Abstract
Emerging evidence suggests that the gaseous molecules hydrogen sulfide (H2S) and nitric oxide (NO) enhances plant acclimation to stress; however, the underlying mechanism remains unclear. In this work, we explored if pretreatment of citrus roots with NaHS (a H2S donor) or sodium nitroprusside (SNP, a NO donor) for 2 days (d) could elicit long-lasting priming effects to subsequent exposure to PEG-associated drought stress for 21 d following a 5 d acclimation period. Detailed physiological study documented that both pretreatments primed plants against drought stress. Analysis of the level of nitrite, NOx, S-nitrosoglutahione reductase, Tyr-nitration and S-nitrosylation along with the expression of genes involved in NO-generation suggested that the nitrosative status of leaves and roots was altered by NaHS and SNP. Using a proteomic approach we characterized S-nitrosylated proteins in citrus leaves exposed to chemical treatments, including well known and novel S-nitrosylated targets. Mass spectrometry analysis also enabled the identification of 42 differentially expressed proteins in PEG alone-treated plants. Several PEG-responsive proteins were down-regulated, especially photosynthetic proteins. Finally, the identification of specific proteins that were regulated by NaHS and SNP under PEG conditions provides novel insight into long-term drought priming in plants and in a fruit crop such as citrus in particular.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drought stress is among the most significant environmental factors that negatively affect plant survival, growth, distribution and productivity (Golldack et al. 2014). There are numerous cellular changes during drought acclimation, ranging from accumulation of osmolytes (Mahajan and Tuteja 2005) to disruption of reactive oxygen species (ROS) and nitrogen species (RNS) homeostasis (Valderrama et al. 2007; Miller et al. 2010). Accumulating evidence also revealed the role of gaseous signal transmitters in orchestrating plant responses to stress. In this context, nitric oxide (NO), a redox-active small molecule, is gradually established as a prominent gaseous signal molecule (Baudouin and Hancock 2014). It has been suggested that abscisic acid (ABA) accumulates in Arabidopsis leaves in response to drought, leading to NO synthesis from nitrate reductase (NR) and nitric oxide associated 1 (AtNOA1); NO was then proposed to activate mitogen-activated protein kinase (MAPK) which drives stomatal closure (Zhang et al. 2007). Evidence also indicates that NO under drought operates partly through post-translational modifications (PTMs) of proteins, notably via S-nitrosylation [addition of a NO group to the thiol side chain of the Cysteine (Cys) amino acid] and tyrosine nitration [addition of a nitro group to the ortho position of the aromatic ring of the Tyrosine (Tyr) residue] (Ziogas et al. 2013). These protein redox modifications alter the function of a broad spectrum of proteins and therefore the analysis of NO-targeted proteome serves as a tool to investigate the mechanism through which NO impacts their functions (Tanou et al. 2010, 2012b).
The important life-supporting role of hydrogen sulfide (H2S) has evolved from bacteria to plants, invertebrates, vertebrates, and finally to mammals. Over the centuries H2S had only been known for its toxicity; however, the physiological significance of H2S has been appreciated for about a decade (Wang 2012). In this sense, H2S has emerged as the third ‘gas signal molecule’ after NO and carbon monoxide (CO) in animals (Tan et al. 2010). Studies in various metazoan systems have established signaling roles for H2S in thermotolerance, atherosclerosis, inflammation, neural diseases, and diabetes (Kabil et al. 2014). H2S has also been a topic of interest in the study of various plant systems, such as stomatal movement under drought (García-Mata and Lamattina 2010). Regarding the enzymatic origin of H2S in plants, two cysteine desulfhydrases (CDes), namely l-cysteine desulfhydrase (LCD, E.C. 4.4.1.1) and d-cysteine desulfhydrase (DCD, EC 4.4.1.15), appear to be involved (Riemenschneider et al. 2005b). However, DES1 (At5g28030), another enzyme having L-Cys desulfhydrase acivity, was identified in Arabidopsis that produces H2S by L-Cys degradation (Álvarez et al. 2010).
Despite long knowing that plants synthesize and release H2S (Wilson et al. 1978), only over the past 10 years has H2S been shown to modulate stress responses in plants. Similar to its roles in metazoans, the overall action of H2S is to relieve stress, (Qiao et al. 2015), including drought (Jin et al. 2011; Christou et al. 2013, 2014; Shen et al. 2013); however the underlying mechanisms remain unknown (Pandey 2014). Limited evidence has demonstrated that H2S and NO have some overlapping physiological functions in plants (García-Mata and Lamattina 2013). Scuffi et al. (2014) showed that the lack of endogenous NO in cysteine desulfhydrase (DES1) knockout Arabidopsis mutant plants (des1) significantly reduces the effects of H2S on stomatal aperture and that NO acts downstream of H2S to close stomata via an ABA-dependent pathway. An interaction between H2S and NO has been noted before, albeit not in plants. In animal systems, it has been shown that H2S inhibits isoforms of nitric oxide synthase (NOS; Kubo et al. 2007) whereas H2S and NO can react together to form new nitrosothiols (Whiteman et al. 2006). Like NO, H2S can also play regulatory roles and modulate protein activity by binding to some protein haemes. Besides, H2S can promote the formation of persulfide groups, a process known as S-sulfhydration, since H2S could perform a nucleophilic attack on oxidized protein cysteine residues (Finkel 2012).
The use of priming techniques (e.g., external application of natural or synthetic compounds in plants to facilitate plant acclimation to environmental stresses) has received much attention in recent years (Tanou et al. 2012b). Recent publications have experimentally addressed that both NO and H2S could trigger priming responses in plants exposed to drought. For example, transgenic Arabidopsis plants that ectopically expressed the rat nNOS gene enhanced the expression of stress-responsive genes (such as AtPYL4 and AtPYL5), resulting in drought tolerance (Shi et al. 2014). Meanwhile, exogenous NaHS treatment, which is a H2S donor, altered the transcriptional reformation of drought associated miRNAs, such as miR167, miR393, miR396 and miR398 and can therefore affect their target gene expressions and thus improve drought tolerance in Arabidopsis (Shen et al. 2013). Moreover, understanding the complex NO- and H2S-priming effects in plants requires a detailed analysis of both physiological and molecular changes. In this study using citrus plants, one of the most important tree crops and also drought sensitive species (García-Sánchez et al. 2007), we found that short-term exposure to sodium nitroprusside (SNP, a NO donor) or to NaHS, elicited long-term drought acclimation in citrus seedlings. Comparative proteomic analysis coupled with NO-targeted proteomic approach identified several drought stress-related protein and revealed related pathways involved in drought acclimation. This study increases our understanding of the physiological roles of NO and H2S in drought acclimation.
Materials and methods
Plant growth and experimental design
Five-month-old sour orange (Citrus aurantium L.) plants were grown in aerated half-strength Hoagland nutrient solution in culture room conditions as described by Tanou et al. (2009). The experimental plants were divided into three groups. The roots of the first, second and third group of plants were incubated for 2 d into distilled water, 100 μΜ SNP or 100 μΜ NaHS, respectively, with the solutions being renewed every 12 h. Afterwards, the three groups of plants were transferred to half-strength Hoagland nutrient solution for 5 d until the initiation of the drought experiment. Finally, these three types of pre-treated plants were exposed to nutrient solutions in the absence or presence of 15 % (w/v) PEG-6000 (polyethylene glycol) (−0.30 MPa) for 21 d to simulate drought stress conditions. Thus, plants were submitted to six treatments as follows: Control (pre-treated with distilled water without drought stress), NaHS (pre-treated with NaHS without drought stress), SNP (pre-treated with SNP without drought stress), PEG (pre-treated with distilled water and then subjected to drought stress), NaHS → PEG (pre-treated with NaHS and then subjected to drought stress) and SNP → PEG (pre-treated with SNP and then subjected to drought stress). Each treatment was independently run in triplicate, and each replicate consisted of ten individual plants. The experimental set up was principally based on our previous studies (Tanou et al. 2009). A diagram of the experimental design is provided in Supplementary Fig. S1. Tissues were also sampled immediately following NaHS or SNP pre-treatments (2 d of the experiment) and after 21 d of PEG stress exposure (day 28 of the experiment). All samples were frozen immediately in liquid nitrogen and stored at −80 °C prior to analysis, except those which were used immediately for the physiological measurements [electrolyte leakage and malondialdehyde (MDA) content], the quantification of nitrite, NOx, and hydrogen sulfide and the detection of endogenous NO by confocal laser scanning microscopy. Chlorophyll content index (CCI) and photosynthetic rate (P N) were measured in fully expanded leaves of intact plants.
