Abstract
Two fructan hydrolases were previously reported to exist in Jerusalem artichoke (Helianthus tuberosus) and one native fructan-β-fructosidase (1-FEH) was purified to homogeneity by SDS-PAGE, but no corresponding cDNA was cloned. Here, we cloned two full-length 1-FEH cDNA sequences from Jerusalem artichoke, named Ht1-FEH I and Ht1-FEH II, which showed high levels of identity with chicory 1-FEH I and 1-FEH II. Functional characterization of the corresponding recombinant proteins in Pichia pastoris X-33 demonstrated that both Ht1-FEHs had high levels of hydrolase activity towards β(2,1)-linked fructans, but low or no activity towards β(2,6)-linked levan and sucrose. Like other plant FEHs, the activities of the recombinant Ht1-FEHs were greatly inhibited by sucrose. Real-time quantitative PCR analysis showed that Ht1-FEH I transcripts accumulated to high levels in the developing leaves and stems of artichoke, whereas the expression levels of Ht1-FEH II increased in tubers during tuber sprouting, which implies that the two Ht1-FEHs play different roles. The levels of both Ht1-FEH I and II transcript were significantly increased in the stems of NaCl-treated plants. NaCl treatment also induced transcription of both Ht1-FEHs in the tubers, while PEG treatments slightly inhibited the expression of Ht1-FEH II in tubers. Analysis of sugar-metabolizing enzyme activities and carbohydrate concentration via HPLC showed that the enzyme activities of 1-FEHs were increased but the fructose content was decreased under NaCl and PEG treatments. Given that FEH hydrolyzes fructan to yield Fru, we discuss possible explanations for the inconsistency between 1-FEH activity and fructan dynamics in artichokes subjected to abiotic stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fructans, fructose-based oligo- and polysaccharides, are found in approximately 15 % of flowering plants, including Poaceae species [e.g., wheat (Triticum aestivum) and barley (Hordeum vulgare)], Liliales [onion (Allium cepa) and garlic (Allium sativum)], and Asparagales [chicory (Cichorium intybus) and Jerusalem artichoke (Helianthus tuberosus); (Livingston III et al. 2009; Vijn and Smeekens 1999)]. Inulin-type fructans consist of linear β(2,1)-linked fructofuranosyl units terminated by Glc and are found mostly in Asparagales, while levan-type fructan is a β(2,6)-linked fructose oligomer or polymer that is terminated by Glc and accumulates mainly in moncot species. Graminan-type fructan is both β(2,1)- and β(2,6)-linked, and is mainly produced by Poaceae species. The more complex neoseries-type fructan exists in Liliaceae (Edelman and Jefford 1964; Van Laere and Van den Ende 2002). Fructans are important ingredients in various food items and function as prebiotics that enhance the growth of beneficial colon bacteria (Ritsema and Smeekens 2003a).
The tuber of Jerusalem artichoke (H. tuberosus) stores inulin-type fructan with an intermediate degree of polymerization (DP; 3–35 fructosyl residues), which accounts for 85 % of the dry weight of the tuber (Edelman and Jefford 1968; Marx et al. 1997). In Jerusalem artichoke, fructans are generally synthesized from Suc in reactions catalyzed by sucrose:sucrose 1-fructosyl transferase (1-SST) and fructan:fructan 1-fructosyl transferase (1-FFT). Transgenic petunia (Petunia hybrida Vilm) plants expressing Ht1-SST accumulate more low DP fructans than the control plants (CK), and the offspring of a cross between 35S-1-SST-containing and 35S-1-FFT-containing plants have increased fructan content, showing that both enzymes function in fructan biosynthesis (Van Der Meer et al. 1998). To synthesize the more complex fructans found in other species, additional enzymes, such as fructan:fructan 6G-fructosyltransferase (6G-FFT) and sucrose:fructan 6-fructosyltransferase (6-SFT), are needed (Lasseur et al. 2006; Sprenger et al. 1995). Plant 1-FFT also plays a role in the fructan degradation pathway, since this enzyme can transfer the fructosyl from high DP fructans to low DP fructans. In addition to 1-FFT, fructan hydrolysis is mainly catalyzed by fructan exohydrolase (FEH), which releases Fru sequentially, finally leaving Suc. Suc can be further degraded by invertase (INV) to produce Fru and Glc.
At least three types of plant FEHs have been identified to date. 1-FEHs hydrolyze β(2,1)-linked fructans; 6-FEHs break down β(2,6)-linked fructans; and 6&1-FEHs degrade both β(2,1)- and β(2,6)-linked fructans. 1-FEHs have been extensively studied in plants such as chicory and Jerusalem artichoke, which mainly store inulin-type fructans. The first plant 1-FEH (1-FEH I) to be isolated was cloned from chicory through forward genetic methods in 2000 and two 1-FEH II (1-FEH IIa and IIb) isoforms from chicory were characterized in 2001 (Van den Ende et al. 2000, 2001). Since then, cDNAs of 1-FEHs from ryegrass (Lolium perenne), rusby (Vernonia herbacea), burdock (Arctium lappa), wheat, and Campanula rapunculoides have been reported (Asega et al. 2008; Le Roy et al. 2007b; Lothier et al. 2007; Ueno et al. 2011; Van den Ende et al. 2003a). In timothy (Phleum pratense), a 6-FEH was found to function in high DP levan hydrolysis (Tamura et al. 2011). 6&1-FEHs from wheat (T. aestivum) were found to selectively degrade small graminans (Kawakami et al. 2005). Two more specific 6-kestosidases (6-KEHs), which preferentially break down trisaccharide 6-kestose, were identified in wheat (Van den Ende et al. 2005). Interestingly, FEHs were also often present in some non-fructan plants; for example, 6-FEH was reported to exist in sugar beet (Beta vulgaris) and 1-FEHs in Arabidopsis (De Coninck et al. 2005; Van den Ende et al. 2003b). Even though it was established in the 1960s that at least two hydrolyses (A and B) that degrade linear inulin occur in Jerusalem artichoke (Edelman and Jefford 1964), only one hydrolase was purified to a single band by SDS-PAGE (Marx et al. 1997). No FEH cDNA sequences have hitherto been reported in Jerusalem artichoke.
Plant 1-SSTs, 1-FFTs, and FEHs are generally thought to originate from plant INVs. The primary structures of these proteins are highly similar and they are all assigned to the plant glycoside hydrolase family (GH) 32 (Lammens et al. 2009; Van den Ende et al. 2009). Plant INVs are further divided into two phylogenetic classes, i.e., the cell wall invertases (cwINV) and the vacuolar invertases (vacINV). Intriguingly, in the GH32 family, FEH and INV differ by only a single amino acid (Le Roy et al. 2007a; Van den Ende et al. 2009; Verhaest et al. 2007). Furthermore, vacINV of wheat can be converted into a 1-SST enzyme through the site-directed mutagenesis of one or two amino acids (Schroeven et al. 2008). A three amino acid mutation transformed ryegrass 6G-FFT to 1-SST (Lasseur et al. 2009). Even though FEHs, like their substrates, were often reported to occur in vacuoles, plant FEHs always cluster with cwINVs instead of vacINVs (De Coninck et al. 2005; Wagner and Wiemken 1986).
