Abstract
The AP2/ERFs are one of the most important family of transcription factors which regulate multiple responses like stress, metabolism and development in plants. We isolated PsAP2 a novel AP2/ERF from Papaver somniferum which was highly upregulated in response to wounding followed by ethylene, methyl jasmonate and ABA treatment. PsAP2 showed specific binding with both DRE and GCC box elements and it was able to transactivate the reporter genes in yeast. PsAP2 overexpressing transgenic tobacco plants exhibited enhanced tolerance towards both abiotic and biotic stresses . Real time transcript expression analysis showed constitutive upregulation of tobacco Alternative oxidase1a and Myo-inositol-1-phosphate synthase in PsAP2 overexpressing tobacco plants. Further, PsAP2 showed interaction with NtAOX1a promoter in vitro, it also specifically activated the NtAOX1a promoter in yeast and tobacco BY2 cells. The silencing of PsAP2 using VIGS lead to significant reduction in the AOX1 level in P. somniferum. Taken together PsAP2 can directly bind and transcriptionally activate NtAOX1a and its overexpression in tobacco imparted increased tolerance towards both abiotic and biotic stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants being sessile in nature are regularly encountered by a number of abiotic and biotic stresses. To combat these stressed conditions, plants evolve various adaptive mechanisms regulated through number of signaling pathways. These stresses are never encountered alone and generally occur along with different forms of stress. Therefore, there is a need to understand the mechanism of multiple stress tolerance and develop varieties resistant to multiple form of stresses without compromising the yield (Atkinson and Urwin 2012). Plant transcription factors (TFs) are one of the most important candidates involved in abiotic and biotic stress tolerance in plants (Nuruzzaman et al. 2013). TFs interact with the cis-elements present in the promoter of downstream target genes (Cheng et al. 2013). The AP2/ERF family of TF contains a ~60 amino acid conserved DNA binding domain through which it bind to various cis-elements like DRE/CRT, GCC box and JERE like elements (Okamuro et al. 1997; Fujimoto et al. 2000). Due to the multiple binding ability of AP2/ERFs to different cis-elements, these proteins can be a useful target for providing resistance against multiple stress responses (Wu et al. 2008).
Recent studies have indicated that both abiotic and biotic stresses are closely connected through a common set of genes that regulate under both the conditions (Suzuki et al. 2014). TSI1 an AP2/ERF identified from tobacco can bind strongly to GCC in comparison to DRE/CRT and can provide tolerance to salt stress and pathogen attack (Park et al. 2001). GmERF3 from soybean binds to both DRE and GCC elements and imparts resistance against salt, drought and disease in transgenic tobacco (Zhang et al. 2009). Ethylene responsive TaERF1 from wheat (Triticum aestivum) also binds to GCC and DRE/CRT elements. TaERF1 can activate the stress response genes including pathogen response (PR) and cold response/response to desiccation (COR/RD) constitutively under normal growth condition leading to improved pathogen and abiotic stress response in transgenic plants. (Xu et al. 2007). Over expression of another ethylene responsive wheat ERF protein TaPIE1 in transgenic plant exhibited increased resistance to Rhizoctonia cerealis and freezing response by increasing the transcript level of defense and stress related genes (Zhu et al. 2014). The simultaneous occurrence of biotic and abiotic stresses in plants growing in their natural habitat and different regulatory roles of AP2/ERF proteins under these stresses raises the importance to understand the regulatory role of these AP2/ERF proteins in plants. Though AP2/ERF proteins imparting both abiotic and biotic stress tolerance have been characterized but the molecular mechanisms involved in these cross-talk is still unclear.
In our previous study to understand the effect of wounding (Mishra et al. 2013) in Papaver somniferum, we identified an EST (AAP56252.1) of AP2/ERF domain containing TF showing induced expression in response to wounding, salt and ethylene treatments. We made it full length and named it PsAP2 which was submitted to Genbank with accession no. JN315881. PsAP2 showed specific interaction to DRE/CRT and GCC-box cis elements in vitro. Transgenic tobacco constitutively expressing PsAP2 showed improved tolerance to abiotic and biotic stresses. In addition, transgenic tobacco lines showed induced expression of tobacco alternative oxidase1a and myoinositol phosphate synthase transcripts. Further PsAP2 was able to bind to the promoter of NtAOx1a and activate the transcription of reporter genes in yeast and tobacco BY2 cells. Virus induced gene silencing of PsAP2 downregulated the expression of AOX1 in P. somniferum suggesting the possible conserved mechanism of action of PsAP2 in P. somniferum as well.
Materials and methods
Plant material and treatment
PsAP2 was cloned from Papaver somniferum cv. sampada (CSIR-CIMAP-Genotype). Seedling growth condition, stress treatment and hormonal treatment were performed as described in Mishra et al. (2013). For generating PsAP2 transgenic lines Nicotiana tabacum var. Xanthi was used. PsAP2 was cloned at the BamHI and XbaI site using PsAP2pBI F and PsAP2pBI R primers (Supporting Table 1a) under CaMV35S promoter. The resulting construct was mobilized in tobacco using Agrobacterium tumefaciens (GV3101) mediated transformation according to Gelvin and Schilperoort (1994).