Physiological measurements
The CCI was determined using a CCM-200 chlorophyll meter (Opti-Sciences, Tyngsboro, MA, USA) upon fully expanded leaves and expressed as CCI units. Net photosynthetic rate (P N) was measured under full sunlight with a portable open-path infrared gas analyzer LCpro (ADC Bioscientific Ltd., Hoddesdon, UK). Leaf membrane damage was evaluated by measuring the electrolyte leakage (Rizhsky et al. 2002) and lipid peroxidation, was estimated as MDA content (Heath and Packer 1968).
Treatments with NO scavenger and inhibitor of H2S biosynthesis
The 2-(4-carboxy-phenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide potassium salt (cPTIO) was commonly used as NO scavenger in plants while hypotaurine (HT) as H2S scavenger (Zhou et al. 2005; Shi et al. 2013; Qiao et al. 2015). To study further the effect of NO scavenger and H2S inhibitor on SNP- or NaHS-induced physiological changes of citrus, twelve additional plants per treatment were treated with the combination of 100 μΜ SNP and 200 μΜ cPTIO (SNP + cPTIO treatment) or with 100 μΜ NaHS plus 200 μΜ HT (NaHS + HT treatment) for 2 d and then were transferred to the nutrient solution in the absence or in the presence of 15 % (w/v) PEG-6000 for 21 d following 5 d acclimation period as described above. Physiological parameters were assayed as above-mentioned.
Nitrite, NOx, and hydrogen sulfide quantification
Nitrite (NO2 −) content was measured according to Zhou et al. (2005) based on the Griess reaction. Nitrite plus nitrate (NOx) levels were quantified via electrochemical detection using an ArrowSTRAIGHT™ System (Los Angeles, CA, USA) in conjunction with an ISO-NO Mark II and Duo.18 data acquisition system (WPI, USA). The results are represented as relative NOx (NO2 − + NO3 −) levels, by attributing the value 1 to the control treatment. Hydrogen sulfide content was estimated as described by Nashef et al. (1977).
Confocal laser scanning microscopy (CLSM) detection of endogenous NO
NO imaging was performed by incubating cross-sections of leaves primary veins and roots tips with 4,5-diaminofluorescin diacetate (DAF-2DA) (Tanou et al. 2012a). As negative controls, control tissues were incubated for 30 min with 300 μM c-PTIO prior staining (data not shown). Fluorescent signals were visualized by a Nikon D-Eclipse C1 CLSM and quantified by using the IMAGEJ software.
RNA extraction, cDNA synthesis and real-time RT-PCR analysis
Total RNA was extracted, from both leaves and roots of citrus plants, by using the Trizol reagent (Invitrogen, USA) and following the manufacturer’s instructions, followed by DNAse I treatment (Invitrogen, USA). The quality and integrity of the RNA was estimated spectrophotometrically, followed by gel electrophoresis. Prior to real-time RT-PCR analysis, cDNA was synthesized by 1 μg aliquot of total RNA by using the Primescript 1st strand cDNA synthesis kit according to the manufacturer’s guidelines (Takara, Japan). The transcript levels were analyzed with an iQ5 real-time PCR cycler (BioRad, Hercules, CA, USA) with the usage of SYBR Fast Master mix according to the manufacturer’s protocol (KapaBiosystems, USA) and 1:5 dilution of the cDNAs synthesized as template. All the reactions were performed in triplicate and the thermocycler conditions were: incubation at 94 °C for 2 min, followed by 33 cycles at 94 °C for 30 s, annealing temperature 56 °C for 45 s, 72 °C for 1 min, plate read at 80 °C for 2 s, followed a final extension step of 10 min at 72 °C. The housekeeping gene, elongation factor 1a (EF-1a), was used as an internal control (Mahe et al. 1992). The quantification of the normalized gene expression ratio and the statistical analysis of all real-time RT-PCR data with pair-wise fixed reallocation randomized tests were estimated using the REST software (Pfaffl et al. 2002). The gene-specific primers used are listed in Supplementary Table S1. Primers were designed based on EST collections in the TIGR Plant Transcript Assemblies Database (http://plantta.jcvi.org/index.shtml). In regard with S-nitrosoglutahione reductase (GSNOR), it should be noted that primers were designed based on initial sequences from known GSNOR homologs from plants (pea and Prunus); the Citrus EST clone chosen (CX287192), is highly homologous to GSNORs and alcohol/formaldehyde dehydrogenases, that belong to the same class with GSNOR (Martínez et al. 1996).
Determination of S-nitrosoglutahione reductase (GSNOR) and l-Cysteine desulfhydrase activity (LCD)
The GSNOR activity was assessed as described by Corpas et al. (2008) by recording the oxidation of NADH at 340 nm. Determination of the l-Cysteine desulfhydrase activity was performed according to Riemenschneider and co-workers (Riemenschneider et al. 2005a). Total proteins were estimated according to Bradford (1976).
Assessment of protein carbonylation and nitration by western blotting
Protein extraction and 1DE-SDS-PAGE was performed in samples from leaves and roots according to Tanou et al. (2010). Prior to immunoblot analysis, equal loading of protein samples was verified by staining with Coomassie Brilliant Blue R-250 (BioRad, USA). Afterwards, proteins were electroblotted onto nitrocellulose membranes (0.2 μm) using Mini Trans Blot Cell (BioRad) following manufacturer’s protocol. Detection of carbonylated proteins was achieved by treating the membranes with 0.5 mM 2,4-dinitrophenylhydrazine (Sigma-Aldrich) and then probed by using a primary antibody against 2,4 dinitrophenylhydrazine (DNPH) (AbDSerotec) (1:3000). Immunodetection of tyrosine-nitrated proteins was assessed by incubating the membranes with a primary antibody against 3-nitrotyrosine (NO2-Tyr) (Affiland) (2 μg/mL). Nitrated bovine serum albumin was used as a positive antibody control (BSA, Sigma, St. Louis, MO, USA). Immunoreactive bands were visualized by enhanced chemiluminescence (Novex®, ECLChemiluminescent Substrate Reagent Kit). Blots were scanned with a Bio-Rad GS-800 Calibrated Densitometer and analyzed using QUANTITY ONE software version 4.7 (BioRad) (Tanou et al. 2012b). The results are presented as intergraded density values per lane by attributing the value one to the control.
Purification of S-nitrosylated proteins by biotin–switch assay
Leaf tissue was homogenized in HEN buffer (25 mM HEPES, pH 7.7, 1 mM EDTA, 0.1 M Neocuproine) (Sigma) containing a Complete™ Mini protease inhibitor cocktail tablet (Roche Molecular Biochemicals) in the dark (Tanou et al. 2009). The specificity of the biotin-switch assay was tested by incubating proteins with the S-nitrosylating agent S-nitrosoglutathione (GSNO; positive control) or the reducing agent dithiothreitol (DTT; negative controls) (data not shown). Detection of S-nitrosylated proteins were performed after their biotinylation and purification of biotin-coupled proteins by affinity chromatography on a neutravidin agarose matrix and then separated by SDS-PAGE (Tanou et al. 2009). Densitometric quantification of nitrosylated bands was conducted as above. Protein S-nitrosylated bands were excised from the gel, digested with trypsin and identified by peptide mass fingerprinting according to Tanou et al. (2014).
Two-dimensional electrophoresis and image analysis
The total proteins were extracted and protein profile was analyzed with 2DE-PAGE as described (Tanou et al. 2009). Following silver nitrate staining, 2DE-gels were analyzed with PDQuest Advanced 2D Gel Analysis Software. In brief, spots were detected, background subtracted, matched and quantitative determination of the spot volumes (normalization mode: total quantity in valid spots) was performed. Statistical analysis was contacted by one-way analysis of variance significance (P < 0.05) and individual means were compared using Student’s t test (significance level 95 %). Significant differences were reported only when the statistical significant differences were further combined by the quantitative twofold change in spot volume.
Mass spectrometry analysis and database searching
Protein zones of interest were cut from the gel and underwent trypsin digestion as follows: destained with destaining solution (15 mM potassium ferricyanide (K3Fe(CN)6), 50 mM sodium thiosulphate (Na2S2O35H2O) 1:1 v/v). After washed with HPLC-grade water and lyophilized, the gel pieces were rehydrated in 25 mM NH4HCO3 with 10 ng sequencing grade-modified trypsin (Promega, Madison, WI, USA). After digestion, the protein peptides were collected, and the gels were washed with 0.1 % formic acid in 50 % ACN followed by 0,1 % formic acid in 100 % ACN to collect the remaining peptides. LC–MS/MS was conducted as reported previously (Tanou et al. 2014). The identified proteins were annotated based on the National Center for Biotechnology Information (NCBI) databases using BLASTp analysis. All identified proteins had a MASCOT score greater than the significance level corresponding to P < 0.05. Moreover, among the positive matches, only protein identifications based on at least two different peptide sequences of more than six consecutive amino acids with an individual score above 20 were accepted. To validate protein identification with one single peptide (e.g., as in protein zone 8 at Supplementary Table 3 and spots 513, 8118, 8121 and 9509 at Supplementary Table 5) additional information, are provided; Supplementary Fig. S4). The threshold score was set above the identity threshold (value of 32) with at least a series of five consecutive y or b fragments in the spectrum. In some cases, the peptide masses and sequences obtained were blasted manually against the current databases.