Fructans function not only as reserve carbohydrates in plants, but also play important roles in osmotic adjustments and the abiotic or biotic stress response, especially under cold conditions and defoliation (i.e., cutting or grazing). Fructans are largely degraded and FEH activity is dramatically increased in the petals of C. rapunculoides, and these changes are assumed to provide the driving force for osmotic adjustments during flowering (Le Roy et al. 2007b). Fructan breakdown is frequently associated with abiotic or biotic stresses, and is thought to supply the carbohydrates or energy needed to respond to these challenges. In the low temperatures that prevail in the late growing season or during prolonged storage, chicory and Jerusalem artichoke fructans are gradually hydrolyzed by FEHs (Marx et al. 1997; Van den Ende et al. 2000). Cold treatment increased the transcription of wheat 6&1-FEH w1 but not 1-FEH w2 at the crown, while defoliation caused chicory 1-FEH II, but not 1-FEH I, to accumulate, revealing that different FEHs played distinct roles (Kawakami et al. 2005; Van den Ende et al. 2001). In some cases, FEHs were even found to be involved in fructan biosynthesis instead of degradation, since they functioned as trimming enzymes that form special types of fructans (Bancal et al. 1992; Lothier et al. 2007; Van den Ende et al. 2003a).
In this study, two Ht1-FEHs were cloned and shown to have high levels of similarity with chicory 1-FEH I and 1-FEH II, based on their deduced amino acid sequences. Both Ht1-FEHs had a substrate preference for β(2,1)-linked fructans and demonstrated common FEH enzyme properties. Spatial–temporal expression analysis showed that Ht1-FEH I transcripts accumulated to high levels in developing leaves, stems, and roots, whereas Ht1-FEH II was up-regulated during tuber sprouting, implying that the two Ht1-FEHs have distinct functions. The transcripts of both Ht1-FEHs were significantly induced by NaCl, but only moderately induced or inhibited by PEG treatments. The enzyme activities of 1-SST and INV in tubers were slightly induced under salinity and drought conditions, but those of 1-FEH increased substantially. Interestingly, the level of Fru was reduced while that of Suc was increased under salinity or drought treatments compared to the untreated controls. We provide a possible explanation for this inconsistency between FEH enzyme activity and fructan dynamics.
Materials and methods
Plant material, growth conditions, and stress treatments
To study relative gene expression during the entire growth period, Jerusalem artichoke (Nanyu No. 1) tubers were planted in the beginning of March in experimental fields with soil of pH 7.3. The soil contained the following nutrient concentrations: organic matter, 8.36 g/kg; total nitrogen, 0.024 g/kg; total phosphorus, 0.024 g/kg; potassium, 0.134 g/kg; and salinity, 0.17 g/kg. Each seedling was provided with 100 g of organic cultivation matrix (Zhenjiang Xing Nong Organic Fertilizer Co., Ltd.) in late May. Six harvests were performed, with three replicates, at 5, 12, 50, 80, 110, 150, and 210 days after planting.
To determine the relative gene expression of growing plants exposed to abiotic stress, Jerusalem artichoke was vegetatively propagated as described (Huang et al. 2013). Seven days after tuber sprouting, Jerusalem artichoke plantlets were transferred to 12-cm pots containing moist acid-washed quartz sand, irrigated with 1/2 Hoagland solution (Hoagland and Arnon 1950), and grown in the greenhouse with the conditions of 16 h light (25 °C) and 8 h darkness (20 °C). Four-week-old plantlets of uniform size with three fully-expanded leaves and one expanding leaf were selected for salt and drought stress treatments. For the salinity treatments, the plantlets were each irrigated with 500 ml 1/2 Hoagland solution plus 50 or 100 mM NaCl, and for the drought treatments, the plantlets were supplied with 500 ml 1/2 Hoagland solution plus 15 % PEG6000. Plantlets irrigated with 500 ml 1/2 Hoagland solution were used as the control (CK). Roots, stems, and leaves were collected after 0, 2, 5, or 24 h of treatment. Tissue samples were harvested, rapidly frozen in liquid nitrogen, and stored at −80 °C until use. Each treatment was performed in biological triplicate.
To study relative gene expression, carbohydrate content, and enzyme activity in Jerusalem artichoke tubers exposed to abiotic treatments, the same sized artichoke tubers were selected, planted into pots containing moist acid-washed quartz sand, irrigated with 1/2 Hoagland solution in a greenhouse as above and then treated once with saturated 100 mM NaCl and 15 % PEG6000 at the beginning of tuber sprouting (5 days after planting). The tubers were incubated for one week (in the absence of further watering) under conditions of 16 h light (25 °C) and 8 h darkness (20 °C). Samples sprayed with the same volume of water at the start of the week were used as the CK. Tuber samples were collected at 0, 0.5, 1, 2, 3, 5, and 7 days after treatment (DAT) for analysis. Each treatment was performed in biological triplicate.
Total RNA isolation and cDNA synthesis
Total RNA was extracted from each sample using an E.Z.N.A. Plant RNA Kit (Omega Biotek, R6827-02) according to the manufacturer’s instructions. Then, total RNA samples were treated with an RNase-Free DNase Set (Omega Biotek, E1091-02) to remove residual genomic DNA. cDNA was synthesized using a PrimeScript@ 1st Strand cDNA Synthesis Kit (Takara Code, D6110A), following the manufacturer’s instructions.
Cloning of 1-FEH cDNA and bioinformatic analysis
ESTs (Expressed Sequence Tags) of Jerusalem artichoke were downloaded from the NCBI website to construct a local database as described previously (Huang et al. 2013). Previously identified 1-FEH cDNA sequences from chicory and V. herbacea were used as queries to local BLAST this EST database. EST sequences were assembled to yield two candidate genes, which were cloned for further bioinformatic analysis.
Jerusalem artichoke 1-FEH sequences were deposited into NCBI GenBank, under accession numbers KJ946352 and KJ946353 for Ht1-FEH I and Ht1-FEH II, respectively. 1-FEHs from chicory, Vernonia, and wheat were retrieved from GenBank for sequence alignment analysis using the ClustalW method in the MegAlign program (DNASTAR. Lasergene.v. 7.1), and then crested with CLC Sequence Viewer 6.8 software (http://www.clcbio.com/products/clc-sequence-viewer/; Liang et al. 2014). Phylogenetic analysis of the 1-FEH protein sequences was then performed using the Neighbor-Joining method in the MEGA (version 5.1) program (Liang et al. 2012).