Cloning and transcript expression analysis of PsAP2
Full length clone of PsAP2 was obtained after 3′ and 5′ RACE using the specific primer sequences given in Supporting Table 1(a). Both the fragments of 3′ and 5′ RACE were cloned in pGEMT and sequenced. The sequences were aligned and a full length ORF was obtained. The full length PsAP2 was amplified using gene specific primers (GSPs) mentioned in Supporting Table 1(a). The semi quantitative PCR was performed for 28 cycles (94 °C for 45 s, 58 °C for 45 s and 72 °C for 60 s) using GSPs. Expression analysis of transcripts was analyzed by qRT-PCR using SYBR green mix according to the protocol described in Mishra et al. (2013). To investigate the tissue specific expression of PsAP2, P. somniferum seedlings were grown for three and half months in green house with 18–22 °C day/10–15 °C at night with 50 % relative humidity and a photoperiod of 8–10 h with appropriate watering. Straw, capsule and floral buds were harvested under control growing conditions after three and half months. The capsule was harvested after 135 days under similar green house growth condition. The relative expression of PsAP2 in these harvested tissues was expressed relative to the expression in leaf tissues. The relative expression of PsAP2 in response to different stresses like salt (250 mM NaCl), cold (4 °C), dehydration, wounding and hormone treatments like ethylene, ABA, methyl jasmonate (MeJA) were monitored in 2 months old P. somniferum seedlings. Dehydration stress was performed by removing seedlings carefully from the pots and by keeping them in between 3 mm paper (Whatman Cligton, NJ). Cold treatment was given by keeping the seedlings at 4 °C. For salt treatment seedlings were removed from the soil and roots were dipped into aerated water with 250 mM NaCl. Wounding was done with sterile pins avoiding any major injuries as described in Mishra et al. (2013). Hormone treatment such as MeJA (50 µM) and ABA (100 µM) were performed as described in Mishra et al. (2013). For ethylene treatment 10 µM solution of ACC was prepared in sterile water and was sprayed on the leaves of 8 week old seedlings, the control was sprayed with only sterile water. The control plants for salt and ABA treatments were removed carefully from soil and kept in sterile water. For both stress and hormone treatments the samples were harvested after 1, 3 and 5 h, RNA was isolated and qRT-PCR expression analysis was performed. Each qRT-PCR result represents the mean of three biological as well as experimental replicates. The error bars represent standard deviation.
Gel mobility shift and transactivation assay
PsAP2 was cloned using PsAP2pGEXF and PSAP2pGEXR primers with restriction sites BamHI and EcoRI maintaining the reading frame with GST in pGEX4T2 vector (Supporting Table 1a). GST-PsAP2 construct was transformed in E. coli BL21 (DE3) cells and induced with 0.3 mM IPTG. The recombinant protein was purified from bacterial lysate using GST-Sepharose beads (Amersham). Only GST purified protein was used as a negative control for Gel retardation assay (GRA). Respective probes for GRA and their mutated oligos were designed as described in Shukla et al. (2006) and their sequences are mentioned in Supporting Table 1(c). For transactivation assay, PsAP2 was cloned in Saccharomyces cerevisiae expression vector pGBKT7 using PsAP2pGB F and PsAP2pGB R primers under NdeI and BamHI sites in fusion with GAL4-DNA-BD (Supporting Table 1a). The construct was transformed into an auxotrophic yeast strain AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4D, gal80D, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UASGAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1) and colonies were screened on SD-His-Trp media (Cagney et al. 2000). The Y1H assay was performed as described in Mishra et al. (2013).
Southern hybridization analysis of transgenic lines
The genomic DNA isolated from transgenic and vector control plants were isolated and digested (20 µg/lane) overnight at 37 °C with BamHI and EcoRI. The digested products were fractionated on 0.8 % agarose gel in 1× TBE, pH 8.3 at low voltage (40 V) for 12 h. The DNA fragments were transferred onto a nylon membrane (Hybond-N, Amersham) by capillary transfer method with 20× SSC (3 M NaCl, 0.3 M sodium citrate, pH 7.0) (Southern 1975). Before transfer, the gel was treated with depurination solution (0.2 N HCl) for 10 min, denaturation solution (1.5 M NaCl, 0.5 N NaOH) for 20 min, two times, and neutralizing solution (1 M Tris–HCl, pH 7.4, 1.5 M NaCl) for 20 min two times. The DNA fragments were UV crosslinked and blot was pre-hybridized for 6 h at 60 °C in a buffer containing 5× SSC, 5× Denhardt’s reagent (0.5 % Ficoll, 0.5 % PVP, 0.5 % BSA), 0.1 % SDS, 100 µg/ml denatured salmon sperm DNA, 10 % dextran sulphate, 50 mM Na-phosphate buffer, 5 mM EDTA (Sambrook et al. 2001). Thereafter, to the pre-hybridization solution, labelled probe (denatured by boiling for 5 min, and chilled on ice for 5 min) was added. After hybridization for 18 h at 60 °C, the blot was washed twice with 2× SSC, 0.1 % SDS and 1× SSC, 0.1 % SDS for 10 min each at 50 °C. Finally, the blot was exposed to Kodak BioMax MR film and kept at −80 °C using intensifying screens.
Analysis of transgenic plants for abiotic stress response
For salt stress, seedlings were first grown for 2 weeks after germination in MS media, then were transferred to MS media containing 250 mM NaCl and allowed to grow for 1 week. For dehydration stress, transgenic and vector transformed seeds were geminated directly on MS media containing 0.4 M mannitol. The respective seeds were grown for 3 weeks. For leaf disc assay full grown healthy leaves from vector transformed and transgenic plants were detached and briefly washed in distilled water. Leaf discs of 1 cm diameter were cut and floated in 250 and 400 mM NaCl solution. The chlorophyll content was measured as described by Aono et al. (1993), after 7 days.
