, DAG) and at the three-leaf stage (15 DAG), respectively. a Quantitative RT-PCR (qRT-PCR) analysis of transcript abundances of endoplasmic reticulum (ER) stress-related genes in both the WT and er-ant1 mutant. b qRT-PCR analysis of expression levels of ER-stress-induced genes, OsbZIP39, OsbZIP60 and OsIRE1 in both the WT and er-ant1 mutant. c RT-PCR analysis of OsbZIP50 transcripts showing that the ER-ANT1 defect does not lead to splicing of OsbZIP50 mRNA in er-ant1 plants. Asterisk marks the splicing form of OsbZIP50 mRNA related to ER stress caused by 2 mM DDT treatment for 3 h. RT-PCR of 25 S rRNA is shown below as an internal control
The UPR in plants is reported to be mediated by two pathways: ATF6-like and IRE1 (Deng et al. 2013). Rice possesses an IRE1-like protein, OsIRE1 (Os07g0471000), which plays an essential role in the UPR by splicing OsbZIP50/OsbZIP74 (Os06g0622700) (Hayashi et al. 2012; Lu et al. 2012). In rice, OsbZIP39 (Os05g0411300) and OsbZIP60 (Os07g0644100) are the counterparts of ATF6 (Hayashi et al. 2013). To investigate the manner in which ER-ANT1 is involved in the UPR, the expression profiles of OsbZIP39, OsbZIP60 and OsIRE1 were analyzed in the er-ant1 mutant and WT (Fig. 6b). The expressions of OsbZIP39 and OsbZIP60 in the er-ant1 mutant were enhanced at the two- and three-leaf stages compared to WT. In contrast, expression levels of OsIRE1 were decreased at the two-leaf stage and up-regulated at the three-leaf stage. To assay IRE1 activity, IRE1-dependent splicing of its substrate, the OsbZIP50 mRNA, was determined by reverse transcription (RT)-PCR (Fig. 6c). As a positive control, the WT seedlings were treated with DTT (2 mM) for 3 h. As anticipated, when RT-PCR was conducted using RNA extracted from WT roots treated with DTT, a spliced product was clearly detected. However, the spliced product was not detected even after 35 cycles of PCR in the er-ant1 mutant. These results indicate that a deficiency in ER-ANT1 does not lead to appreciable activation of OsIRE1, as measured by splicing of OsbZIP50 mRNA. This result is in agreement with previous reports of IRE1 activation in response to glucose deprivation in pancreatic β-cells (Gomez et al. 2008).
Effects of CO2 on gene expression
To investigate the effect of CO2 on gene expression involved in sugar metabolism and ER stress signaling. We conducted the gene expression analysis when plants are transferred from high CO2 to normal air (so as to see the effect of having impaired photorespiration). Under high CO2 condition, the er-ant1 mutant showed mRNA levels similar to wild-type plants (Fig. 7). Under ambient air, however, the transcript abundances of several interest genes in er-ant1 mutant, such as GRP94 and CIPK15, were significant higher compared to wild-type plant (Fig. 7). The results showed that sugar metabolism and ER stress-related genes were up-regulated in the er-ant1mutant under photorespiratory conditions, further confirming the relationship between the absence of ER-ANT1 and ER-stress responses and sugar metabolism.
Effects of CO2 on expression of genes involved in sugar metabolism and ER stress response. 13-day old seedlings were grown under 1.5 % CO2 and a light/dark cycle of 13/11 h for 1 week and subsequently shifted to ambient air. Samples were collected 12 or 24 h of light after plants were shifted to ambient air
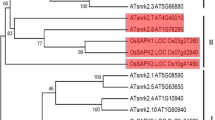
Phylogenetic analyses of mitochondrial ADP/ATP carrier family with RRRMM motif in plants
Database similarity searching identified 29 putative homologues of OsER-ANT1 from rice (O. sativa L.), Arabidopsis, poplar (Populus trichocarpa), grape (Vitis vinifera), maize (Zea mays), and sorghum (Sorghum bicolor). Phylogenetic analysis of mitochondrial ADP/ATP carrier (AAC) family in plants showed that the AAC family is divided into three major groups: mitochondrial ADP/ATP carrier (Mito-AAC), endoplasmic reticulum adenine nucleotide transporter (ER-ANT) and plasma membrane adenine nucleotide transporter (PM-ANT) (Fig. 8). OsER-ANT1 is a member of the ER-ANT subgroup along with AtER-ANT1 from Arabidopsis, an ER-localized ATP/ADP transporter (Leroch et al. 2008). Interestingly, members of the ER-ANT subgroup possess the conserved RRRMML motif, while members of the other subfamilies contain the RRRMMM motif (Supplementary Fig. 1). The phylogenetic analysis also suggested that most AAC genes from monocots (rice) and dicots (Arabidopsis) are not clustered in species-specific clades, indicating non-species-specific expansion in plants. In other words, these interspecies orthologs are more similar to each other than to their intraspecies paralogs.