Statistical analysis
Significance was evaluated by one-way ANOVA and the results were expressed as the mean values ±SE of three independent experiments. Significant differences are indicated with different letters according to Duncan’s multiple range test (P < 0.05).
Results
Phenotypic observation and physiological analysis
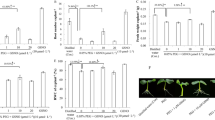
PEG treatment for 21 d resulted in visible foliar injury, manifested as leaf wilting and necrosis (Fig. 1a). In contrast, SNP- or NaHS-pre-treated plants exposed to PEG stress after 5 d of acclimation period exhibited obvious mitigating effect (Fig. 1a). Apparent negative effects of direct PEG stress on chlorophyll content (Fig. 1b) photosynthesis rate (Fig. 1c), ion leakage (Fig. 1d) and lipid peroxidation (Fig. 1e) were observed. These negative effects were alleviated by pre-treatment with SNP (SNP → PEG treatment) or NaHS (NaHS → PEG treatment) before drought stress imposition (Fig. 1b–e). Real time RT-PCR analysis also indicated that the expression of 9-cis epoxycarotenoid dioxygenase (NCED), which cleaves 9-cis xanthophylls to xanthoxin, a precursor of ABA biosynthesis, was remarkably up-regulated by PEG treatment (Fig. 1f), underlining the severe drought stress status of the PEG alone-treated plants. Furthermore, plants exposed to SNP or NaHS under physiological conditions (absence of PEG) just after pretreatment (Supplementary Fig. S2) as well as at the end of experimental period maintain their typical phenotype (Fig. 1a) and physiological (Fig. 1b–d) status. Meanwhile, physiological responses induced by the NO and H2S donors was blocked by cPTIO and HT, respectively, confirming the effectiveness of these chemicals as NO scavenger/H2S inhibitor and suggesting that H2S and NO are actively involved in the drought acclimation signaling pathway. In addition, cPTIO and HT treatment alone (in the absence of SNP or NaHS) had no significant physiological effect on plants under non-stress or PEG stress conditions (data not shown).
Pre-treatment with NaHS or SNP leads to priming response in citrus plants towards PEG-induced drought stress. Roots of plants were incubated into SNP (100 μM for 2 d) or NaHS (100 μM for 2 d) or distilled water (control). The three groups of plants were then transferred to nutrient solution for an additional 5 d acclimation period. Afterwards, all plants were maintained in Hoagland solution with or without PEG (15 % w/v) for 21 d. Control: plants pre-treated with water and not drought-stressed; NaHS: plants pre-treated with NaHS and not drought-stressed; SNP: plants pre-treated with SNP and not drought-stressed; PEG: plants pre-treated with water and drought-stressed; NaHS → PEG: plants pre-treated with NaHS and drought-stressed; SNP → PEG: plants pre-treated with SNP and drought-stressed. a Phenotype of citrus plants pre-treated with NaHS or SNP after 21 d of PEG stress imposition (day 28 of the experiment). b–e Physiological parameters of plants exposed to NaHS or SNP. Chlorophyll content index (b), photosynthetic rate (c), ion leakage (d) and (e) lipid peroxidation. To further characterize the physiological status of citrus, SNP- and NaHS-pretreated plants were also incubated into cPTIO (a NO scavenger; SNP + cPTIO treatment) or into HT (an inhibitor of H2S biosynthesis; NaHS + HT treatment) and then were subjected to PEG as described above (SNP + cPTIO → PEG and NaHS + HT → PEG treatments, respectively). Leaf samples were analyzed after 21 d of PEG treatment (day 28 of the experiment). Different letters above bars indicate significant differences among treatments (P < 0.05, Duncan’s test). Values are mean ± SE (n = 6). f Relative gene expression of 9-cis epoxycarotenoid dioxygenase (NCED) by real-time RT-PCR using three biological repeats. Asterisk indicates statistically different values according to the pairwise fixed reallocation randomization test (P < 0.05). EF-1a was used as the housekeeping control gene
Effects of SNP and NaHS on the nitrosative status of citrus plants
Since at least two distinct methods, based upon different chemical reactions, are recommended for the estimation of NO levels (Rümer et al. 2012), NO status was investigated in citrus plants using electrochemical study, Griess reaction and CLMS analysis. Contents of nitrite and NOx along with DAF-2DA fluorescence were found to increase in leaves and roots exposed to PEG treatment compared with control (Fig. 2a–h). Moreover, SNP → PEG or NaHS → PEG treatments minimized nitrite content, NOx accumulation and DAF-2DA fluorescence, compared with PEG treatment. Generally, plants pre-treated with SNP (SNP treatment) or NaHS (NaHS treatment) under physiological conditions presented nitrite, NOx and DAF-2DA fluorescence levels similar to control plants at the 28th day of the experiment (Fig. 2a–h).
Pre-treatment with SNP or NaHS regulates the nitrosative status of citrus plants under drought stress. Electrochemical measurements of NOx (nitrite plus nitrate) (a, e), nitrite content (b, f), representative images illustrating the CLSM detection of endogenous NO production using DAF-2DA fluorescence probe (c, g) and quantification of DAF-2DA fluorescence (expressed relative to control) in leaves (d) and roots (h) of citrus plants at the day 28 of the experiment. Scale bar 60 μm. Additional experimental details as described in Fig. 1. Different letters above bars indicate significant differences between treatments (P < 0.05; Duncan’s test). Values are mean ± SE (n = 3)
S-nitrosoglutathione reductase (GSNOR) catalyzes the NADH-dependent reduction of S-nitrosoglutathione to oxidized glutathione and NH3, which is considered as a reaction of denitrosylation (Chaki et al. 2011). In this study, GSNOR activity was higher in roots compared with leaves (Fig. 3a) whereas it was also increased in PEG alone-treated leaves compared with control (Fig. 3a). In leaves, SNP → PEG treatment remarkably enhanced GSNOR activity while NaHS → PEG treatment diminished its activity relative to PEG treatment (Fig. 3a). Also, leaf GSNOR activity was increased by SNP or NaHS treatment (Fig. 3a). In roots, both SNP → PEG and NaHS → PEG treatments decreased GSNOR activity compared to PEG treatment (Fig. 3a). Meanwhile, the current data confirmed previous observations (Wünsche et al. 2011) showing that there is no direct correlation between transcript levels of GSNOR and its activity (Fig. 3b).
Pre-treatment with SNP or NaHS modifies the enzymatic activity (a) and the relative gene expression (b) of GSNOR in leaves and roots of citrus plants. Additional experimental details as described in Fig. 1. For enzymatic activities, the different letters above bars indicate significant differences among treatments (P < 0.05; Duncan’s test). For gene expression analysis, asterisk indicates statistically different values according to the pairwise fixed reallocation randomization test (P < 0.05). EF-1a was used as the housekeeping control gene. Values are mean ± SE (n = 3)
Transcription pattern of putative NO-generation enzymes
The relative temporal mRNA expression ratio of a diverse set of specific genes involved in NO-generation (Tanou et al. 2012a) is presented in the heat diagram of Fig. 4. This gene expression analysis showed that levels of NOS-like and NADHox in leaves as well as NiR and NADHde in roots remained unchanged (Fig. 4). Significantly induced expression of NR and NADHde was observed in leaves exposed to PEG treatment. In roots NR expression was up-regulated whereas the expression of NOS-like, NADHox and AOX were down-regulated (Fig. 4). Furthermore, SNP → PEG treatment caused the down-regulation of NADHde and AOX expressions in leaves. Furthermore, NaHS → PEG resulted in the down-regulation of NOS-like expression in roots (Fig. 4). SNP treatment induced NR expression in leaves and depressed the expression of NOS-like and AOX in roots. Finally, NaHS treatment resulted in the down-regulation of NiR, NADHde and AOX transcripts in leaves as well as the expression of NOS-like, NADHox and AOX in roots (Fig. 4).