Analysis of transgenic plants for biotic stress response
To check the biotic stress response bacterial strain Pseudomonas syringae pv. tabaci was used to infect the leaves of transgenic and vector transformed tobacco plants. The strain was grown in LB-Tetracycline at 28 °C. The over night grown culture was pelleted, washed and resuspended in 10 mM MgCl2. The bacterial suspension was infiltrated into fully expanded and healthy leaves using 5 ml plastic syringe without needle as described in Park et al. (2001). Approximately 0.5 ml of inoculum was infiltrated per panel forming an equal infiltrated area. The lesion diameter was calculated and plotted. The total bacterial population was measured by grinding six leaf discs containing the infection from three independent lines in 10 mM MgCl2. The serial dilution of ground leaf samples was made in Kings B medium (Martin et al. 1993) and colony forming units were counted. To identify the genes regulated through PsAP2 we screened 26 stress marker genes in control and transgenic plants using qRT-PCR. The list of selected genes along with their primer sequences and accession numbers are given in Supporting Table 1(b).
Antioxidant analysis of transgenic plants
Sample extracts from transgenic as well as vector transformed plants were prepared by grinding 100 mg tissue in 1 ml of 50 % ethanol, centrifuged at 10,000g at 4 °C for 10 min. Supernatant was used within 4 h for antioxidant assays. For DPPH radical scavenging activity extracts were made to total volume of 3 ml using methanol. To it 0.15 ml of freshly prepared DPPH solution (98 µg/ml) was added, stirred and left to stand at RT for 30 min in dark. The control contains only DPPH solution in methanol while methanol served as the blank (negative control). The reduction capability of DPPH radical was determined by the decrease in its absorbance at 517 nm (Patel et al. 2011). The estimation was done for 50 µg and 100 µg extracts and calculation was done as follows:
For NO scavenging assay, to the test sample (50 µg and 100 µg extracts) added 200 μl of 10 mM Sodium Nitroprusside and incubated at RT for 2 h. From it 50 μl of the sample was taken and to it added 100 μl of Griess reagent (1 part of the 0.1 % Naphthalene Diamine Dihydrochloride in water and 1 part 1 % sulphanilamide in 5 % concentrated phosphoric acid). The absorbance of chromophore formed was measured at 546 nm on UV–visible spectrometer (Patel et al. 2011). NO scavenging was calculated using following formula similarly using 50 µg and 100 µg of individual extracts:
All spectrophotometric analysis was conducted at room temperature on a UV/visible spectrophotometer (Eppendorf BioSpectrometer (Basic).
Relative fresh weight measurement after salt and mannitol stress
For salt stress seedlings were transferred to MS media containing 250 mM NaCl after 2 weeks of germination and allowed to grow for 6 days. Relative fresh weight of the seedlings was measured after 2, 4 and 6 days of the treatment by comparing with the seedlings grown in the control.
Similarly for dehydration stress, transgenic and vector transformed wild type seeds were germinated directly on MS media, and 2 weeks after germination, seedlings were transferred to 0.4 M mannitol supplemented MS media. Relative fresh weight of the seedlings was measured after 2, 4 and 6 days of the treatment by comparing with the seedlings grown in the control.
Genome walking and cloning of NtAOX1a promoter
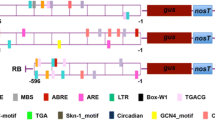
The primers for genome walking were designed from the first exon of NtAOX1a transcript (AB281425.1) (Supporting Table 1a). The isolated fragment of 826 bp was cloned in pGEMT vector and sequenced using M13 forward and reverse primers (Supporting Fig. 4). Analysis of cis-acting elements in NtAOX1a promoter was done using the Plant cis acting regulatory element (PLACE) program. The predicted transcription start site was indicated within the promoter sequence of NtAOX1a. Further 651 bp of NtAOX1a promoter carrying both DRE and GCC box cis element was cloned into pHIS2.0 vector using pHIS AOX F and pHIS AOX R primers with EcoRI and SacI restriction sites (Supporting Table 1a). Also truncated promoter fragments carrying only DRE (297 bp) and GCC cis elements (351 bp) were also cloned under EcoRI and Sac I sites in pHIS2.0 vector with specific primers (Supporting Table 1a). The NtAOX1a promoter containing the DRE, GCC and predicted transcription start site is shown in Supporting Fig. 5a.
NtAOX1a promoter activation and GRA with NtAOX1a promoter fragments
GRA was performed with 41 bp dimeric region of NtAOX1a promoter containing both the elements. We also used 39 bp complementary oligos from NtAOX1a promoter carrying only DRE or GCC box cis element (Supporting Table 1c). For Y1H PsAP2 was cloned in NdeI and BamHI sites of pGAD vector with Leu as a selection marker with the primers PsAP2pGAD F and PsAP2pGAD R (Supporting Table 1a). All pHIS2-NtAOX1a promoter constructs (carrying both or single cis element) and pGAD-PsAP2 constructs were co-transformed into yeast AH109 strain and the resulting transformants were selected on SD-His-Leu-Trp medium supplemented with 3-AT. For the reporter assay, same 651 bp 5′-UAS sequence of NtAOX1a gene (containing both DRE and GCC box element) was cloned between the Hind III and NcoI sites of pCAMBIA1305.1 with primers AOX1BY2F and AOX1BY2R (Supporting Table 1a). For construction of effector plasmid PsAP2 was cloned in XbaI and BamHI sites of binary vector pBI121 with primers PsAP2BY2F and PsAP2BY2R. PEG mediated protoplast co-transformation in BY2 cells was performed according to Mishra et al. (2013). Fluorometric GUS assays was performed as described by Berger et al. (2007). The results are based upon three independent protoplast co-transfection experiments.