Phylogenetic analyses of putative homologs of OsER-ANT1. Phylogenetic analysis of putative OsER-ANT1 homologs using MEGA with neighbor-joining method; bootstrap analysis was performed with 1000 replicates and excluding positions with gaps. The numbers at the branches are confidence values (percent). The proteins organized in three main clusters, Mito-AAC, ER-ANT and PM-ANT. Homologous genes are with the following accession numbers: Arabidopsis thaliana (At3g08580, At4g28390, At5g13490, At5g17400, At5g56450); Oryza sativa L (Os02g48720, Os05g23720, Os05g46220, Os11g43960); Sorghum bicolor (Sb04g008410, Sb04g029750, Sb01g048660, Sb10g012150, Sb09g026900); Vitis vinifera (GSVIVG00000009001, GSVIVG00018612001, GSVIVG00024630001, GSVIVG00032194001); Populus trichocarpa (POPTR_0007 s01010, POPTR_0009 s06690, POPTR_0001 s274801, POPTR_0003 s20360, POPTR_0012 s05990, POPTR_0006 s24360); and Zea mays (ACF88121, ACR35796, ACG34816, GRMZM2G111017, ACF88062). Carriers characterized by direct transport measurements are marked with a black triangle
There are at least two members in Mito-AAC subfamily, while there is single copy of the other two subfamilies. Interestingly, the members from the Mito-AAC subfamily are putatively located in mitochondria of plant cells, based on the mitochondrial localization of AtAAC1 (At3g08580) (Millar and Heazlewood 2003), while ones from the ER-ANT subfamily are putatively located in endoplasmic reticulum according to the ER-localization of AtER-ANT1 (Leroch et al. 2008) and OsER-ANT1 (Fig. 4c). In addition, members from the PM-ANT subfamily are putatively plasma membrane-localized based on the fact of subcellular localization of AtPM-ANT1 (Rieder and Neuhaus 2011). These indicate that the members from different subfamilies probably have different subcellular localization and biological functions, but those within the same subfamily may be functionally similar.
Discussion
Members of ER-ANT subfamily are functionally conserved in photorespiratory metabolism
The ATP/ADP carrier (AAC) system was discovered four decades ago (Bruni et al. 1964) and has been investigated extensively (Fiore et al. 1998). Typical AAC proteins reside in the mitochondria and catalyze a strictly counter exchange of cytosolic ADP with matrix ATP. Interestingly, AAC-type adenine nucleotide transporters in plant cells are not restricted to mitochondria, but are also present in other membranes, such as the plasma membrane (Rieder and Neuhaus 2011). The phylogenetic tree of AAC proteins shows that AAC-type transporters can be divided into three subfamilies, Mito-AAC, ER-ANT and PM-ANT (Fig. 8). As for the ER-ANT subfamily, amino acid sequences are highly homologous to AAC proteins. However, ER-ANTs lack the typical N-terminal sequence extension of mitochondrial AACs in plants (Supplementary Fig. 2). Similar to AtER-ANT1 (Leroch et al. 2008), our results showed that OsER-ANT1 is localized to the ER (Fig. 4c). In addition, OsER-ANT1, as well as AtER-ANT1, is demonstrated to be able to transport ATP and ADP (Hoffmann et al. 2013).
Photorespiration encompasses four sub-cellular compartments: chloroplast, mitochondrion, peroxisome and cytoplasm (Hodges et al. 2013). More recently, a study of an Arabidopsis mutant lacking AtER-ANT showed that the ER is involved in photorespiration in this model dicotyledonous plant (Hoffmann et al. 2013). In this study, we provide further evidence that the ER is associated with photorespiration in grasses. These findings suggest that ER-localized plant ATP/ADP carriers are functionally conserved in photorespiratory metabolism.