Construction of expression vector and expression of recombinant protein in Pichia pastoris X-33
The isolated cDNA was expressed in the methylotrophic yeast P. pastoris X-33 with the secretory expression vector pPICZαC (EasySelect Pichia Expression Kit, Invitrogen, USA; Wang et al. 2011). Pichia pastoris X-33 was grown and maintained in YPD or YPDS medium (Resina et al. 2004). The DNA sequence corresponding to the putative mature protein region predicted by SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) was amplified using the primers Pic-Ht1-FEH-F/R (Supplementary Table S1). The amplified fragment was digested with Xho I and Xba I (TaKaRa), and ligated into pPICZαC behind the α-factor signal sequence. To express the protein with a native N-terminus, the Xho I site was used to clone Ht1-FEH genes flush with the Kex2 cleavage site (AAAAGA). Proper construction was confirmed by restriction digestion and DNA sequencing, and the resulting vector was designated as pPICZαC-FEH. P. pastoris strain X-33 was transformed by electroporation (Gene Pulser Xcell™ Electroporation System 165-2660; Bio-Rad, Richmond, CA) using 5–10 µg of the SacI-linearized construct, and transformants were selected on YPDS plates containing 100 μg ml−1 Zeocin™. P. pastoris X-33 transformed with pPICZαC was used as a CK.
A 50-ml preculture medium (BMGY) was inoculated with a freshly prepared single colony and cultured at 30 °C with vigorous shaking (200 rpm) until the culture reached an OD600 of 2–6 (after approximately 20–24 h). The cells were collected by centrifugation at 3,000g for 5 min at room temperature, the cell pellet was resuspended to an OD600 of 1.0 in 100 ml of inoculation medium (BMMY), and the resulting suspension was incubated at 30 °C with shaking at 200 rpm. Methanol was added (to a final concentration of 2 %) to the culture medium every day to maintain induction. After 120 h of induction, the culture was centrifuged at 5,000g for 10 min at room temperature and the resulting supernatant was recovered and tested for enzyme activity. The supernatants were rapidly frozen in liquid N2 and stored at −80 °C until ready to assay.
Purification of recombinant protein and SDS-PAGE and LC–MS/MS analyses
Ht1-FEHs were expressed in yeast P. pastoris X-33 with a 6 × His-tag at the C-terminus to facilitate the purification of recombinant protein through Ni2+ -charged column chromatography. Recombinant proteins were purified as described (Paula de Mattos Arêas et al. 2002). Briefly, the expressed 6 × His-tagged proteins were purified through an affinity chromatography column filled with 1 cm Ni–NTA resin (TransGen, Beijing, China). After adsorption of the protein, the resin was washed with five volumes of binding buffer containing 200 mM imidazole. The fractions collected after purification were concentrated by ultrafiltration on a Vivaspin 500 with a cut-off of 30 kDa (Sartorius Stedim Biotech). All purification steps were performed at 0–4 °C.
The recombinant crude enzymes and concentrated proteins were analyzed by SDS-PAGE, on an 8 % polyacrylamide gel that was stained with Coomassie Brilliant Blue-R250. For LC–MS/MS analysis, protein bands were excised from the gel, washed, S-alkylated, and in-gel digested using a protocol based on a previous report (Liu et al. 2013). The samples were desalted by C-18 ZipTip (Millipore, Brussels, Belgium) chromatography. A liquid chromatography–mass spectrometry (LC–MS) system consisting of a Dionex Ultimate 3000 Nano-LC unit (nano UHPLC, Sunnyvale, CA, USA) connected to a Linear Quadrupole Ion Trap Orbitrap (LTQ Orbitrap XL) Mass Spectrometer (ThermoElectron, Bremen, Germany), and equipped with a nanoelectrospray ion source, was used. For LC separation, an Acclaim PepMap 100 column (C18, 3 μm, 100 l) (Dionex, Sunnyvale, CA, USA) capillary with a 15-cm bed length was used with a flow rate of 300 nl/min. Two solvents, A (0.1 % formic acid) and B (aqueous 80 % acetonitrile in 0.08 % formic acid), were used to elute the peptides from the nanocolumn. The gradient was increased from 4 to 50 % B in 85 min and from 50 to 98 % B in 5 min, with a total run time of 125 min. The mass spectrometer was operated in the data-dependent mode so as to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. Electrospray voltage and the temperature of the ion transfer capillary were 2.2 kV and 200 °C, respectively. Survey full-scan MS spectra (from m/z 350 to 1,800) were acquired in the Orbitrap with a resolution of r = 60,000 at m/z 400, allowing for the sequential isolation of the top ten signal intensity ions for collision-induced dissociation at a collision energy of 35 V. A dynamic exclusion mode was enabled to exclude the previously selected ions during the repeated cycle of 60 s. The external mass calibration of the Orbitrap was performed once every three days to ensure a working mass accuracy of <5 ppm. The obtained MS/MS spectra were searched against self-defined databases (the deduced amino acid sequences of Ht1-FEH I and Ht1-FEH II) using the SEQUEST algorithm run in Proteome Discoverer 1.3 software (Thermo Scientific, San Jose, CA, USA). Search results were filtered for a false discovery rate of 1 % using a decoy search strategy and a reverse database.
Characterization of purified recombinant enzymes
To determine the substrate specificity and the effects of pH and temperature, the enzyme amount and incubation time were adjusted to achieve the linear production of Fru during the incubation period (Asega et al. 2008; Lothier et al. 2007; Tamura et al. 2011). A reaction mixture (500 µl) consisting of purified recombinant enzymes and substrates in 50 mM sodium phosphate buffer (pH 6.0) was incubated at 30 °C in a water bath. The reaction was stopped by heating at 95 °C for 5 min. To determine the optimal pH for recombinant enzyme activity, the activity was evaluated at pH 4.0–6.0 (50 mM sodium acetate buffer) and pH 6.0–7.0 (50 mM sodium phosphate buffer) at 30 °C for 10 h. To determine the optimal temperature for enzyme activity, the activity was monitored at pH 6.0 from 4 to 50 °C overnight. Inulin (Sigma, St Louis, MO), levan (Sigma, from Erwinia herbicola), 1-kestose, nystose, 1F-fructofuranosylnystose (Wako Pure Chemical Industries, Ltd), and sucrose were used as substrates. The reaction products were analyzed with an Agilent 1200 HPLC (Agilent Technologies, PaloAlto, CA, USA), a Prevail™ Carbohydrate ES column-W250*4.6 mm 5um (Alltech), and an ELSD 3300 detector (Alltech) at a flow rate of 1.0 ml/min (Li et al. 2014). The mobile phases (A and B) were acetonitrile and distilled water, respectively. The column was equilibrated in 75 % mobile phase A as described in Li et al. (2014) with some modifications, and the system was as follows: 0–10 min linear gradient to 65 % A, 10-20 min linear gradient to 50 % A, 20–25 min of 50 % A, 25–26 min linear gradient to 75 % A, and 26–35 min of 75 % A. The activity of recombinant enzyme was determined as µmol product mg−1protein min−1. Purified enzyme solutions were used to determine the protein concentration using a BCA Protein Assay Kit (AURAGENE, P001B-1) with BSA as standard, according to the manufacturer’s instructions.