Virus induced gene silencing of PsAP2 in P. somniferum
A fragment of 495 bp from PsAP2 was cloned in pTRV2 vector with primers PSTRVF and PSTRVR (Supporting Table 1d). The resulting construct and pTRV1 vector were transformed into Agrobacterium GV3101 strain. pTRV1vector and pTRV2-PsAP2 transformed Agrobacterium cultures were mixed in a ratio of 1:1 in presence of 0.05 % silwet. For mock treatment pTRV1 and pTRV2 vector transformed Agrobacterium cultures were used. The poppy seedlings were infiltrated at 6-8 leaf stage. Eight seedlings were infiltrated at a time. The agroinfiltration and VIGS was performed using the experimental procedure described in Hileman et al. (2005); Desgagné-Penix and Facchini (2012). Total RNA was isolated and cDNA was prepared from newly emerged leaflets after 2 weeks of agroinfiltration. Agroinfiltration was confirmed by qRT-PCR analysis using vector specific primers TRV2F, TRV2R, TRV1F and TRV1R. The level of PsAP2 and AOX1 was measured using primer pairs PsAP2RTF, PsAP2RTR, AOXPRTF and AOXPRTR, respectively. Actin was used as an internal control. The expression levels were monitored in five experimental plants with three different agroinfiltration.
Results
PsAP2 encodes an AP2/ERF class of plant transcription factor
The EST identified initially from the wound and salt stress library (Accession number AAP56252.1) was made full length by 3′/5′ RACE and named as PsAP2. The identified sequence of 1800 bp with an ORF of 1248 bp encodes a protein of 416 amino acids and a conserved AP2 domain of 57 amino acids. The nucleotide sequence aligned with its amino acid sequence is shown in Fig. 1a. Amino acid analysis of PsAP2 showed maximum homology with Populus trichocarpa AP2/ERF followed by DREB6 from Malus domestica. Phylogenetic analysis with other AP2/ERF proteins showed that PsAP2 falls in the same lineage of RAP2-4-like from Vitis vinifera and DREB from Populus euphratica (Fig. 1b).
The complete cDNA of PsAP2 with deduced protein sequence and phylogenetic tree. a The nucleotide and amino acid sequence is shown, translation start site is indicated by ‘M’ (methionine) and termination by ‘stop’. The AP2 domain is indicated by underline. b Maximum likelihood phylogenetic analysis of PsAP2 based on the alignment of their full-length coding sequences generated by MEGA5 software. Scale bar indicates 0.2 amino acid substitutions per site
Expression of PsAP2 in response to various stress and hormone treatments
The transcript level of PsAP2 under normal growth condition as analyzed by qRT-PCR was found to be maximum in floral bud followed by capsule and root (Fig. 2a). The expression pattern of PsAP2 was analyzed under various stress and hormone treatments at different time intervals. PsAP2 showed induction of 24 fold after 1 h of wounding and gradually decreased to 8 fold after 5th hour (Fig. 2b). After MeJA treatment it was showing induction of 2.8 and 2.7 fold at 3rd and 5th hour of treatment. Ethylene treatment lead to induction of 3.7 and 4.9 fold after 1 and 3 h of treatment followed by a decline in 5th hour with a fold induction of 3.4 fold (Fig. 2b). In dehydration and salt stress PsAP2 showed an induction of more than twofold after 5th hour. ABA treatment also induced PsAP2 expression more than 3 fold after 5th hour. PsAP2 showed initial induction of more than twofold after 1 h of cold stress followed by a gradual decrease in its level (Fig. 2b).
Relative expression of PsAP2. a The relative expression of PsAP2 in different tissues in comparison with leaf. b Expression of PsAP2 in 2 months old seedings of Papaver somnifreum after various stress treatments such as cold, dehydration, salinity, wounding, ABA and MeJA treatment. Transcript abundance was compared relative to control plant. Mean values were calculated from three biological replicates, error bars represent the standard deviation. The data presented were significant and represents mean of three independent experiments with SD having P value <0.01
PsAP2 binds to DRE and GCC box elements in vitro and transactivates the reporter gene in yeast
GRA was performed to study the binding preferences of PsAP2 protein towards DRE/CRT and GCC-box elements. Recombinant GST-PsAP2 purified protein showed gel shift with the fragment having DRE2 (core sequence-ACCGAC) and GCC box (core sequence-GCCGCC) unlike DRE1 cis-element (core sequence-GCCGAC). The results showed that PsAP2 was specific towards ACCGAC motif of DRE cis-element. Also PsAP2 was not able to produce gel shift with mutated DRE2 or GCC box demonstrating that PsAP2 binds specifically to DRE/CRT and GCC box as shown in (Fig. 3b). The binding of PsAP2 with both DRE and GCC box was competed out with (50×) excess cold probe. The oligos used for gel retardation assay with their consensus cis elements are shown in Fig. 3a.
PsAP2 binds with DRE and GCC box elements in in vitro and in vivo assays. a Consensus and mutated sequences of the cis elements are shown in the figure with mutated nucleotides represented in red color. b GRA representing the binding of PsAP2 with core DRE2 and GCC box cis elements. PsAP2 didn’t show any binding with mutated probes. c Transactivation assay of PsAP2. The colonies carrying pGBKT7-GAL4:PsAP2 construct can grow on YPD and SD-His-Trp minimal media while the vector transformed or wild type strains could not grow. LacZ reporter activation was also observed in pGBKT7-GAL4:PsAP2 transformed colonies. All the transformants could grow similarly on YPD media. d The relative assay of β-galactosidase reporter in strain, vector transformed strain and pGBKT7-GAL4:PsAP2 transformants. The β-galactosidase assay was performed with three independent transformants and were represented here with their standard deviation
Transactivation property of PsAP2 was demonstrated by the activation of reporter genes in yeast strain AH109 carrying His and Trp auxotrophic markers and lacZ reporter gene under GAL4 promoter. Gal4 DNA binding fused pGBKT7-PsAP2 construct was transformed in the yeast strains and growth of colony was observed on the minimal auxotrophic SD-His-Trp medium. Colonies containing PsAP2 were able to grow on medium lacking Trp and His unlike the wild type and vector transformed colonies (Fig. 3c). PsAP2 transformed colonies also showed β-galactosidase activity (Fig. 3d).