Heat map showing the expression profile of several genes associated with NO generation in leaves and roots of citrus plants. Additional experimental details as described in Fig. 1. Relative mRNA abundance was evaluated by quantitative real-time RT-PCR using three biological repeats. Up-regulation is marked with green while down-regulation is indicated in red. A scale of colour intensity is presented as a legend. EF-1a was used as the housekeeping control gene. Actual relative expression data are shown in Supplementary Table S2
Effects of SNP and NaHS on the H2S level of citrus plants
Under PEG treatment at the end of the experimental period, citrus plants exhibited elevated H2S content in leaves and roots in respect to control (Fig. 5a). Compared with PEG treatment, leaves exposed to either SNP → PEG or NaHS → PEG showed lower H2S content (Fig. 5a) whereas NaHS → PEG enhanced H2S level in roots (Fig. 5a). l-Cysteine desulfhydrase (LCD), that decomposes cysteine into H2S, NH3 and pyruvate, participate to the overall H2S level in plants (Bloem et al. 2004). In this study, LCD activity was increased in leaves and roots of citrus plants exposed to all PEG treatments (Fig. 5b) whereas SNP → PEG treatment showed lower level of LCD activity in roots compared to PEG treatment (Fig. 5b). SNP and NaHS treatments under non-stress conditions demonstrated minor alternation in H2S content (Fig. 5a) and LCD activity (Fig. 5b) in citrus tissues in respect to control.
Pre-treatment of citrus plants with NaHS or SNP change the levels of H2S (a, c) and l-cysteine desulfhydrase activity (LCD) (b, d) in leaves (a, b) and roots (c, d) of citrus plants. Additional experimental details as described in Fig. 1. Different letters above bars indicate significant differences among treatments (P < 0.05; Duncan’s test). Values are mean ± SE (n = 3)
Effects of SNP and NaHS on PTMs
Protein carbonylation and Tyr-nitration represent biological markers of oxidative and nitrosative stress in plants (Tanou et al. 2012a). To study whether citrus plants could be affected by these redox modifications, leaf and root protein samples were assayed by western blotting using well-characterized specific antibodies against 2,4-dinitrophenylhydrazine and 3-nitrotyrosine, respectively (Tanou et al. 2012a). A representative western blot detecting protein carbonyls (i.e., oxyblot) is shown in Fig. 6b. Quantitative analysis of the oxyblots showed that protein carbonyl levels were increased in leaves and roots exposed to PEG treatment (Fig. 6c). Furthermore, NaHS treatment suppressed protein carbonylation in leaves compared with control (Fig. 6b, c, g, h) whereas leaf carbonylation was also decreased by NaHS → PEG treatment compared with PEG (Fig. 6b, c). On the other hand, SNP suppressed nitration in roots (Fig. 6i, j) whereas SNP → PEG treatment exhibited extended nitration profile in both leaves and roots (Fig. 6b, e, i, j).
Profile of protein carbonylation and Tyr-nitration in leaves and roots of citrus plants under non-stress and PEG stress conditions. Additional experimental details as described in Fig. 1. Protein extracts (17 μg) from leaves (a) and roots (f) were staining with Coomassie Brilliant Blue R-250 following 1DE-PAGE. The position of molecular mass markers is indicated in the left. Carbonylation profile in leaves (b) and roots (g), followed by Western blotting using an anti-2,4-dinitrophenylhydrazine antibody (dilution 1:3000). Detection of 3-nitrotyrosine in leaves (d) and roots (i), followed by Western blotting using a monoclonal antibody against 3-nitrotyrosine (2 μg/ml). Positive control: commercial nitrated bovine serum albumin (NO2-BSA, 2 μg protein). Lane 1 Control, Lane 2 SNP, Lane 3 NaHS, Lane 4 PEG, Lane 5 SNP → PEG, Lane 6 NaHS → PEG. The red arrows on the right of the blots indicate the position of carbonylated and nitrated bands, while the number indicates the arithmetical sequence of the band. Each experiment was repeated three times and the relative intensity of the carbonylation (c leaves; h roots) and Tyr-nitration (e leaves; j roots) profile per total lane was estimated for each treatment and expressed as arbitrary units (AU)
To find S-nitrosylated proteins arising as a consequence of chemical treatments, we purified leaf citrus proteins with affinity chromatography to harvest the biotin-labelled peptides using the procedure previously described (Jaffrey and Snyder 2001). An increase in S-nitrosylation was observed in leaves exposed to NaHS and particularly to SNP treatments under physiological conditions (Fig. 7a, b). PEG treatment resulted in reduced S-nitrosylation. By contrast, both SNP → PEG and NaHS → PEG treatments largely increased S-nitrosylation (Fig. 7a, b). These results suggest that S-nitrosylation is a pivotal mediator for SNP- and NaHS-induced drought acclimation in citrus.
A Purification of S-nitrosylated proteins in leaves of citrus plants at 28 d of the experiment. M: Molecular mass marker, CBB: Soluble proteins from control leaves stained with Coomassie Brilliant Blue (CBB), Lane 1 Control, Lane 2 SNP, Lane 3 NaHS, Lane 4 PEG, Lane 5 SNP → PEG, Lane 6 NaHS → PEG. The selected S-nitrosylated proteins that submitted to mass spectrometry analysis are given with arrow. B Relative quantity of S-nitrosylation intensity of total lane per treatment expressed in arbitrary unit (AU). C Functional categories of S-nitrosylated proteins. The list of the of S-nitrosylated proteins is presented in Supplementary Table S3
To further investigate the S-nitrosylation in citrus leaves, 19 distinct protein bands, marked in Fig. 7a, were digested with trypsin and the resulting peptides were subjected to nano LC–MS/MS analysis. Following this approach, 25 proteins have been successfully identified as targets of S-nitrosylation. All the identification parameters (peptide sequences, accession number, matching criteria etc.) are given in Supplementary Table S3. Grouping these S-nitrosylated proteins according to their functional category established by Bevan et al. (1998), showed that the largest group was associated with photosynthesis (58 %) (Fig. 7c). The second largest group was related with transport ATPases (6 %), while the other groups, namely glycolysis, nucleotide metabolism, storage protein and detoxification where all equally represented by a portion of 3 % (Fig. 7c). More specifically, this proteomic analysis indicates that RubisCO large subunit was a potent target of S-nitrosylation (Supplementary Table S3) under non-stress conditions (control). The potent S-nitrosylation targets following SNP treatment were involved in photosynthesis, glycolysis, nucleotide metabolism, storage proteins and transport ATPases (Supplementary Table S3). Under NaHS treatment, all S-nitrosylated proteins identified were involved in photosynthesis. In response to PEG treatment, only one protein related to photosynthesis (chlorophyll a-b binding protein) was identified. The SNP → PEG treatment, pinpointed S-nitrosylated proteins involved in photosynthesis, detoxification and transport ATPases (Supplementary Table S3). Finally, the identified S-nitrosylated proteins in leaves exposed to NaHS → PEG treatment were mainly related to photosynthesis (Supplementary Table S3).
Global protein changes in citrus plants exposed to drought conditions
This study aimed to gain a better understanding on the physiological and molecular roles of SNP and NaHS in drought acclimation and therefore a 2-DE-PAGE coupled with mass spectrometry was used to obtain an overall view of the protein changes associated with PEG stress in leaves of citrus plants. The 2-DE maps are presented in Fig. 8 and the specific protein changes are given in Fig. 9 (for more details see also Supplementary Table S5). The Venn diagrams in Fig. 10 also summarize the functional classification and the number of proteins whose the accumulation of which increased or decreased in response to various PEG treatments. Proteomic analysis revealed a total of 42 proteins that showed differences in protein abundance (according to the criteria described in the “Materials and methods” section) between control and PEG-treated samples, among them 16 exhibited increased abundance in response to PEG treatment while 26 were less abundant (Fig. 10; Supplementary Table S5). The largest group (23.8 %) of PEG-responsive proteins, which consisted of ten proteins (two up-regulated and eight down-regulated) was related to photosynthesis and the Calvin–Benson cycle. The disease/defense/detoxification protein cluster was the second most abundant category (11.9 %) followed by the energy/glycolysis cluster (9.5 %) (Fig. 10b; Supplementary Table S5).