Gene expression analysis by quantitative real-time RT-PCR
Quantitative PCR (qPCR) assays and data analysis were performed as described (Huang et al. 2013). The levels of Ht1-FEH I and Ht1-FEH II transcripts were quantified by real-time RT-PCR using a 7500 Real Time PCR System (Applied Biosystems) and SYBR Premix Ex Taq™ (Takara, DRR420A) according to the manufacturer’s instructions. Data were processed using the 2−ΔΔCT method; that is, the expression level of Ht1-FEH I and II relative to the Jerusalem artichoke housekeeping gene actin (the internal reference gene) was calculated (Huang et al. 2013) and adjusted against the expression levels of untreated samples, which were set to 1.0. Three biological and technical repeats were performed for each sample.
Protein extraction and measurements of sugar-metabolizing enzyme activity
Ten grams of fresh tuber was crushed in a mortar and pestle in liquid nitrogen and mixed with three volumes of ice-cold 50 mM Na-acetate buffer (pH 5.0, also containing 1 mM phenylmethylsulfonyl fluoride, 10 mM NaHSO3, 1 mM β-mercaptoethanol, 100 mM mannitol, 0.02 % (w/v) Na-azide, and 0.1 % (w/v) PVPP) for 2–3 h on ice, and the homogenate was centrifuged at 10,000g for 5 min at 4 °C (Joudi et al. 2012). The supernatant was then mixed with 516 mg solid (NH4)2SO4 per ml, gently stirred, and kept on ice overnight. The precipitates were collected by centrifugation at 10,000g for 5 min, and washed two times with 20 ml of 80 % (NH4)2SO4 (v/v). After a second centrifugation at 10,000g for 5 min, the precipitate was dissolved in 2.5 ml of 50 mM Na-acetate buffer, pH 5.0, containing 0.02 % (w/v) Na-azide.
The enzyme extracts obtained were used to determine the protein concentration using a BCA Protein Assay Kit. as described above. The activities of 1-FEH (EC 3.2.1.153), INV (EC 3.2.1.26), 1-SST (EC 2.4.1.99), and 1-FFT (EC 2.4.1.100) were measured as described by Joudi et al. (2012) and Vandoorne et al. (2012) with some modifications. The enzymes 1-SST and INV were assayed in the same reaction mixture, which contained 500 µl enzyme solution, Na-acetate buffer (pH 5.0), and 100 mM Suc at 30 °C. The 1-FEH enzyme was incubated with 1 % inulin at 30 °C and 1-FFT enzyme was incubated with 10 mM 1-K on ice. All reactions were incubated overnight and stopped by incubating the samples at 95 °C for 5 min. The enzyme activities were determined as nmol product gFW−1 min−1 (1-K for 1-SST, Nys for 1-FFT, and Fru for 1-FEH and INV) with HPLC, according to the procedures described above.
Analysis of carbohydrate content
Fresh Jerusalem artichoke tubers were dried in a thermomixer (CIMO, DHG-9143BS-III) for 48 h at 80 °C, and 0.5 g of dry sample was ground into powder, mixed with 7 ml distilled water, and incubated in a water bath at 90 °C for 60 min. After cooling to room temperature, the extract was passed through filter paper and the effluent was collected through the 0.45-μm membrane filter. Fructans in the extract were separated and a 10-µl solution was used for HPLC analysis as above (Li et al. 2014). Quantification of the peak areas was performed using the external standards method for Fru, Glc, Suc, 1-K, Nys, and 1F-F.
Primers
Primers for gene cloning, construction of the expression plasmid, and qPCR are listed in Supplementary Table S1.
Statistical analysis
One-way ANOVA using Dunnett’s t test was performed on gene expression levels using SPSS 19.0 (SPSS Corp., Chicago, IL, USA) to evaluate internal significant differences among treatments. Figures displaying enzyme activities, gene expression levels, and fructan contents were created using SigmaPlot 10.0 (Systat Software, Inc., Germany).
Results
Cloning of candidate 1-FEH cDNAs from Jerusalem artichoke
The previously reported coding sequences of chicory and V. herbacea 1-FEHs were used to BLASTN the Jerusalem artichoke EST database and several ESTs with high levels of amino acid sequence similarities were retrieved. The ESTs were assembled by SeqMan™ (DNASTAR) into two contigs. Contig 1 consisted of 1,825 bases and was assembled from six ESTs (GE493228, GE497420, BU028179, DQ012383, GE497421, and GE493229) that included an ORF (open reading frame) of 1,683 bases. We named this clone Ht1-FEH I (KJ946352) and deduced that it encoded a protein of 560 amino acids. The second contig, which was 2,061 bp long and was assembled from twelve ESTs (DY909202, EL414360, DY907342, DY910053, DY910436, DY910407, EL415379, EL420098, EL439056, EL458987, EL461979, and EL433220), was designated Ht1-FEH II (KJ946353). This clone included an ORF of 1,746 bases and was deduced to encode a protein of 581 amino acids. According to the SignalP4.1 program, the first 25 amino acids of both Ht1-FEH I and Ht1-FEH II were predicted to be the signal peptide for targeting the proteins to the endoplasmic reticulum. Also, both ORFs contained three conserved amino acid motifs (S/NDPNG, RDP, and EC) that are essential for β-fructosidase activity (Verhaest et al. 2005; Fig. 1).
Alignments of the deduced amino acid sequences of Ht1-FEH I and Ht1-FEH II from H. tuberosus with FEHs from C. intybus (Ci I, AJ242538; Ci IIa, AJ295033; and Ci IIb, AJ295034) and V. herbacea (AM231149). The β-fructosidase NDPNG, FRDP, and WECPD motifs are boxed and the single amino acid ‘P’ is marked in red bold in the conserved WECPD motif. Parallelograms indicate a tryptophan residue in the GSAT region and a serine in the YTG region. Potential N-glycosylation sites are also boxed (ovals for Ht1-FEH I and diamonds for Ht1-FEH II). The first amino acid of the putative mature protein of Ht1-FEH I and Ht1-FEH II are indicated with an arrow and black inverted triangle, respectively. In the consensus line, uppercase letters represent identity in more than 50 % of sequences and X represents less than 50 % identity
The deduced Ht1-FEH I protein was predicted using DNASTAR software (Lasergene.v7.1) to have a pI of 6.65, molecular mass of 62.8 kDa, and seven potential N-glycosylation sites (N-X-S/T; http://www.cbs.dtu.dk/services/NetNGlyc/), with the 73rd and 399th amino acid positions having the highest possibility of glycosylation, i.e., 0.7141 and 0.6383, respectively. Ht1-FEH II was predicted to have a pI of 4.87 and a molecular mass of 65.1 kDa, and to contain four N-glycosylation sites (N-X-S/T; the first site is irrelevant, because it is removed when the presequence is excised; Fig. 1). The full-length deduced amino acid sequences of Ht1-FEH I and II were aligned with 1-FEHs from other species (Fig. 2). The sequence of Ht1-FEH II exhibited high levels of identity with that of C. intybus 1-FEH IIa (81.4 %), C. intybus 1-FEH IIb (80.3 %), and V. herbacea 1-FEH (80.8 %), but only 51.1 % identity with C. intybus 1-FEH I. Ht1-FEH I showed 76.0 % identity with C. intybus 1-FEH I.