PsAP2 overexpression in tobacco increases tolerance to abiotic and biotic stresses
To establish the functional role of PsAP2 tobacco transgenic lines were developed. We have grown 11 independent lines up to maturity. However, only two of them were further taken up for analysis namely PsAP2-L7 and PsAP2-L2 containing a single copy of the gene as confirmed by Southern blotting (Fig. 4b). Constitutive induced expression of PsAP2 transcript was determined by semi-qPCR (Fig. 4a). For dehydration treatment PsAP2 T1 seeds were germinated on MS as well as MS + 0.4 M mannitol for 3 weeks. Growth and germination rate of transgenic as well as vector transformed seeds was similar on MS media but in mannitol added media the germination rate was significantly higher in transgenic lines (Fig. 4c and supporting Fig. 3). The percentage germination of seeds was analyzed after 21 days. It was 54 % for PsAP2-7 line and 42 % for PsAP2-2 line while the vector transformed seeds showed only 10 % in mannitol supplemented media (Fig. 4d). To analyse the salt stress response 2 weeks old PsAP2 along with vector transformed seedlings were transferred to MS + 250 mM of NaCl media and were allowed to grow for 1 week. Chlorosis appeared in vector transformed plants after 3 days of transfer and started appearing in transgenic lines from 5th day. More than 50 % of the vector transformed seedlings showed complete bleaching in comparison to PsAP2 overexpressing lines (Fig. 5a). Salt tolerance of vector transformed and PsAP2 overexpressing tobacco plants was additionally assessed by chlorophyll estimation in salt treated leaf discs (Supporting Fig. 2). The average chlorophyll content of vector transformed leaf discs was found to be less than PsAP2 overexpressing transgenic lines in both 250 and 400 mM NaCl solutions (Fig. 5b). Thus PsAP2 overexpressing transgenic plants were showing enhanced tolerance against osmotic shock of NaCl solution by reducing chlorosis.
PsAP2 over-expressing tobacco seedlings are more tolerant to dehydration stress. a Semi quantitative PCR analysis of PsAP2 overexpressing transgenic lines along with vector transformed tobacco plant. Actin was used as an internal control (accession number EB740770). b Southern blot analysis showing the single copy number of PsAP2 in both transgenic lines. c Survival rates of vector transformed and PsAP2 overexpressing transgenic seedlings in MS and MS + 400 mM mannitol. d Percentage germination of vector transformed as well as PsAP2 overexpressing transgenic tobacco lines in MS + 400 mM mannitol media. e Relative fresh weight of vector transformed and transgenic lines under 2, 4 and 6 days of dehydration treatment. Each data point is the mean of three independent experiments with their SD. The values are significantly different for PsAP2 transgenic plants in comparison with vector control with P value <0.05
PsAP2 over-expressing tobacco seedlings are more tolerant to salt stress. a Survival of vector transformed and PsAP2 overexpressing transgenic seedlings in MS + 250 mM NaCl. b Chlorophyll contents (mg/g fresh weight) were measured from NaCl-treated leaf discs of transgenic and vector transformed tobacco plants. c Relative fresh weight of vector transformed and transgenic lines under 2, 4 and 6 days of salt treatment. Each data point is the mean of three independent experiments with their SD. The values are significantly different in comparison with vector control with P value <0.05
Fresh weight of transgenic as well as vector transformed seedlings was measured after 2, 4 and 6 days of mannitol (0.4 M) and salt (250 mM) treatment. It was observed that relative fresh weight of transgenic lines was higher than the vector transformed lines (Fig. 4f). Upon 6 days of salt stress the relative fresh weight of vector transformed control seedlings was 51 % of those grown in stress free medium were as relative fresh weight of transgenic seedlings remained high i.e. approximately 80 % when compared with normal grown seedlings. Transgenic seedlings exposed to mannitol stress had almost 2 fold more relative fresh weight when compared with vector transformed seedlings. High relative fresh weight content of transgenic plants under stress condition highlights the better response of PsAP2 overexpressing plants in salt and mannitol stress compared with vector transformed plants.
To check the response of PsAP2 towards biotic stress, P. syringae cultures were infiltrated on the leaves of vector transformed and transgenic lines. The bacterial symptoms appeared after 2nd day post inoculation in vector transformed plants while in PsAP2 overexpressing lines it appeared on 4th day post-inoculation. The lesion area of bacterial infection was significantly less in PsAP2 overexpressing lines in comparison to vector transformed plants. Photographs were taken after 8th day post-inoculation of P. syringae (Fig. 6a). The relative percentage of lesion size was found to be more than 4 fold in vector transformed plants in comparison to PsAP2 overexpressing lines (Fig. 6b). The bacterial count CFU/cm2 was also found to be 3 fold less in PsAP2 overexpressing lines (Fig. 6c). The above results suggested that the PsAP2 overexpressing lines confer enhanced resistance towards bacterial pathogen P. syringae tabaci.