The effect of SNP or NaHS pre-treatments on the proteome of citrus leaves exposed to drought (PEG) treatments at 28 d of the experiment. Additional experimental details as described in Fig. 1. Equal amount of protein extract (50 μg) was loaded in each gel. a Representative silver-stained 2-DE-PAGE of total proteins from control plants. b Enlarged from (1–5) of 2-DE gels as shown in (a) for control plants versus plants exposed only to drought stress conditions (PEG), and (c) plants exposed only to drought stress conditions (PEG) versus plants pre-treated with SNP (SNP → PEG) or NaHS (NaHS → PEG) prior to PEG exposure. Protein spot quantification was contacted as described in Experimental procedures, from at least three gels for each leaf sample and presented in the Supplementary Table S4. The number assigned to the protein spot correspond to those listed in Supplementary Table S5. Red or green arrows indicate only the protein spots that were down-regulated or up-regulated, respectively, in leaves of plants exposed directly to drought stress (PEG) compared with control plants
Protein changes heat-map profile in citrus leaves exposed to drought treatments. Proteins the abundance of which increased or decreased in response to directly PEG treatment compared with control (a) and in response to SNP → PEG or NaHS → PEG treatments compared with directly PEG treatment (b). Each colorized cell represents the averaged spot quantity, according to the colour scale given in the Figure. Detailed list of identified proteins is given in Supplementary Table S5
Analysis of differently expressed proteins under PEG-induced drought stress. a Venn diagram indicating the number of PEG-affected proteins whose abundance were unaffected or specifically changed by either SNP → PEG or NaHS → PEG treatments. The ‘+’ or ‘−’ symbols indicate the number of up-regulated or down-regulated proteins, respectively. b, c Functional classification of PEG-affected proteins following direct PEG stress or pre-treatment with SNP or NaHS prior to PEG stress. The total number of proteins in each leaf samples is indicated
The fact that PEG-responsive proteins that are differently accumulated following SNP → PEG or NaHS → PEG treatments could be involved in drought acclimation led us to apply a comparative proteomic analysis between leaves subjected to PEG-alone, SNP → PEG and NaHS → PEG treatments. This approach revealed the existence of five protein groups. The first group, consisted of 35 proteins, which were only PEG-alone responsive and their accumulation was unaffected by either SNP → PEG or NaHS → PEG (Fig. 10a; Supplementary Table S5). Meanwhile, 15 of these PEG-alone affected proteins were up-regulated while 20 were down-regulated. The largest group of those 35 PEG-responsive proteins was associated with photosynthesis followed by glycolysis and detoxification (Fig. 10b; Supplementary Table S5). The second group was composed of 2 proteins that changed (down-regulated) by both PEG and SNP → PEG (Fig. 10a; Supplementary Table S5). The third group contained four proteins the accumulation of which was modified only after SNP → PEG treatment (SNP specific). Among these SNP specific proteins, only one was up-regulated while three were down-regulated. These SNP specific proteins were related to energy metabolism (photosynthesis, glycolysis and TCA cycle) and cell structure (chloroplast) (Fig. 10b; Supplementary Table S5). The fourth group includes five proteins that were affected by both PEG and NaHS → PEG, containing one protein that was up-regulated and four that were down-regulated. Among these five proteins, two were related to protein synthesis while the other proteins were related to photosynthesis and detoxification. The fifth group consisted of ten proteins the accumulation pattern of which was specifically affected by NaHS → PEG treatment (NaHS specific), with six proteins being up-regulated and four proteins being down-regulated (Fig. 10a; Supplementary Table S5). These NaHS specific proteins were predominantly related to photosynthesis, TCA cycle and detoxification processes (Fig. 10b; Supplementary Table S5).
Discussion
Since drought stress seriously affects crop growth and productivity throughout the world (Xiong et al. 2006), studies on plant priming against drought are important. Herein, pre-treatment of citrus plants with SNP or NaHS induced acclimation to subsequently drought stress produced by PEG treatment, as documented by the phenotypic observations (Fig. 1a) coupled with physiological data, including chlorophyll content (Fig. 1b), photosynthesis rate (Fig. 1c), ion leakage (Fig. 1d), lipid peroxidation (Fig. 1e) and NCED expression (Fig. 1f). It is noted that NCED was induced by individual NaHS and particularly by PEG application but depressed by NaHS-PEG treatment (Fig. 1f) denoting difference of NaHS-derived ABA signaling under normal and drought stress conditions (Jin et al. 2013). One possible explanation for this is the different NaHS regulation mechanisms under stress and physiological conditions. Reports on plant stress response have shown that cell priming involves accumulation of latent signaling components that are not used until challenged by exposure to stress (Tanou et al. 2012b). Hence, an exposure to PEG may alter the physiological status of act of NaHS-pretreated plants and thus partially impair the NaHS-derived ABA signaling. Meanwhile, the fact that SNP negatively regulate NCED expression (Fig. 1f) agreed with recent data obtained in Arabidopsis guard cells (Wang et al. 2015) showing that ABA-induced NO functions are achieved via inactivation/S-nitrosylation of sucrose non fermenting 1 (SNF1)-related protein kinase 2.6 (SnRK2.6) to negatively feedback regulate ABA signaling. Overall, these data clearly suggest that pre-exposure to SNP or to NaHS could prime citrus plants against drought and support the signaling role of these compounds, possibly in conjunction with other molecules, such as ABA, under abiotic stress (Tanou et al. 2009; Jin et al. 2011; Christou et al. 2013). Interestingly, the fact that the local application of NaHS or SNP in roots for 2 d followed by 5 d of acclimation induced drought acclimation in leaves after approximately 1 month further suggests that these chemical treatments elicit long-distance and long-term drought signaling possibly via regulation of several pathways.
Despite the signaling function of NO, NO-derived RNS are toxic at high concentrations in plants (Molassiotis and Fotopoulos 2011). In this study, exposure to SNP or NaHS to prior to PEG lowered the PEG-induced nitrite and NOx content as well as DAF-2DA fluorescence (Fig. 2a–h) in leaves and roots, as previously observed in strawberry plants (Christou et al. 2013). In addition, among the genes associated with NO biosynthesis (i.e., NR, NiR, NOS-like, NADHox, AOX) only NR expression was stimulated in leaves and roots exposed to PEG alone condition (Fig. 4), supporting the role of NR as a principal NO generation system in plants under drought stress (Gupta et al. 2011). In line with recent evidence that H2S steady-level was increased by PEG (Christou et al. 2013), we further observed that both H2S concentration and LCD activity were enhanced in leaves and roots exposed to all drought treatments applied (Fig. 5). The observation that H2S content in citrus’s tissues was not affected after 28 d of NaHS pre-treatment (Fig. 5a, c), indicative of H2S signaling function, is consistent with the relative short burst of H2S given by the NaHS (Wang 2012) Whether endogenously accumulated H2S in PEG alone-treated leaves (Fig. 5a) may reflect a cytotoxic situation is not clear, but it possibly that this PEG-elicited H2S accumulation would inhibit the effects of endogenous H2S arising from exogenous NaHS application, thereby altering the physiognomy of H2S signaling. Furthermore, the fact that leaf NO and H2S content were decreased by NaHS or SNP exposure before PEG stress imposition in relation to direct PEG treatment (Figs. 2a–h, 5a), could be linked to the (1) quenching of NO by H2S leading to the formation of nitrosothiol-like intermediates (Whiteman et al. 2006) (2) feedback inhibition of NO generation by NaHS (Lisjak et al. 2010) that is further strengthened by the fact that NO-linked genes (e.g., NR, NOS-like, NADHox, NADHde, AOX) in citrus either remain unaffected or strongly suppressed (e.g., NiR) by the NaHS treatments (Fig. 4). It is also interesting to note that the different transcriptional patterns (Fig. 4) and H2S content (Fig. 5a, c) observed in roots compared with leaves in pre-treated and non pre-treated plants under PEG stress are in agreement with the view that citrus tissues function in an autonomous manner during stress acclimation (Tanou et al. 2012a).
Evidence defines the ROS- and RNS-based PTMs as a regulatory system for stress signaling in plants (Lounifi et al. 2013). In this regard, another interesting finding that emerged from this work is the fact that NaHS and SNP may prime citrus plants against drought via PTMs. More precisely, NaHS treatment under non-stress and stress conditions diminished leaf protein carbonylation (Fig. 6b, c), indicating that NaHS attenuated protein oxidation, as already proposed in mammalian tissues (Esechie et al. 2008). Although this observation along with the fact that SNP plus PEG stimulated Tyr-nitration in leaves and roots compared with PEG (Fig. 6b, e, i, j) suggests that some PTM responses in citrus are differentially regulated by NaHS and SNP, there is also evidence that these molecules act on common stress signaling pathways. For instance, both chemical pre-treatments before drought lowered nitration to levels close to unstressed control roots (Fig. 6i, j) that were commonly accompanied with the disappearance of specific nitrated bands (Fig. 6i; e.g., 11, 12 and 13). Other significant common functions between NaHS and SNP occur at the level of S-nitrosylation. The current data show that the extent of S-nitrosylation in leaves was considerably induced by both chemical treatments under physiological and drought stress conditions (Fig. 7a, b). In particular, the S-nitrosylation (Fig. 7a, b) and Tyr-nitration (Fig. 6d, e) responses induced by SNP under PEG stress were consistent with the stimulation of GSNOR activity (Fig. 3a) and the lower amount of NO2 and NOx (Fig. 2), indicating that SNP could regulate drought response by controlling the levels of nitrosative stress.