Phylogenetic tree of protein sequences of plant fructan metabolism genes and vacuolar and cell wall invertases. Group I, fructan exohydrolases (FEH) and cell wall invertases (cwINV) and Group II, fructosyltransferases (SST, FFT) and vacuolar invertases (vacINV). Ht1-FEH I and Ht1-FEH II are in red bold font. GenBank accession numbers of the genes used in this analysis are given after the gene and species abbreviation. Abbreviations for the species are: Ta Triticum aestivum, Lp Lolium perenne, Bp Bromus pictus, Pp Phleum pratense, Ci Cichorium intybus, Vh Vernonia herbacea, Ht Helianthus tuberosus, At Arabidopsis thaliana, Cr Chenopodium rubrum, Vf Vicia faba, Ps Pisum sativum, Dc Daucus carota, Vr Vigna radiata, and Bo Brassica oleracea
Purification, SDS-PAGE, and LC–MS analysis of the recombinant proteins
Two coding sequences containing the predicted mature protein region of Ht1-FEH I and II (Fig. 1) but lacking the signal peptides were ligated in-frame behind the α-signal sequence and heterologously expressed in the methylotrophic yeast P. pastoris X-33 to yield a recombinant protein with a 6 × His-tag at its C-terminus. This expression system is now recognized as being a valid technique for determining the functionality of plant FEHs (De Coninck et al. 2005). Hydrolytic activity was detected in the culture medium of both of the transformed P. pastoris lines when using 1-K as a substrate. However, no 1-FEH activity was detected in the culture medium of the CK, which was transformed with the empty vector pPICZαC under the same culture conditions (data not shown).
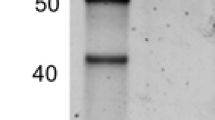
SDS–PAGE revealed that both recombinant Ht1-FEH I and Ht1-FEH II proteins had a mass of approximately 90 kDa (data not shown), thus exceeding the predicted molecular masses (62.9 and 65.1 kDa of recombinant Ht1-FEH I and Ht1-FEH II, respectively, without the signal sequence length, but including the length of the c-myc epitope and polyhistidine tag) and the 79 kDa of the native 1-FEH purified from Jerusalem artichoke tubers (Marx et al. 1997). Discrepancies between observed and predicted molecular masses were commonly found for other plant FEHs (Tamura et al. 2011; Ueno et al. 2011; Van den Ende et al. 2000). LC–MS/MS analysis showed that 59 and 47 peptides passed the XCorr Score versus Charge criteria and exhibited 79.46 and 51.98 % coverage of the deduced Ht1-FEH I and II sequence, respectively (Table S2 and S3). Moreover, no higher-score fragment matches were found in the NCBI database. From these results, the 90-kDa bands were determined to be the recombinant Ht1-FEHs.
General properties of recombinant Ht1-FEH I and II enzymes
The optimal pH for recombinant Ht1-FEH I and Ht1-FEH II activity using 1-K as substrate was 6.0 (Fig. 3a), which agreed well with the optimal pH of native 1-FEHs from Jerusalem artichoke tubers (Edelman and Jefford 1964; Marx et al. 1997). The optimal temperature for recombinant Ht1-FEH I and Ht1-FEH II activity was 35 °C (Fig. 3b). Substrate specificities were investigated using different types of fructan to characterize the properties of the recombinant Ht1-FEHs in detail (Table 1). Ht1-FEH I and II showed high levels of activity against β(2,1)-type fructans, such as 1-K, Nys, 1F-F, and inulin. None or very low levels of hydrolysis activity were detected towards Suc and high DP levan, which are mainly linked by β(2,6) bonds. When a mixture of 1-K and Suc was used as a substrate, the activities of Ht1-FEH I and II were significantly lower than when 1-K alone was used as the substrate. The addition of more Suc increasingly inhibited the activities of the recombinant enzymes, with 13 % of Ht1-FEH I inhibition occurring at 10 mM and 49 % at 100 mM Suc, and 21 % inhibition of Ht1-FEH II occurring at 10 mM and 65 % at 100 mM Suc. The substrate specificities were in accordance with the results of previous studies of native Ht1-FEHs (Edelman and Jefford 1964; Marx et al. 1997); hence, we concluded that the enzymes were neither a classic INV nor a 6-FEH, but were 1-FEHs.
Effect of pH and temperature on the activities of the purified recombinant Ht1-FEHs. Results are shown as values relative to the maximum rate of fructose release after incubation of recombinant Ht1-FEH I or Ht1-FEH II with 10 mM 1-kestose and treatment with various pHs (a) and temperatures (b). Data are mean ± SD, n = 3
While measuring Ht1-FEH activity by HPLC, Fru and Suc were produced in the reaction mixture of 1-K and the individual 1-FEH enzyme. When Nys was incubated with the enzyme, Fru and 1-K were produced and Suc was also produced by prolonged incubation. Moreover, Fru and Nys were produced when 1F-F was incubated with the enzyme and 1-K was also produced after prolonged incubation. In reaction mixtures with inulin, Fru and some low DP fructans were simultaneously produced. These results demonstrated that Ht1-FEH I and Ht1-FEH II are capable of degrading each β(2,1) type substrate via an exo-type of cleavage, thereby sequentially releasing terminal fructosyl residues, as are C. intybus 1-FEH I (Van den Ende et al. 2000), C. intybus 1-FEH IIa/b (Van den Ende et al. 2001), and A. lappa L. AlEH1 (Ueno et al. 2011).
Relative expression of Ht1-FEH I and II throughout the growth period
Since the fructan-β-fructosidase activities in tubers of Jerusalem artichoke are low in growing tubers but increase during sprouting (Marx et al. 1997), Ht1-FEH I and II transcription was analyzed in Jerusalem artichoke at various growth stages, including tuber sprouting and seedling development (Fig. 4). In the period of tuber sprouting (about 5–50 days after planting, DAP), Ht1-FEH II had relatively higher levels of expression in the tubers than at other stages, while Ht1-FEH I had low levels of expression. Ht1-FEH I showed strongest expression in the stems at 80 DAP and in the source leaves at 110 DAP, and its expression was higher in the stems and source leaves at 150 DAP than in other tissues. By contrast, the expression levels of Ht1-FEH II were low during the growing period compared to the tuber-sprouting period. Both Ht1-FEH I and Ht1-FEH II expression levels were relatively low in mature tubers at 210 DAP.