PsAP2 enhances tolerance to pathogen infection in tobacco. a Disease symptoms in vector transformed and PsAP2 transgenic plant leaves after 8 days post inoculation. b The average lesion area of each independent transgenic line was calculated and their relative lesion areas are shown in columns after comparison with the average lesion area on vector transformed plants. c CFU count in transgenic in comparison to vector transformed plants. d Relative expression of NtAOX1a and MIPS in transgenic and vector transformed plants. e Percent DPPH inhibition in vector transformed and overexpressing lines. f Percent NO scavenging in vector transformed and overexpressing lines. The data is significant and is represented from three independent experiment, with P value <0.05
PsAP2 significantly enhances NtAOX1a and MIPS transcripts and regulates antioxidant activity in transgenic tobacco plants
To identify the possible target genes of PsAP2, we analyzed expression level of 26 stress inducible marker genes of tobacco under control condition. Interestingly, only 2 genes were showing significant induced expression namely MIPS with an induction of more than 6 fold and NtAOX1a with an induction of more than 20 fold in PsAP2 overexpressing lines in comparison to vector transformed tobacco lines (Fig. 6d). As AOX1a is involved in ROS regulation we estimated the antioxidant properties of PsAP2 transgenic as well as vector transformed plants. When we measured the total antioxidant activity through DPPH percent scavenging assay, PsAP2 transgenic lines were showing enhanced activity in comparison to vector transformed lines (Fig. 6e). Similar result was obtained for NO scavenging activity (Fig. 6f). These results indicated that PsAP2 lead to enhanced abiotic and biotic stress tolerance may be by regulating antioxidant enzymes and associated genes.
PsAP2 transcriptionally regulates NtAOx1a by interacting with the stress responsive cis-elements present in the promoter
To understand the mechanism of induced expression of NtAOX1a transcript in PsAP2 overexpressing lines, we isolated the promoter of NtAOX1a gene by genome walking (Supporting Fig. 4 and 5). When both the constructs (pHIS-NtAOX1a and pGAD-PsAP2) were co-transformed in AH109 strain of S. cerevisiae, only those colonies that contained both the constructs were able to grow on SD-His-Leu-Trp minimal media supplemented with 3-AT. The cells carrying only pGAD-PsAP2 or pHIS-NtAOX1a or their truncated fragments (pHIS-NtAOX1a with only DRE or GCC element) or wild type strain could not grow on minimal medium (Fig. 7b). On the other hand all the transformants containing pGAD-PsAP2 alone or pHIS-NtAOX1a promoter regions and strain were able to grow on YPD media. The results lead to the confirmation that PsAP2 was able to activate the NtAOX1a promoter in in vivo condition.
PsAP2 binds with NtAOX1a promoter in in vivo and in vitro. a The sequences of the oligos from the promoter of NtAOX1a containing DRE and/or GCC cis elements used for GRA. b GRA showing binding of PsAP2 to the full length probe containing both DRE and GCC box element along with fragment of Nt AOX1P + GCC (Contains only GCC box), and NtAOX1P + DRE (contains only DRE element). The binding of PsAP2 with individual probes of NtAOx1a was competed out with excess of cold probe suggesting the interaction of PsAP2 with NtAOX1a promoter was specific. PsAP2 antisense clone was used as a negative control during GRA. c PsAP2 transactivates the His reporter gene by binding to the upstream promoter element of Nt-AOX1a (fragment carrying both the elements as well as the fragment carrying only DRE or GCC). Only PsAP2 or promoter fragments or strain could not grow on minimal media while all the transformants carrying both PsAp2 + NtAOX1a promoter fragments were able to grow on YPD
To further examine the direct interaction of PsAP2 with NtAOX1a promoter in vitro, we used one oligo dimer of 41 bp containing both the cis elements (DRE and GCC) and two oligo dimers of 39 bp having only single cis element (DRE or GCC) designed from NtAOX1a promoter (Fig. 7a, Supporting Fig. 5b). PsAP2 showed interaction with all the 3 probes, containing DRE and/or GCC box element/s but the interaction was stronger with the probe containing both DRE and GCC box cis elements (Fig. 7b). Specific binding of PsAP2 to NtAOX1a promoter was checked by excess 50× concentration of cold probe. Both Y1H and GRA results suggested that PsAP2 was able to bind specifically to NtAOX1a promoter and higher expression of NtAOX1a in PsAP2 transgenic plants might be because of transcriptional activation of NtAOX1a promoter.
To further validate the activation of NtAOX1a by PsAP2 we examined GUS reporter activity in tobacco BY2 cells. 651 bp promoter region of NtAOX1a was cloned in fusion with GUS reporter gene by replacing the CaMV35S promoter of pCAMBIA1305.1. This reporter construct was co-transformed with effector construct pBI121-PsAP2 in tobacco BY2 cells. For control pBI vector and NtAOX1a-pCAMBIA constructs were also co-transformed. The schematic representation of effector and reporter fragments is shown in Fig. 8a. GUS activity in the NtAOX1a-pCAMBIA and PsAP2 co-transformed BY2 protoplasts was more than 18 fold than that of the vector control and NtAOX1a-pCAMBIA transformed BY2 cells, showing that PsAP2 can activate transcription from the NtAOX1a-5′-UAS (Fig. 8b).
Reporter activation and VIGS of PsAP2. a The pictorial representation of effector and reporter constructs used for GUS reporter activation. b Activity of GUS reporter construct fused to NtAOx1a promoter sequence in tobacco BY2 cells harboring empty vector or PBI121-PsAP2. c qRT-PCR analysis of P. somniferum AOX1 and PsAP2 transcripts in leaves of plants subjected to silencing of PsAP2 using VIGS. d Percent NO scavenging in vector transformed and VIGS plants. The data is significant and is represented from three independent experiment, with P value <0.05
VIGS of PsAP2 downregulates the homologous AOX1 transcript in P. somniferum
Due to lack of functional mutant in P. somniferum, we have used VIGS to silence the PsAP2 transcript. We identified an another EST (accession number-GO238748) showing homology to the the tobacco AOX1 within the same wound inducible library from where we identified PsAP2 (Mishra et al. 2013). The relative expression level of PsAP2 along with the endogenous poppy AOX1a was found to be significantly reduced in PsAP2 silenced plants in comparison to only vector transformed plants (Fig. 8c). To further study whether silencing of PsAP2 and downregulation of AOX1a has any effect on the antioxidant properties of PsAP2-VIGS poppy plants, we estimated percent NO scavenging in vector control and VIGS plants. There was a decrease in the percent NO scavenging in the VIGS poppy plants in comparison to the control whereas there was a higher antioxidant activity in the PsAP2 over expressed tobacco plants (Fig. 8d). These results highlighted the significant role of PsAP2 in imparting abiotic and biotic stress tolerance through AOX1a and antioxidant regulation. It also indicated an conserved mechanism of action of PsAP2 and AOX1a in different plant species.