S-nitrosylation leads to changes not only in protein activity but also in protein–protein interactions (Puyaubert and Baudouin 2014) and therefore the distribution of S-nitrosoproteome is biological significant for stress acclimation situations. Using LC–MS/MS analysis, 25 leaf S-nitrosylated proteins were identified (Supplementary Table S3) which are mainly associated with photosynthesis (Fig. 7cC), confirming that the photosynthetic system is targeted by S-nitrosylation (Lindermayr et al. 2005). Most of the presented S-nitrosylated proteins were also identified previously in various plant systems (e.g., chlorophyll a-b binding protein, elongation factor 1, glycolate oxidase, carbonic anhydrase; Lindermayr et al. 2005; Abat et al. 2008; Ortega-Galisteo et al. 2012; Tanou et al. 2009, 2012b), however this study pinpoints 2 S-nitrosylated proteins, namely aspartic proteinase nepehthesin-1 and stem-loop binding protein, that represent novel targets of S-nitrosylation in plants. Of particular interest is also the fact that the S-nitrosylated protein targets can differ among treatments. Several proteins, such as, photosystem II CP43 protein, glycolate oxidase, hydroxypyruvate reductase, aspartic proteinase nepenthesin-1, sedoheptulose-1,7-bisphosphatase and stem-loop binding protein are specifically S-nitrosylated in SNP-treated plants. Also, photosystem II 32 kDa protein was specifically S-nitrosylated in plants exposed to NaHS. Proteins exclusively S-nitrosylated in SNP- or NaHS-pretreated and subsequently stressed plants includes catalase, serine hydroxymethyl transferase and photosystem II 22 kDa protein, respectively. In addition, several S-nitrosylated proteins, such as RubisCO large/small subunit and ATP synthase subunit beta were identified following chemical treatments under both control and drought conditions, disclosing that these proteins are universal markers of SNP and NaHS signaling.
Proteomic analysis of PEG-alone stressed plants compared with controls, revealed the involvement of several pathways leading to phenotypic defects (Fig. 1a). As a result of direct PEG treatment, we found that several proteins were down-regulated, which are involved in CO2 fixation (e.g., carbonic anhydrase) photosystem II (e.g., photosystem II oxygen-evolving complex protein, oxygen-evolving enhancer protein 2 and the cytochrome b6/f complex (Fig. 9a), as previously noted in several drought studies (Zadražnik et al. 2013). Therefore a decreased accumulation of these proteins could contribute to the photosynthetic disturbances observed in plants directly stressed with PEG (Fig. 1c). Low abundance of several enzymes involved in ROS detoxification, such as GDP-mannose-3,5-epimerase (Fig. 9a), a key enzyme in ascorbic acid biosynthesis, contributed further to drought sensitivity of PEG-treated plants. Meanwhile, the observed down-regulation of the pyruvate dehydrogenase, an enzyme that catalyzes the formation of acetyl coenzyme A (acetyl-CoA) from pyruvate, possibly represents an attempt for energy regulation since acetyl-CoA serves as a hub in cell metabolism. This regulation is also strengthened by the down-regulation of NADP-dependent malic enzyme, which catalyzes the conversion of malate to pyruvate in TCA cycle, minimizing the amount of pyruvate, thus affecting the activity of pyruvate dehydrogenase.
In the present study, a set of chaperones were induced by PEG treatment (Fig. 9a), indicating that these proteins play a critical role in cells by preventing irreversible protein aggregation/insolubilization and maintaining denatured proteins in a folding-competent state under drought stress (Fink 1999). In response to PEG-alone stress, the down-regulation of FK506-binding protein (Fig. 9a), which belongs to the peptidyl-prolyl cis/trans isomerase (PPIase) superfamily, could be related with the decrease of the photosynthetic rate and chlorophyll content, since FK506-binding proteins are physiologically important to chloroplast function (Yu et al. 2012) and biogenesis (Kurek et al. 2002). The finding that a number of ribosomal proteins, including 60S ribosomal protein, 40S ribosomal protein and nascent polypeptide-associated complex subunit were up-regulated in citrus exposed to PEG (Fig. 9a) may be connected to the fact that ribosomal proteins contribute to the biosynthesis of stress-defense proteins as part of the overall acclimation strategy against stress (Zadražnik et al. 2013). The expression of other proteins involved in specific metabolic pathways was also up-regulated in leaves subjected to PEG treatment. Thus, the up-regulation of cysteine synthase, a key enzyme in cysteine synthesis, which is required for the biosynthesis of glutathione, along with the up-regulation of SOUL-heme binding protein that participates in heme transfer/binding, could prevent oxidative damage (Taketani et al. 1998). Furthermore, the up-regulation of nucleoside diphosphate kinase (NDPK), an enzyme that uses ATP to maintain cellular levels of CTP, GTP and UTP, could be associated with a general stress response mechanism, as previously proposed by Zadražnik et al. (2013).
Another interesting finding that emerged from this work is the fact that SNP or NaHS exposure prior to PEG modified protein profiles differently compared with PEG treatment. Primarily, we observed that two PEG-induced proteins, namely nascent polypeptide-associated complex subunit-alpha like (α-NAC) and 14-3-3 protein, showed a down-regulation following SNP pre-treatment (Fig. 9). This suppression of α-NAC by SNP could be associated with a general repression of translation under drought stress (Riccardi et al. 1998), as also supported by the decreased abundance of the 14-3-3 protein since this protein acts as a mediator of transcription factors under abiotic stress (Deeba et al. 2012). In addition, several PEG-affected proteins were specifically changed following SNP pre-treatment before PEG. More specifically, the decreased accumulation of glycolate oxidase, a peroxisomal FMN-depended oxidase that catalyzes the oxidation of a-hydroxy acids to a-ketoacids, indicates that photorespiration is suppressed under SNP plus PEG conditions (Xu et al. 2011). The observed decrease in abundance of NADP-isocitrate dehydrogenase could be attributed to a metabolic interconnection of this enzyme with molecules involved in RNS metabolism, as already proposed by Leterrier et al. (2012). The increased abundance of UPF0603 protein Atlg54780, which is involved in photosystem II repair cycle, could be associated with the elevated level of chlorophyll and photosynthetic rate (Fig. 1b, c) under SNP plus PEG treatment, justifying the protective role of SNP in the recovery of photosystem II (Chen et al. 2013).
The drought acclimation function of NaHS is clearly illustrated by the finding that 5 PEG-affected citrus proteins were changed following this pre-treatment. The fact that a set of these proteins participating in protein synthesis (60S and 40S ribosomal proteins) and in ROS-detoxification (catalase and SOUL heme-binding protein) were down-regulated (Fig. 9b) led us to hypothesize that NaHS exposure resulted in minimal requirement of stress-defense proteins due to alleviation of the negative impact of PEG stress. In this specific group of NaHS-affected proteins, the up-regulation of photosystem II oxygen-evolving complex protein, RubisCO large subunit and glutamate glyoxylate aminotransferase indicates the beneficial role of NaHS towards maintaining the functionality of the photosynthetic apparatus under PEG stress. Apart from photosynthetic proteins, various citrus proteins were differentially accumulated following NaHS treatment, such as epidermis-specific secreted protein EP1. Notably, the up-regulation of epidermis-specific secreted protein EP1, which is a protein involved in the limitation of water flow through the outer epidermal cell wall (van Engelen et al. 1993), might also reflect a drought acclimation response in citrus plants by restricting water loss across the cell wall leading to the maintenance of the cell turgor. Furthermore, the up-regulation of dihydrolipoyl dehydrogenase (DLD) could by linked with the suppressed level of nitrite and NOx in citrus plants exposed to combined NaHS and PEG treatments (Fig. 2), because it was suggested that DLD catalyzes the NADH-dependent scavenging of NO (Igamberdiev et al. 2004).
Conclusion
The evidence discussed presently suggests an important priming role for SNP and NaHS against drought that is associated, among others, with a specific proteome reprogramming. However, our limited knowledge of NO and H2S-associated source and metabolism in plants is still a major gap in our understanding of the function of these signaling molecules under drought conditions. Therefore, the application of comprehensive systems biology approaches will contribute to further understanding of SNP and NaHS priming mechanism in plants.