Relative gene expression of Ht1-FEH I and II in various organs throughout development. Values represent mean ± SE of three biological replicates. The expression level of Ht1-FEH I (a) and Ht1-FEH II (b) in 5-DAP tubers was set to 1.0. Significant differences between different developmental samples and 5-DAP tubers are indicated by •(P < 0.05) or *(P < 0.01)
Relative expression of Ht1-FEHs in growing plants under abiotic stresses
Numerous reports show that fructan dynamics are altered during the plant’s abiotic or biotic stress response (Hendry 1993; Livingston and Henson 1998; Pilon-Smits et al. 1995; Tamura et al. 2011; Valluru and Van den Ende 2008; Vandoorne et al. 2012). To determine if FEHs are involved in the plant’s response to abiotic stresses, we monitored the expression profiles of two 1-FEHs in growing plants subjected to abiotic stress for 24 h when all mother tubers had been absorbed (Fig. 5). Ht1-FEH I transcripts were abundant in NaCl-treated stems and leaves, but not in roots, and the expression levels peaked in stems after 24 h of treatment with 100 mM NaCl. Similarly, a 100 mM NaCl treatment caused a significant increase in the expression of Ht1-FEH II in stems at 2 h and in roots at 5 h compared to the CK. Furthermore, Ht1-FEH I showed higher expression in roots and stems after 24 h of 15 % PEG6000 treatment. Ht1-FEH II expression was up-regulated in all tissues after 2 or 5 h of drought treatment.
Relative gene expression of Ht1-FEH I and II the roots, stems, and leaves of plants subjected to a 24-h salt or drought stress treatment. Ht1-FEH I (b, c) and Ht1-FEH II (b, d) expression in the roots, stems, and leaves under 50 mM, 100 mM NaCl (a, b) and 15 % PEG6000 (c, d) treatments for 24 h at the stage when tubers were completely absorbed. Samples not subjected to treatment were used as the control (CK) and tissue samples were collected after 0, 2, 5, and 24 h of treatment. The expression levels were calculated by dividing the expression level of each gene by the corresponding gene expression level in the untreated control at each time point. Significant differences between stress-treated and control plants are indicated by •(P < 0.05) or *(P < 0.01)
We also monitored Ht1-FEH I and II expression in sprouting Jerusalem artichoke tubers subjected to a one-week treatment with 100 mM NaCl and 15 % PEG6000 (Fig. 6). Ht1-FEH I expression was slightly increased from 3 days after treatment (DAT) in tubers, and peaked at 5 DAT under 100 mM NaCl treatment. Ht1-FEH I expression was slightly increased under drought treatment. Ht1-FEH II expression was slightly increased in tubers under NaCl conditions, but had low levels of expression after two and three of treatment with 15 % PEG6000 when compared to the CK.
Relative gene expression of Ht1-FEH I and Ht1-FEH II in tubers subjected to one week of stress treatment. Five-day-old Jerusalem artichoke tubers were treated with 100 mM NaCl or 15 % PEG6000 for 0, 0.5, 1, 2, 3, 5, and 7 days, respectively. Samples lacking treatment at each time point were used as the control (CK). Values represent mean ± SE of three biological replicates. The expression levels were calculated by dividing the expression level of each gene by the corresponding gene expression level in the untreated control at each time point. Significant differences between stress treatments and control plants are indicated by •(P < 0.05) or *(P < 0.01)
Sugar-metabolizing enzyme activities and carbohydrate concentration during tuber sprouting
Previous studies demonstrated that 1-FEH enzyme activities were highly correlated with their gene expression, suggesting that 1-FEHs are mainly subjected to transcriptional regulation (Asega et al. 2011; Chalmers et al. 2005; Ritsema and Smeekens 2003b; Van Laere and Van den Ende 2002). We thus measured the activities of sugar-metabolizing enzymes and the sugar content during salinity and drought treatments.
Since the ratio of dry weight/fresh weight (DW/FW) of Jerusalem artichoke tubers subjected to various stress treatments (the ratio ranges between 15 and 22 %) did not vary much, all enzyme activities were expressed as the yield of product per unit of fresh mass.
The activities of fructan-metabolizing enzymes in Jerusalem artichoke tubers all increased significantly at 1 DAT in the unstressed control group, with the exception of 1-SST (Fig. 7). The activity of 1-FEH was relatively stable under both abiotic conditions, while 1-FEH activity in the CK gradually decreased after 1 DAT, and reached a minimum value at 7 DAT (Fig. 7a). INV activity significantly increased at 1 DAT in the CK, but greatly decreased after that point. Under drought stress, INV activity gradually decreased. Under salinity treatment, INV activity fluctuated during a 7-d period, but also generally decreased compared to the 0-d samples (Fig. 7b). Both 1-FEH and INV activities were lower in the CK than in salinity- or drought-treated tubers after 3 DAT. For fructosyltransferase activities, 1-SST (Fig. 7c) and 1-FFT (Fig. 7d) showed different trends. The activity of 1-SST started to decrease after 2 DAT both under CK conditions and abiotic stress compared to 0-d CK, but 1-SST activity was slightly higher than the CK under drought conditions. The 1-SST activities were highest in the CK at 1 DAT. 1-FFT activity showed no marked differences in all experimental plants during 2-7 DAT, except that it increased transiently at 1 DAT in salinity-treated and untreated CK samples.
1-FEH, INV, 1-SST, and 1-FFT activities in sprouting tubers subjected to treatment with 100 mM NaCl or 15 % PEG6000 for one week. Samples without any treatment were used as the control (CK). Tuber samples were collected at 0, 0.5, 1, 2, 3, 5, and 7 days after treatment. The enzyme reaction products were detected by HPLC and the enzyme activities of 1-FEH (a), INV (b), 1-SST (c), and 1-FFT (d) were determined as nmol product gFW−1 min−1 (the product: 1-K for 1-SST, Nys for 1-FFT, and Fru for 1-FEH and INV). Values are mean ± SE (n = 3)
HPLC analysis of fructan during the 7-d stage was performed for each time-point, but results of the sugar profiles are only shown at 0 and 7 DAT (Fig. 8) since the sugar profiles in the other days were similar to 7 DAT. The content of six sugars at each time point is quantified in Supplementary Fig. S1. During tuber sprouting, a significant increase in Fru and reduction in Suc and intermediate DP fructans (e.g., 1-K, Nys, 1F-F) were observed at 7 DAT in the CK (Fig. 8b and Supplementary Fig. S1), when compared to the CK at 0 DAT (Fig. 8a). Interestingly, the Suc content increased at 7 DAT under stress treatment, especially in plants treated with 15 % PEG6000 (Fig. 8d and Supplementary Fig. S1C) when compared to CK plants at both 0 and 7 DAT. The Glc content was very low compared to that of other sugars throughout the experiment (Supplementary Fig. S1B).