Link between ER and sucrose metabolism
The analysis of sugar metabolites indicated that mutation of OsER-ANT1 largely affected sugar metabolism, especially sucrose (Fig. 5a). However, the transcription of sucrose synthesis-related genes was induced in er-ant1 seedlings (Fig. 5c), indicating feedback regulation of transcription by the low levels of sucrose. In addition, enzymatic assays indicated that the ER-ANT1 deficiency had no direct effect on SPS and cyFBPase activity (Supplementary Fig. 3). Thus, the decrease in sucrose likely resulted from reduced levels of sucrose synthesis substrates, such as fructose 6-phosphate, probably caused by the disturbance of photorespiration, thereby disrupting photosynthesis. The protein kinase CIPK15 plays a critical role in the response to sucrose deficiency and acts through the SnRK1A-mediated sugar-signaling pathway to regulate carbohydrate catabolism in rice (Lee et al. 2009). Interestingly, an increase in the abundance of CIPK15 and SnRK1A mRNAs was observed in the er-ant1 mutant along with the sucrose deficiency (Fig. 5a, c). The results indicate that the deficiency of ER-ANT1 resulted in sugar starvation and triggered the sucrose-signaling pathway.
Accumulating evidence has suggested that the strength of the photorespiratory phenotype could be modulated by alterations of several factors including organic carbon supply (Timm and Bauwe 2013). An external sucrose supply can alleviate the photorespiratory phenotype, but never completely recover the photorespiratory mutants (Timm and Bauwe 2013). The partial complementation of photorespiratory mutants by sucrose is most likely explained by the reduced carbon recycling resulting from disruption of the photorespiratory cycle. Similarly, the growth of the er-ant1 mutant was observably recovered in the presence of external sucrose in normal air (Fig. 5b), indicating that ER-ANT1 deficiency resulted in photorespiratory disturbance and may subsequently trigger sucrose starvation. Remarkably, some of the photorespiratory mutants exhibit high sugar concentrations in normal air. One example comes from mitochondrial serine hydroxymethyltransferase gene (SHM1)-knockout plants. The leaf concentration of glucose in shm1 was considerably higher compared to that in WT plants (Eisenhut et al. 2013). Another example is the recently isolated knockout mutant defective in the mitochondrial BOUT DE SOUFFLE (BOU) gene. Similar to the shm1 mutant, the bou mutant exhibited elevated levels of glucose and sucrose (Eisenhut et al. 2013). Unlike the shm1 and bou mutants, the er-ant1 mutant shows markedly reduced levels of sugars, including glucose, fructose and sucrose, indicating the complex modulation of carbohydrate metabolism in plants by photorespiration.
Recent findings have significantly advanced our understanding of the interconnection between ER function and metabolism, including lipogenesis, gluconeogenesis, lipoprotein production and lipid catabolism (Tyra et al. 2012). Our findings suggest that the ER plays an important role in sucrose metabolism and signaling pathways in plants, likely via the photorespiration pathway.
Convergent energy and stress signaling
In eukaryotes, protein N-glycosylation is catalyzed by the heteromeric, multisubunit oligosaccharyltransferase (OST) complex in the lumen of the ER (Yan and Lennarz 2005). It has been reported that defects in protein N-glycosylation are closely associated with the UPR (Liu and Howell 2010). In this study, analysis of gene expression at the two-leaf stage showed that homologs to these OST subunits in rice, such as Ribophorin I, Ribophorin II and STT3, were induced in the er-ant1 mutant (Fig. 6a). In addition, representative ER stress-responsive genes were upregulated. Taken together, these data demonstrate that a Ds insertion in ER-ANT1 resulted in induction of the UPR due to putative ER energy deficiency.
The widespread down-regulation of genes in response to prolonged and/or severe ER stress, which is referred to as regulated IRE1-dependent decay (RIDD) of specific mRNAs, was observed in yeast and animals (Hollien and Weissman 2006). Recent studies suggested that the down-regulation of genes via the RIDD pathway also occurs in plants and is dependent on IRE1 RNase activity, but not on OsbZIP50 (Wakasa et al. 2012). In this study, in contrast to the up-regulation of genes in response to ER stress at the two-leaf stage, genes were down-regulated at the three-leaf stage (Fig. 6a), suggesting the occurrence of prolonged ER stress. OsIRE1 was induced transcriptionally, but splicing of OsbZIP50 was not apparent at the three-leaf stage (Fig. 6c). The observations suggest that RIDD may occur in the er-ant1 mutant, contributing to reduced levels of ER chaperone genes. It is possible that functional loss of ER-ANT1 initially elicits ER stress in rice, despite the fact that the mRNA levels of ER stress genes were lower in the er-ant1 mutant compared to WT plants at the three-leaf stage. Taken together, our data suggest that ER stress and photorespiration are closely linked.