Discussion
AP2/ERFs regulate various processes like development, stress response and metabolism in plants (Okamuro et al. 1997; Yamaguchi-Shinozaki and Shinozaki 2006; Zhu et al. 2014). This ability of AP2/ERFs might be because of the evolved capacity of these proteins to bind to multiple cis elements present within the promoter of stress responsive genes (Park et al. 2001). We have isolated a novel AP2/ERF from P. somniferum and named it as PsAP2. At the protein level PsAP2 only showed maximum homology with AP2/ERF protein identified from Populus trichocarpa and it showed significant sequence homology within the EREBP/AP2 domain with other proteins, otherwise the amino acid sequences outside the DNA binding domain was quite unique. PsAP2 showed maximum expression in flower followed by capsule and root in comparison with leaf. Apart from stress response many AP2/ERFs are involved in plant developmental response also, so based on the PsAP2 tissue specific and basal level expression we assume it might have some direct or indirect role in normal plant growth or development. Stress induced transcript expression study has shown that, PsAP2 transcript is responsive to various stress and hormonal treatment and early induction highlighted the role of PsAP2 in first line of defense regulation. Interestingly, the induced expression of PsAP2 in response to wounding, ethylene and MeJA was much higher in comparison to dehydration, salt and ABA. This indicated the probable role of PsAP2 in the regulation of multiple simultaneous responses in plants. Unlike TSI1 from tobacco and ERF1 from Arabidopsis (Park et al. 2001; Cheng et al. 2013), PsAP2 showed late transcript accumulation in response to salt and dehydration stress. The transcript of PsAP2 showed induced expression even after 5th hour of MeJA, ABA and ethylene treatment showing that mode of action of PsAP2 transcript is different from TSI1 or AtERF1.
PsAP2 encodes a conserved AP2/EREBP DNA binding domain which binds to both DRE/CRT or GCC box elements. The DRE and GCC box are involved in abiotic and biotic stress responsive gene expression in plants which explains the better response of PsAP2 overexpressed lines under different stresses (Fujimoto et al. 2000; Cheng et al. 2013). We did not observe any difference in the affinity of binding of PsAP2 towards both DRE/CRT and GCC box cis element unlike TSI1 from Tobacco which showed stronger binding affinity towards GCC box cis element. This binding affinity may explain the involvement of PsAP2 in the regulation of both biotic as well as abiotic stresses. But PsAP2 showed preferential binding with the DRE core element having cis-element ACCGAC, however it was not able to produce gel shift with a core sequence of GCCGAC. For GCC-box element PsAP2 showed gel retardation with GCCGCC whereas it cannot produce a gel shift with ACCGCC. This specific and preferential binding of PsAP2 for DRE/CRT with core element ACCGAC and GCC box element with GCCGCC suggested that PsAP2 might function by same or different mechanism as compared to other characterized AP2/ERF proteins. Most of the AP2/ERF proteins characterized in response to both biotic and abiotic stresses in plants have induced level of stress related marker genes like ERD, PDF1.2, LEA and P5CS1 (Shukla et al. 2009; Zhu et al. 2014). To further understand the possible mechanism of stress tolerance of these PsAP2 transgenic lines we analyzed 26 stress inducible marker genes of tobacco. However the tobacco transgenic plants expressing PsAP2 did not show induced expression of ERD’s, which contains DRE/CRT box unlike it showed in CAP2, an ERF from chickpea, under normal growth condition (Shukla et al. 2006). Also like TSI1, ERF1 and other characterized AP2/ERF proteins, PsAP2 overexpressing lines did not show any induced expression of other stress related marker genes like LEA proteins. Out of 26 transcripts analyzed only Nicotiana tabacum AOX1a and MIPS showed constitutive induced expression in PsAP2 overexpressing tobacco transgenic plants. MIPS catalyzes the rate limiting step in myoinositol synthesis that is required for cell metabolism, plant growth and it acts as precursor for a variety of compounds (Kaur et al. 2008). MfMIPS1 transcript cloned from cold tolerant Medicago falcata showed induced expression in response to cold, dehydration and salt stress, in addition it also showed induced expression in response to H2O2 and NO. Over expression of MfMIPS1 in tobacco enhances the resistance of transgenic lines towards chilling, drought and salt stress (Tan et al. 2013). On the other hand NtAOX1a is nuclear encoded gene and generally induced rapidly in response to both abiotic and biotic stresses (Borecky et al. 2006). A number of studies in different plant species revealed that induced expression of NtAOX1a leads to different physiological response that are related to abiotic and biotic stress (Atkinson and Urwin 2012). During stress, reduced capacity of electron transport leads to slow relative ATP turnover which finally results in increased ROS production (Moller 2001). It is now evident that apart from ROS, reactive nitrogen species (RNS) are also generated in mitochondria. Alternative oxidase maintains both the ROS and RNS level under different stress condition in plants. Induced expression of MIPS and NtAOX1a in PsAP2 overexpressing transgenic tobacco plants, leads to increase in antioxidant activity which in tern provides better response of transgenic lines under different stress conditions. While in PsAP2-VIGS poppy plants there was a decrease in AOX1a transcript level leading to decrease in antioxidant property indicating similar mode of action of PsAP2 in poppy also, as it was observed in transgenic tobacco. So by maintaining the level of antioxidant enzymes, AOX1a and MIPS, PsAP2 might be able to regulate the level of RNS and ROS under stress condition leading to enhanced resistance against both abiotic and biotic stress responses. The induced expression of NtAOX1a and MIPS in PsAP2 transgenic tobacco plants, specific interaction and activation of NtAOX1a promoter highlights a new possible direction and involvement of an AP2/ERF protein in regulating the alternative oxidase gene and varied stress response.