References
Abat JK, Saigal P, Deswal R (2008) S-Nitrosylation—another biological switch like phosphorylation? Physiol Mol Biol Plants 14:119–130. doi:10.1007/s12298-008-0011-5
Álvarez C, Calo L, Romero LC, García I, Gotor C (2010) An o-acetylserine(thiol)lyase homolog with l-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol 152:656–669. doi:10.1104/pp.109.147975
Baudouin E, Hancock JT (2014) Nitric oxide signaling in plants. Front Plant Sci 4:553. doi:10.3389/fpls.2013.00553
Bevan M, Bancroft I, Bent E et al (1998) Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391:485–488. doi:10.1038/35140
Bloem E, Riemenschneider A, Volker J et al (2004) Sulphur supply and infection with Pyrenopeziza brassicae influence l-cysteine desulphydrase activity in Brassica napus L. J Exp Bot 55:2305–2312. doi:10.1093/jxb/erh236
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254. doi:10.1016/0003-2697(76)90527-3
Chaki M, Valderrama R, Fernández-Ocaña AM et al (2011) High temperature triggers the metabolism of S-nitrosothiols in sunflower mediating a process of nitrosative stress which provokes the inhibition of ferredoxin-NADP reductase by tyrosine nitration. Plant, Cell Environ 34:1803–1818. doi:10.1111/j.1365-3040.2011.02376.x
Chen K, Chen L, Fan J, Fu J (2013) Alleviation of heat damage to photosystem II by nitric oxide in tall fescue. Photosynth Res 116:21–31. doi:10.1007/s11120-013-9883-5
Christou A, Manganaris GA, Papadopoulos I, Fotopoulos V (2013) Hydrogen sulfide induces systemic tolerance to salinity and non-ionic osmotic stress in strawberry plants through modification of reactive species biosynthesis and transcriptional regulation of multiple defence pathways. J Exp Bot 64:1953–1966. doi:10.1093/jxb/ert055
Christou A, Filippou P, Manganaris G, Fotopoulos V (2014) Sodium hydrosulfide induces systemic thermotolerance to strawberry plants through transcriptional regulation of heat shock proteins and aquaporin. BMC Plant Biol 14:42. doi:10.1186/1471-2229-14-42
Corpas FJ, Chaki M, Fernández-Ocaña A et al (2008) Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol 49:1711–1722. doi:10.1093/pcp/pcn144
Deeba F et al (2012) Physiological and proteomic responses of cotton (Gossypium herbaceum L.) to drought stress. Plant Physiol Biochem 53:6–18. doi:10.1016/j.plaphy.2012.01.002
Esechie A, Kiss L, Olah G, Horvath EM, Horvath E, Szabo C, Traber DL (2008) Protective effect of hydrogen sulfide in a murine model of acute lung injury induced by combined burn and smoke inhalation. Clin Sci 115:91–97. doi:10.1042/CS20080021
Fink AL (1999) Chaperone-mediated protein folding. Physiol Rev 79:425–449. doi:10.1038/nrm1492
Finkel T (2012) From sulfenylation to sulfhydration: what a thiolate needs to tolerate. Sci Signal 5(215):pe10. doi:10.1126/scisignal.2002943
García-Mata C, Lamattina L (2010) Hydrogen sulphide, a novel gasotransmitter involved in guard cell signaling. New Phytol 188:977–984. doi:10.1111/j.1469-8137.2010.03465.x
García-Mata C, Lamattina L (2013) Gasotransmitters are emerging as new guard cell signaling molecules and regulators of leaf gas exchange. Plant Sci 201–202:66–73. doi:10.1016/j.plantsci.2012.11.007
García-Sánchez F, Syvertsen JP, Gimeno V, Botía P, Perez-Perez JG (2007) Responses to flooding and drought stress by two citrus rootstock seedlings with different water-use efficiency. Physiologia Plant 130:532–542. doi:10.1111/j.1399-3054.2007.00925.x
Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5:151. doi:10.3389/fpls.2014.00151
Gupta KJ, Fernie AR, Kaiser WM, van Dongen JT (2011) On the origins of nitric oxide. Trends Plant Sci 16:160–168. doi:10.1016/j.tplants.2010.11.007
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198. doi:10.1016/0003-9861(68)90654-1
Igamberdiev AU, Bykova NV, Ens W, Hill RD (2004) Dihydrolipoamide dehydrogenase from porcine heart catalyzes NADH-dependent scavenging of nitric oxide. FEBS Lett 568:146–150. doi:10.1016/j.febslet.2004.05.024
Jaffrey SR, Snyder SH (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci Signal 86:pl1. doi:10.1126/stke.2001.86.pl1
Jin Z, Shen J, Qiao Z, Yang G, Wang R, Pei Y (2011) Hydrogen sulfide improves drought resistance in Arabidopsis thaliana. Biochem Biophys Res Commun 414:481–486. doi:10.1016/j.bbrc.2011.09.090
Jin Z, Xue S, Luo Y, Tian B, Fang H, Li H, Pei Y (2013) Hydrogen sulfide interacting with abscisic acid in stomatal regulation responses to drought stress in Arabidopsis. Plant Physiol Biochem 62:41–46. doi:10.1016/j.plaphy.2012.10.017
Kabil O, Vitvitsky V, Banerjee R (2014) Sulfur as a signaling nutrient through hydrogen sulfide. Annu Rev Nutr 34:171–205. doi:10.1146/annurev-nutr-071813-105654
Kubo S, Kurokawa Y, Doe I, Masuko T, Sekiguchi F, Kawabata A (2007) Hydrogen sulfide inhibits activity of three isoforms of recombinant nitric oxide synthase. Toxicology 241:92–97. doi:10.1016/j.tox.2007.08.087
Kurek I, Stoger E, Dulberger R, Christou P, Breiman A (2002) Overexpression of the wheat FK506-binding protein 73 (FKBP73) and the heat-induced wheat FKBP77 in transgenic wheat reveals different functions of the two isoforms. Transgenic Res 11:373–379. doi:10.1007/s00438-008-0318-5
Leterrier M, Barroso JB, Palma JM, Corpas FJ (2012) Cytosolic NADP-isocitrate dehydrogenase in Arabidopsis leaves and roots. Biol Plant 56:705–710. doi:10.1007/s10535-012-0244-6
Lindermayr C, Saalbach G, Durner J (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137:921–930. doi:10.1104/pp.104.058719
Lisjak M et al (2010) A novel hydrogen sulfide donor causes stomatal opening and reduces nitric oxide accumulation. Plant Physiol Biochem 48:931–935. doi:10.1016/j.plaphy.2010.09.016
Lounifi I, Arc E, Molassiotis A, Job D, Rajjou L, Tanou G (2013) Interplay between protein carbonylation and nitrosylation in plants. Proteomics 13:568–578. doi:10.1002/pmic.201200304
Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444:139–158. doi:10.1016/j.abb.2005.10.018
Mahe A, Grisvard J, Dron M (1992) Fungal and specific gene markers to follow the bean-anthracnose infection process and mormalize the bean chitinase rnRNA induction. Mol Plant Microbe Interact 5:242–248
Martínez MC, Achkor H, Persson B et al (1996) Arabidopsis formaldehyde dehydrogenase. Molecular properties of plant class III alcohol dehydrogenase provide further insights into the origins, structure and function of plant class P and liver class I alcohol dehydrogenases. Eur J Biochem 241:849–857. doi:10.1111/j.1432-1033.1996.00849.x
Miller GAD, Suzuki N, Ciftci-Yilmaz S, Mittler RON (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33:453–467. doi:10.1111/j.1365-3040.2009.02041.x
Molassiotis A, Fotopoulos V (2011) Oxidative and nitrosative signaling in plants: two branches in the same tree? Plant Signal Behav 6:210–214. doi:10.4161/psb.6.2.14878
Nashef AS, Osuga DT, Feeney RE (1977) Determination of hydrogen sulfide with 5,5′-dithiobis-(2-nitrobenzoic acid), N-ethylmaleimide, and parachloromercuribenzoate. Anal Biochem 79:394–405. doi:10.1016/0003-2697(77)90413-4
Ortega-Galisteo AP, Rodríguez-Serrano M, Pazmiño DM, Gupta DK, Sandalio LM, Romero-Puertas MC (2012) S-nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. J Exp Bot 63:2089–2103. doi:10.1093/jxb/err414
Pandey S (2014) Hydrogen sulfide: a new node in the abscisic acid-dependent guard cell signaling network? Plant Physiol 166:1680–1681. doi:10.1104/pp.114.251686