Fructan profiles in sprouting tubers subjected to stress conditions. Tubers were analyzed by HPLC at 0 (a CK) and 7 (b CK; c 100 mM NaCl treatment; d 15 % PEG6000 treatment) days after treatment. Abbreviations for each sugar peak are: F fructose, G glucose, S sucrose, 1-K 1-kestose, Nys nystose, and 1 F -F 1F-fructofuranosylnystose. Fructose, glucose, sucrose, 1-kestose, nystose, and 1F-fructofuranosylnystose were quantified in Fig. S1
Discussion
Ht1-FEH I and Ht1-FEH II have high levels of sequence identity with chicory 1-FEH I and 1-FEH II and similar enzymatic characteristics to Jerusalem artichoke hydrolase A and B
In this study, we cloned Ht1-FEH I and Ht1-FEH II cDNAs that consisted of ORFs of 1,683 and 1,746 nucleotides, respectively, from Jerusalem artichoke. Both 1-FEH sequences include a short N-terminal signal peptide of 25 amino acids that resembles the 20-amino acid signal of Lp1-FEHa (Lothier et al. 2007), which is required for cotranslational insertion in the endoplasmic reticulum. Both Ht1-FEHs were highly similar to chicory 1-FEH I and 1-FEH II. Plant fructan-metabolizing enzymes (i.e., 1-FEH, 1-SST, 1-FFT, 6-FEH, and 6-SFT), and invertases (cwINV and vacINV) belong to the GH32 family and show high levels of similarity at the amino acid sequence level. Genes encoding plant 1-FEHs seem to have evolved from an ancestral cwINV gene (Fig. 2[I]; De Coninck et al. 2005; Van den Ende et al. 2000, 2001), although fructans are predominantly found in vacuoles (Darwen and John 1989; Wagner and Wiemken 1986). Interestingly, the different substrate specificity between vacuolar invertases (containing the conserved WECVD motif) and extracellular invertases (containing the conserved WECPD motif) was fully determined by a single amino acid difference (V to P) in the conserved sequence (Goetz and Roitsch 1999). The amino acid sequences of 1-FEH from various plant species, including the Ht 1-FEHs cloned here, all contain a P in this conserved motif (Fig. 1). However, like all other FEHs characterized to date, the predicted pI of Ht1-FEH I and Ht1-FEH II was lower than that of typical cwINVs (Van den Ende et al. 2003b, 2004).
As the methylotrophic yeast P. pastoris does not produce any invertases or fructosyl transferases, it is suitable for generating recombinant FEH for functional analysis (Asega et al. 2008). The P. pastoris X-33 system was chosen to express recombinant Ht1-FEH I and II in this study, since this expression system could secrete the fusion protein. The molecular masses of the Ht1-FEH I and II purified fusion proteins from the recombinant yeasts were both about 90 kDa (determined by SDS-PAGE), instead of the expected 62.9 and 65.1 kDa, respectively. Also, the molecular masses of the fusion proteins exceeded that of the native 1-FEH purified from tubers of Jerusalem artichoke by SDS-PAGE (Marx et al. 1997). Our results were similar to those observed for recombinant proteins from P. pastoris, including 1-FEH from A. lappa L (Ueno et al. 2011) and exoinulinase from Penicillium janthinellum (Wang et al. 2011), strongly suggesting that the recombinant proteins were glycosylated. Furthermore, Ueno et al. (2011) reported that the molecular mass of the purified enzyme was in agreement with the calculated mass of the recombinant protein after deglycosylation by N-glycosidase F (PNGase F). Our LC–MS/MS results indicated that the heterologously expressed Ht1-FEH I (KJ946352) and Ht1-FEH II (KJ946353) were indeed recombinant Ht1-FEHs.
Although the activity of recombinant Ht1-FEHs from P. pastoris and hydrolase A and B from Jerusalem artichoke tubers (Edelman and Jefford 1964; Marx et al. 1997) is optimal at acidic pH, the values differed slightly between them. The pH optimum of recombinant Ht1-FEH I and II was determined to be 6.0, whereas that of native 1-FEH from Jerusalem artichoke tubers was pH 5.2 after a 1-h pre-incubation at 40 °C, followed by a 1-h incubation with the substrate Nys (10 mM) at 30 °C (Marx et al. 1997). The optimal temperature of both Ht 1-FEHs was close to 35 °C after incubation with 10 mM 1-K for 10 h (Fig. 3). Slight differences in optimum pH were probably due to inconsistencies in reaction times and reaction conditions. In addition, Guo et al. (2008) reported that glycosylation affects the biochemical characterization of a recombinant phytase expressed in P. pastoris and that the pH and thermal stability improved when the recombinant phytase was glycosylated. However, the properties of 1-FFT were not affected by glycosylation when compared with the native enzyme (Van den Ende et al. 2006).
Since recombinant Ht1-FEH I and II showed very low or no activity against Suc and levan-type fructans (Table 1), the enzymes were not INVs or 6-FEHs. When the enzymes were incubated with 1-K, free Fru and Suc were released. These results demonstrated that Ht1-FEH I and II preferentially hydrolyze the β(2,1) linkage, which further confirmed that the recombinant enzymes were indeed 1-FEHs. Suc acted as a inhibitor of 1-FEHs and this inhibitory effect was concentration-dependent, since treatment with 100 mM Suc reduced 1-FEH enzyme activity to a greater extent than did treatment with 10 mM. This inhibition by Suc was observed for most plant FEHs purified to date (Marx et al. 1997; Tamura et al. 2011; Van den Ende et al. 2003a). It was previously reported that 86 % of recombinant Lp 1-FEHa enzyme activity was inhibited by 10 mM Suc and 90 % of Lp 1-FEHa was inhibited by 20 mM Suc (Lothier et al. 2007). According to Verhaest et al. (2007), enzymes that were strongly inhibited by Suc contain a S or G in the YTG region and a W in the GSAT region. These authors also observed that W82 and S101 are important for binding Suc as an inhibitor. In agreement with this, Vernonia 1-FEH (Asega et al. 2008), chicory 1-FEH II (De Roover et al. 1999; Van den Ende et al. 2001), and our Ht1-FEH II, which were strongly inhibited by Suc, contain these amino acids in these positions (Fig. 1). However, ryegrass Lp1-FEHa (Lothier et al. 2007) and Ht1-FEH I, which were also strongly inhibited by Suc, do not contain W82 and S101, while chicory 1-FEH I, which was weakly inhibited by Suc, similarly lacks W82 and S101 (Claessens et al. 1990; Van den Ende et al. 2000). The substrate specificity of Ht1-FEH I and II were similar to that of the natural hydrolases A and B extracted from Jerusalem artichoke, which were assumed to be mainly 1-FEHs (Edelman and Jefford 1964). In addition, Ht1-FEH I and II correspond well to hydrolases A and B based on their enzyme characteristics (Edelman and Jefford 1964). In 1997 (Marx et al. 1997), a hydrolase was purified to homogeneity in SDS-PAGE; however, it is difficult to discriminate between Ht1-FEH I and II and the hydrolase, since both Ht1-FEHs had essentially the same enzyme features in terms of optimal pH, temperature, and substrate specificity.