As sessile organisms, plants must have evolved sensitive mechanisms to rapidly perceive environmental cues and modify growth and developmental patterns for a successful adaptive response. Most environmental perturbations have direct or indirect deleterious impacts on photosynthesis and/or respiration and, in consequence, the overall energy status of the cell. Two hours of hypoxic stress, for example, are enough to cause a nearly 50 % decrease in the levels of cellular ATP (Branco-Price et al. 2008). Thus, it is postulated that various types of stress are decoded, at least partly, as energy-deficiency signals triggering convergent responses independent of the original cause. Despite our increased understanding of convergent energy and stress signals (Baena-González 2010; Baena-González and Sheen 2008), the connections between these pathways through putative common nodes are still unclear. Increasing evidence has suggested that oxidative stress and reactive oxygen species (ROS) generation are integral components of ER stress and not merely consequences of ER stress induction (Bhandary et al. 2012). Our findings, together with the generation of ROS in the absence of AtER-ANT1, an ortholog of rice ER-ANT1 (Hoffmann et al. 2013), indicate the possibility that an ER-localized ATP/ADP transporter may be a common node for converging energy and stress signals in plants.
References
Baena-González E (2010) Energy signaling in the regulation of gene expression during stress. Mol Plant 3:300–313
Baena-González E, Sheen J (2008) Convergent energy and stress signaling. Trends Plant Sci 13:474–482
Bhandary B, Marahatta A, Kim H, Chae H (2012) An involvement of oxidative stress in endoplasmic reticulum stress and its associated diseases. Int J Mol Sci 14:434–456
Braakman I, Helenius J, Helenius A (1992) Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature 356:260–262
Branco-Price C, Kaiser KA, Jang CJH, Larive CK, Bailey-Serres J (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56:743–755
Bruni A, Luciani S, Contessa A (1964) Inhibition by atractyloside of the binding of adenine-nucleotides to rat-liver mitochondria. Nature 201:1219–1220
Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G (2014) Identification of a plant receptor for extracellular ATP. Science 343:290–294
Coello P, Hey SJ, Halford NG (2011) The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J Exp Bot 62:883–893
Deng Y, Srivastava R, Howell S (2013) Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int J Mol Sci 14:8188–8212
Eichmann R, Schäfer P (2012) The endoplasmic reticulum in plant immunity and cell death. Front Plant Sci 3:200
Eisenhut M, Planchais S, Cabassa C, Guivarc’h A, Justin A, Taconnat L, Renou J, Linka M, Gagneul D, Timm S et al (2013) Arabidopsis A BOUT DE SOUFFLE is a putative mitochondrial transporter involved in photorespiratory metabolism and is required for meristem growth at ambient CO2 levels. Plant J 73:836–849
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Fiore C, Trézéguet V, Le Saux A, Roux P, Schwimmer C, Dianoux AC, Noel F, Lauquin GJ, Brandolin G, Vignais PV (1998) The mitochondrial ADP/ATP carrier: structural, physiological and pathological aspects. Biochimie 80:137–150
Gomez E, Powell ML, Bevington A, Herbert TP (2008) A decrease in cellular energy status stimulates PERK-dependent eIF2α phosphorylation and regulates protein synthesis in pancreatic β-cells. Biochem J 410:485–493
Hayashi S, Wakasa Y, Takahashi H, Kawakatsu T, Takaiwa F (2012) Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J 69:946–956
Hayashi S, Wakasa Y, Takaiwa F (2013) Recent advances in understanding the control of secretory proteins by the unfolded protein response in plants. Int J Mol Sci 14:9396–9407
Hodges M, Jossier M, Boex-Fontvieille E, Tcherkez G (2013) Protein phosphorylation and photorespiration. Plant Biol 15:694–706
Hoffmann C, Plocharski B, Haferkamp I, Leroch M, Ewald R, Bauwe H, Riemer J, Herrmann JM, Neuhaus HE (2013) From endoplasmic reticulum to mitochondria: absence of the Arabidopsis ATP antiporter ER-ANT1 perturbs photorespiration. Plant Cell 25:2647–2660
Hollien J, Weissman JS (2006) Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313:104–107
Jefferson RA (1989) The GUS reporter gene system. Nature 342:837–838
Kozutsumi Y, Segal M, Normington K, Gething M, Sambrook J (1988) The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332:462–464
Kudahettige NP, Pucciariello C, Parlanti S, Alpi A, Perata P (2011) Regulatory interplay of the Sub1A and CIPK15 pathways in the regulation of α-amylase production in flooded rice plants. Plant Biol 13:611–619
Lee K, Chen P, Lu C, Chen S, Ho TD, Yu S (2009) Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal 2:ra61