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi:10.1093/nar/30.9.e36
Puyaubert J, Baudouin E (2014) New clues for a cold case: nitric oxide response to low temperature. Plant Cell Environ 37:2623–2630. doi:10.1111/pce.12329
Qiao Z, Jing T, Liu Z, Zhang L, Jin Z, Liu D, Pei Y (2015) H2S acting as a downstream signaling molecule of SA regulates Cd tolerance in Arabidopsis. Plant Soil 393:137–146. doi:10.1007/s11104-015-2475-8
Riccardi F, Gazeau P, de Vienne D, Zivy M (1998) Protein changes in response to progressive water deficit in maize: quantitative variation and polypeptide identification. Plant Physiol 117:1253–1263. doi:10.1104/pp.117.4.1253
Riemenschneider A, Nikiforova V, Hoefgen R, De Kok LJ, Papenbrock J (2005a) Impact of elevated H2S on metabolite levels, activity of enzymes and expression of genes involved in cysteine metabolism. Plant Physiol Biochem 43:473–483. doi:10.1016/j.plaphy.2005.04.001
Riemenschneider A, Wegele R, Schmidt A, Papenbrock J (2005b) Isolation and characterization of a d-cysteine desulfhydrase protein from Arabidopsis thaliana. FEBS J 272:1291–1304. doi:10.1111/j.1742-4658.2005.04567.x
Rizhsky L et al (2002) Double antisense plants lacking ascorbate peroxidase and catalase are less sensitive to oxidative stress than single antisense plants lacking ascorbate peroxidase or catalase. Plant J 32:329–342. doi:10.1046/j.1365-313X.2002.01427.x
Rümer S, Krischke M, Fekete A, Mueller MJ, Kaiser WM (2012) DAF-fluorescence without NO: elicitor treated tobacco cells produce fluorescing DAF-derivatives not related to DAF-2 triazol. Nitric Oxide 27:123–135. doi:10.1016/j.niox.2012.05.007
Scuffi D, Álvarez C, Laspina N, Gotor C, Lamattina L, García-Mata C (2014) Hydrogen sulfide generated by l-cysteine desulfhydrase acts upstream of nitric oxide to modulate abscisic acid-dependent stomatal closure. Plant Physiol 166:2065–2076. doi:10.1104/pp.114.245373
Shen J, Xing T, Yuan H, Liu Z, Jin Z, Zhang L, Pei Y (2013) Hydrogen sulfide improves drought tolerance in Arabidopsis thaliana by microRNA expressions. PLoS ONE 8:e77047. doi:10.1371/journal.pone.0077047
Shi H, Ye T, Chan Z (2013) Exogenous application of hydrogen sulfide donor sodium hydrosulfide enhanced multiple abiotic stress tolerance in bermudagrass (Cynodon dactylon (L). Pers.). Plant Physiol Biochem 71:226–234. doi:10.1016/j.plaphy.2013.07.021
Shi H, Ye T, Zhu J-K, Chan Z (2014) Constitutive production of nitric oxide leads to enhanced drought stress resistance and extensive transcriptional reprogramming in Arabidopsis. J Exp Bot 65:4119–4131. doi:10.1093/jxb/eru184
Taketani S, Adachi Y, Kohno H, Ikehara S, Tokunaga R, Ishii T (1998) Molecular characterization of a newly identified heme-binding protein induced during differentiation of urine erythroleukemia cells. J Biol Chem 273:31388–31394. doi:10.1074/jbc.273.47.31388
Tan BH, Wong PTH, Bian J-S (2010) Hydrogen sulfide: a novel signaling molecule in the central nervous system. Neurochem Int 56:3–10. doi:10.1016/j.neuint.2009.08.008
Tanou G, Job C, Rajjou L, Arc E, Belghazi M, Diamantidis G, Molassiotis A, Job D (2009) Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J 60:795–804. doi:10.1111/j.1365-313X.2009.04000.x
Tanou G, Job C, Belghazi M, Molassiotis A, Diamantidis G, Job D (2010) Proteomic signatures uncover hydrogen peroxide and nitric oxide cross-talk signaling network in citrus plants. J Proteome Res 9:5994–6006. doi:10.1021/pr100782h
Tanou G, Filippou P, Belghazi M, Job D, Diamantidis G, Fotopoulos V, Molassiotis A (2012a) Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J 72:585–599. doi:10.1111/j.1365-313X.2012.05100.x
Tanou G, Fotopoulos V, Molassiotis A (2012b) Priming against environmental challenges and proteomics in plants: update and agricultural perspectives. Front Plant Sci 3:216. doi:10.3389/fpls.2012.00216
Tanou G, Ziogas V, Belghazi M, Christou A, Filippou P, Job D, Fotopoulos V, Molassiotis A (2014) Polyamines reprogram oxidative and nitrosative status and the proteome of citrus plants exposed to salinity stress. Plant Cell Environ 37:864–885. doi:10.1111/pce.12204
Valderrama R, Francisco JC, Carreras A et al (2007) Nitrosative stress in plants. FEBS Lett 581:453–461. doi:10.1016/j.febslet.2007.01.006
van Engelen FA, Hartog MV, Thomas TL, Taylor B, Sturm A, van Kammen A, de Vries SC (1993) The carrot secreted glycoprotein gene EP1 is expressed in the epidermis and has sequence homology to Brassica S-locus glycoproteins. Plant J 4:855–862. doi:10.1046/j.1365-313X.1993.04050855.x
Wang R (2012) Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev 92:791–896. doi:10.1152/physrev.00017.2011
Wang P, Du Y, Hou Y-J, Zhao Y, Hsu C-C, Yuan F, Zhu X, Tao WA, Song C-P, Zhu J-K (2015) Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc Natl Acad Sci USA 112:613–618. doi:10.1073/pnas.1423481112
Whiteman M, Li L, Kostetski I, Chu SH, Siau JL, Bhatia M, Moore PK (2006) Evidence for the formation of a novel nitrosothiol from the gaseous mediators nitric oxide and hydrogen sulphide. Biochem Biophys Res Commun 343:303–310. doi:10.1016/j.bbrc.2006.02.154
Wilson LG, Bressan RA, Filner P (1978) Light-dependent emission of hydrogen sulfide from plants. Plant Physiol 61:184–189. doi:10.1104/pp.61.2.184
Wünsche H, Baldwin IT, Wu J (2011) S-Nitrosoglutathione reductase (GSNOR) mediates the biosynthesis of jasmonic acid and ethylene induced by feeding of the insect herbivore Manduca sexta and is important for jasmonate-elicited responses in Nicotiana attenuata. J Exp Bot 62:4605–4616. doi:10.1093/jxb/err171
Xiong L, Wang R-G, Mao G, Koczan JM (2006) Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiol 142:1065–1074. doi:10.1104/pp.106.084632
Xu C, Jiang Z, Huang B (2011) Nitrogen deficiency-induced protein changes in immature and mature leaves of creeping bentgrass. J Am Soc Hortic Sci 136:399–407
Yu Y, Zhang H, Li W, Mu C, Zhang F, Wang L, Meng Z (2012) Genome-wide analysis and environmental response profiling of the FK506-binding protein gene family in maize (Zea mays L.). Gene 498:212–222. doi:10.1016/j.gene.2012.01.094
Zadražnik T, Hollung K, Egge-Jacobsen W, Meglič V, Šuštar-Vozlič J (2013) Differential proteomic analysis of drought stress response in leaves of common bean (Phaseolus vulgaris L.). J Proteomics 78:254–272. doi:10.1016/j.jprot.2012.09.021
Zhang A, Jiang M, Zhang J, Ding H, Xu S, Hu X, Tan M (2007) Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol 175:36–50. doi:10.1111/j.1469-8137.2007.02071.x
Zhou B, Guo Z, Xing J, Huang B (2005) Nitric oxide is involved in abscisic acid-induced antioxidant activities in Stylosanthes guianensis. J Exp Bot 56:3223–3228. doi:10.1093/jxb/eri319
Ziogas V, Tanou G, Filippou P, Diamantidis G, Vasilakakis M, Fotopoulos V, Molassiotis A (2013) Nitrosative responses in citrus plants exposed to six abiotic stress conditions. Plant Physiol Biochem 68:118–126. doi:10.1016/j.plaphy.2013.04.004
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ziogas, V., Tanou, G., Belghazi, M. et al. Roles of sodium hydrosulfide and sodium nitroprusside as priming molecules during drought acclimation in citrus plants. Plant Mol Biol 89, 433–450 (2015). https://doi.org/10.1007/s11103-015-0379-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0379-x