Abbreviations
- 3-AT:
-
3-Amino-1,2,4-triazole
- AP2/ERF:
-
Apetela 2/ethylene response factor
- bp:
-
Base pair
- GRA:
-
Gel retardation assay
- GST:
-
Glutathione S-transferase
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- MeJA:
-
Methyl jasmonate
- MQ:
-
Milli-Q water
- MS media:
-
Murashige and Skoog media
- TF:
-
Transcription factor
- VIGS:
-
Virus-induced gene silencing
- Y1H:
-
Yeast one hybrid
- MIPS:
-
Myo-inositol-1-phosphate synthase
- AOX1a:
-
Alternative oxidase1a
References
Aono M, Kubo A, Saji H, Tanaka K, Kondo N (1993) Enhanced tolerance to photooxidative stress of transgenic Nicotiana tabacum with high chloroplastic glutathione reductase activity. Plant Cell Physiol 34:129–135
Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63:3523–3543
Berger B, Stracke R, Yatusevich R, Weisshaar B, Flügge UI (2007) A simplified method for the analysis of transcription factor–promoter interactions that allows high-throughput data generation. Plant J 50:911–916
Borecky J, Nogueira FT, de Oliveira KA, Maia IG, Vercesi AE, Arruda P (2006) The plant energy dissipating mitochondrial systems: depicting the genomic structure and the expression profiles of the gene families of uncoupling protein and alternative oxidase in monocots and dicots. J Exp Bot 4:849–864
Cagney G, Uetz P, Fields S (2000) High throughput screening for protein–protein interactions using the two-hybrid assay. Methods Enzymol 328:3–14
Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ethylene response factor1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162:1566–1582
Desgagné-Penix I, Facchini PJ (2012) Systematic silencing of benzylisoquinoline alkaloid biosynthetic genes reveals the major route to papaverine in opium poppy. Plant J 72:331–344
Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 3:393–404
Gelvin SB, Schilperoort RA (1994) Plant molecular biology manual. Kluwer, Dordrecht
Hileman LC, Drea S, Martino G, Litt A, Irish VF (2005) Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 44:334–341
Kaur H, Shukla RK, Yadav G, Chattopadhyay D, Majee M (2008) Two divergent genes encoding L-myo-inositol 1-phosphate synthase1 (CaMIPS1) and 2 (CaMIPS2) are differentially expressed in Chickpea. Plant, Cell Environ 31:1701–1716
Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262:1432–1436
Mishra S, Triptahi V, Singh S, Phukan UJ, Gupta MM, Shanker K, Shukla RK (2013) Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS ONE 1:52784
Moller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52:561–591
Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4:248
Okamuro JK, Caster B, Villarroel R, Montagu MV, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. PNAS 94:7076–7081
Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH (2001) Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 5:1035–1046
Patel RM, Patel NJ (2011) In vitro antioxidant activity of coumarin compounds by DPPH, super oxide and nitric oxide free radical scavenging methods. J Adv Pharm Educ Res 1:52–68
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory Manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Shukla RK, Raha S, Tripathi V, Chattopadhyay D (2006) Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant Physiol 1:113–123
Shukla RK, Tripathi V, Jain D, Yadav RK, Chattopadhyay D (2009) CAP2 enhances germination of transgenic tobacco seeds at high temperature and promotes heat stress tolerance in yeast. FEBS J 18:5252–5262
Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R (2014) Abiotic and biotic stress combinations. New Phytol 203:32–43
Tan J, Wang C, Xiang B, Han R, Guo Z (2013) Hydrogen peroxide and nitric oxide mediated cold- and dehydration-induced myo-inositol phosphate synthase that confers multiple resistances to abiotic stresses. Plant, Cell Environ 36:288–299
Wu L, Zhang Z, Zhang H, Wang XC, Huang R (2008) Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol 148:1953–1963
Xu ZS, Xia LQ, Chen M, Cheng XG, Zhang RY, Li LC, Zhao YX, Lu Y, Ni ZY, Liu L, Qiu ZG, Ma YZ (2007) Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol Biol 6:719–732
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, Ma Y (2009) Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot 13:3781–3796
Zhu X, Qi L, Liu X, Cai S, Xu H, Huang R, Li J, Wei X, Zhang Z (2014) The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol 3:1499–1514
Acknowledgments
The authors acknowledge Director, CSIR-Central Institute of Medicinal and Aromatic Plants, Lucknow, India for providing the necessary facilities. Author’s also acknowledge CIMAP-National Gene bank for providing Papaver somniferum cv. Sampada seeds and Arabidopsis Biological Resource Centre for providing pTRV1 and pTRV2 vectors. RKS acknowledge Department of Science and Technology, INDIA under Fast Track Project Number: SR/FT/LS-189/2009 for financial support. Sonal and Ujjal acknowledge UGC, New Delhi, India for providing fellowship.
Author contributions statement
SM, UJ and VT has performed most of the in vitro and in vivo experiments. DKS and SL has contributed towards antioxidant profiling of transgenic and vector transformed tobacco lines provided to them. RKS has designed and coordinated the work. RKS, SM, UJ, VT wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mishra, S., Phukan, U.J., Tripathi, V. et al. PsAP2 an AP2/ERF family transcription factor from Papaver somniferum enhances abiotic and biotic stress tolerance in transgenic tobacco. Plant Mol Biol 89, 173–186 (2015). https://doi.org/10.1007/s11103-015-0361-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0361-7