Leroch M, Neuhaus HE, Kirchberger S, Zimmermann S, Melzer M, Gerhold J, Tjaden J (2008) Identification of a novel adenine nucleotide transporter in the endoplasmic reticulum of Arabidopsis. Plant Cell 20:438–451
Liu J, Howell SH (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22:2930–2942
Liu Y, Whittier R (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681
Liu F, Zhang X, Zhang Z, Chen Z, Zhu H, Wang J, Zhang J, Zhang G (2007) Transpositional behaviour of the Ds element in the Ac/Ds system in rice. Chin Sci Bull 52:2789–2796
Lu S, Yang Z, Sun L, Sun L, Song Z, Liu J (2012) Conservation of IRE1-regulated bZIP74 mRNA unconventional splicing in rice (Oryza sativa L.) involved in ER stress responses. Mol Plant 5:504–514
Luan S (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14:37–42
Maurino VG, Peterhansel C (2010) Photorespiration: current status and approaches for metabolic engineering. Curr Opin Plant Biol 13:248–255
Millar AH, Heazlewood JL (2003) Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol 131:443–453
Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51:1126–1136
Ostrovsky O, Makarewich CA, Snapp EL, Argon Y (2009) An essential role for ATP binding and hydrolysis in the chaperone activity of GRP94 in cells. Proc Natl Acad Sci USA 106:11600–11605
Oyadomari S, Araki E, Mori M (2002) Endoplasmic reticulum stress-mediated apoptosis in pancreatic beta-cells. Apoptosis 7:335–345
Papa F, Zhang C, Shokat K (2003) Bypassing a kinase activity with an ATP-competitive drug. Science 302:1533–1537
Pick TR, Bräutigam A, Schulz MA, Obata T, Fernie AR, Weber APM (2013) PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc Natl Acad Sci USA 110:3185–3190
Renné P, Dreßen U, Hebbeker U, Hille D, Flügge U, Westhoff P, Weber APM (2003) The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J 35:316–331
Rieder B, Neuhaus HE (2011) Identification of an Arabidopsis plasma membrane–located ATP transporter important for anther development. Plant Cell 23:1932–1944
Sidrauski C, Walter P (1997) The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90:1031–1039
Staehelin LA (1997) The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J 11:1151–1165
Timm S, Bauwe H (2013) The variety of photorespiratory phenotypes—employing the current status for future research directions on photorespiration. Plant Biol 15:737–747
Timm S, Nunes-Nesi A, Pärnik T, Morgenthal K, Wienkoop S, Keerberg O, Weckwerth W, Kleczkowski LA, Fernie AR, Bauwe H (2008) A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell 20:2848–2859
Tyra HM, Spitz DR, Rutkowski DT (2012) Inhibition of fatty acid oxidation enhances oxidative protein folding and protects hepatocytes from endoplasmic reticulum stress. Mol Biol Cell 23:811–819
Wakasa Y, Hayashi S, Ozawa K, Takaiwa F (2012) Multiple roles of the ER stress sensor IRE1 demonstrated by gene targeting in rice. Sci Rep. doi:10.1038/srep00944
Walker AK, Atkin JD (2011) Stress signaling from the endoplasmic reticulum: a central player in the pathogenesis of amyotrophic lateral sclerosis. Iubmb Life 63:754–763
Wu J, Kaufman RJ (2006) From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ 13:374–384
Yan A, Lennarz WJ (2005) Unraveling the mechanism of protein N-glycosylation. J Biol Chem 280:3121–3124
Yoo S, Cho Y, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Zelitch I, Schultes NP, Peterson RB, Brown P, Brutnell TP (2009) High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol 149:195–204
Zheng X, Wu S, Zhai H, Zhou P, Song M, Su L, Xi Y, Li Z, Cai Y, Meng F et al (2013) Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25:115–133
Acknowledgments
We thank Dr. David Pincus (Whitehead Institute at MIT) for careful reading and useful comments and suggestions on the manuscript. We also thank Dr. Yulong Ren (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences) for the editing, suggestions and assistance to this work. This research was supported by grants from the National Natural Science Foundation of China (Nos. 91435207, U1031002, 31671594 and 30900884), and from the Genetically Modified Organisms Breeding Major Projects of China (No. 2014ZX08009-001).
Author contributions
The author(s) have made the following declarations about their contributions: Conceived and designed the experiments: GZ JY. Performed the experiments: XQZ XZ SK HZ FL. Analyzed the data: XP HP SW MS. Contributed reagents/materials/analysis tools: ZZ RZ ZL LG. Wrote the paper: GZ JY XQZ HP. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Xiangqian Zhang and Xu Zheng have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, X., Zheng, X., Ke, S. et al. ER-localized adenine nucleotide transporter ER-ANT1: an integrator of energy and stress signaling in rice. Plant Mol Biol 92, 701–715 (2016). https://doi.org/10.1007/s11103-016-0540-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0540-1