Ht1-FEH I and II may play different roles during plant development and under stress treatments
Fructan content is decreased during tuber sprouting in Jerusalem artichoke due to the activity of a group of enzymes called fructan 1-exohydrolases (1-FEHs; Edelman and Jefford 1964; Marx et al. 1997). In contrast to Ht1-FEH II, which is strongly induced in sprouting tubers, Ht1-FEH I was strongly expressed in stems and leaves, where small amounts of intermediate DP fructans (i.e., around 0.5-6 mg g−1 DW each of 1-K, 1F-F, and Nys, versus around 25.21- 41.43 mg g−1 DW in sprouting tubers) exist throughout the growth period (Li et al. 2014). Chicory 1-FEH II was found to be abundantly expressed in etiolated leaves (Van den Ende et al. 2001). The above result implies that Ht1-FEH I may have roles other than fructan breakdown. Lp1-FEH was reported to be involved in fructan trimming instead of fructan degradation based on its transcript abundance in leaves (Lothier et al. 2007). Interestingly, 1-FEHs were also found in non-fructan species, such as Arabidopsis, where they were thought to participate in the plant’s defense response (De Coninck et al. 2005; Van den Ende et al. 2004).
FEHs have long been known to promote fructan hydrolysis under abiotic or biotic conditions, especially cold and defoliation. In this study, we investigated the expression levels of Ht1-FEH I and 1-FEH II under salinity and drought treatments, since Jerusalem artichoke is widely planted in semi-arid soils (Gao et al. 2013; Zhao et al. 2008, 2006). Previous studies focused on 1-FEH in tissues with a high fructan content, such as tubers of Jerusalem artichoke (Edelman and Jefford 1964; Marx et al. 1997) and roots of chicory (Van den Ende et al. 2000, 2001), but Ht1-FEHs were also expressed in the stems and leaves of growing plants, indicating that Ht1-FEHs play multiple roles during plant development. As shown in Fig. 5, Ht1-FEH I transcripts were abundant in stems and leaves after 24 h of treatment with 100 mM NaCl. Moreover, Ht1-FEH I also showed higher expression in roots and stems after 24 h of 15 % PEG6000 treatment compared to the CK. The expression of Ht1-FEH II increased significantly in stems after 2 h and in roots after 5 h of 100 mM NaCl treatment compared to the CK. During tuber sprouting, NaCl but not PEG greatly induced the transcription of both Ht1-FEHs in the tubers after 5 d of treatment (Fig. 6). Interestingly, the level of Ht1-FEH II transcript declined after 2 d of PEG treatment (Fig. 6b). Previous studies showed that drought stress generally leads to increased levels of 1-FEH transcript in several plant species; however, some exceptions do exist (Kerepesi et al. 1998; Spollen and Nelson 1994). The levels of 1-FEH w3 transcripts in wheat stems gradually increased in plants (at 30 days post anthesis) subjected to water deficit, but not in well-watered plants (Zhang et al. 2009). Interestingly, the levels of 1-FEH w1 and w2 transcripts in wheat remained almost constant under the above treatment. In addition, in accordance with our findings, water stress slightly increased 1-FEH activities in the roots of C. intybus (Vandoorne et al., 2012).
Several studies show that the expression patterns of plant 1-FEHs are coherent with their enzyme activities (Asega et al. 2011; Chalmers et al. 2005; Ritsema and Smeekens 2003b; Van Laere and Van den Ende 2002). In our study, 1-FEH enzyme activities were higher after 2 d of salinity or drought treatment than in the untreated CK (Fig. 7a), which was in agreement with their corresponding expression levels. PEG treatments also increased the INV and 1-SST enzyme activities after 3 d of treatment compared to CK, whereas 1-FFT enzyme activities were almost the same after 1 d of treatment. The enzyme activities of SST and FFT in the tuberous roots of Viguiera discolor Baker decreased after 9 days of water deficit treatments, but increased upon rewatering (Oliveira et al. 2013). Prolonged drought treatments (25 weeks) inhibited the 1-SST and 1-FFT enzyme activities, but slightly increased the 1-FEH enzyme activities from the 19th week onwards in the roots of C. intybus (Vandoorne et al. 2012). According to Ji et al. (2010), the relative expression of fructan biosynthesis genes (1-SST and especially 6-SFT) and fructan content were reduced in drought-stressed wheat anthers and in the ovaries of drought-sensitive genotypes. By contrast, the expression of the vacuolar invertase genes OsVIN2 and Ivr of maize, which may help mobilize stored Suc and fructan for use in metabolic processes, were up-regulated by drought stress (Ji et al. 2007; Kim et al. 2000; Trouverie et al. 2003). These findings show that the enzymes involved in fructan metabolism are affected by abiotic stress treatment.
Fructan (DP ≥ 3) was largely consumed to support the growth of new plantlets when tubers sprouted in the absence of any treatments (Fig. 8 and Supplementary Fig. S1), and was presumably transported out of the tubers in the form of Suc (Edelman and Jefford 1968). We found that salinity and drought treatments increased the Suc content in Jerusalem artichoke tubers. By contrast, the Fru content was much lower than that of the CK at any time-point and the levels of intermediate DP fructans (such as 1-K and Nys) were the same as in untreated tubers under salt and drought treatments except at 1 DAT (Fig. S1). Two scenarios may explain the inconsistency between decreased fructose content and increased FEH activities in artichoke tubers subjected to salinity or drought stress, given that 1-FEHs are mainly responsible for fructan breakdown: (1) FEH may be active in vivo; (2) FEH may be inhibited in vivo. Regarding the first possibility, several authors have suggested that plant 1-FEHs act as trimming enzymes (Bancal et al. 1992; Lothier et al. 2007; Van den Ende et al. 2003a), and the “elongation-trimming” pathway was first proposed for wheat FEHs (Bancal et al. 1992). Also, wheat 1-FEH w2 was purified from young stems that were actively synthesizing graminan-type fructan, which was suggested to function as a trimmer (Van den Ende et al. 2003a). A similar result was found in L. perenne, where Lp1-FEH transcripts and FEH enzyme activities were increased in leaf sheaths and elongating leaf bases, which accumulated fructans (Lothier et al. 2007). However, since Jerusalem artichoke accumulated only β(2,1) linkage fructan, it is not possible that Ht 1-FEHs function as trimmers during tuber sprouting. Regarding the alternative possibility, we found that two Ht 1-FEHs were dramatically inhibited by Suc and that Suc content was increased after 3 DAT under the two abiotic stresses examined. Similar to our findings, Suc content was significantly higher in the anthers of water-stressed rice plants than in those of the CK (Nguyen et al. 2010). Suc content is determined by Suc transport (mainly via Suc transporters), biosynthesis, and degradation. The rice Suc transporters OsSUT1, OsSUT4, and OsSUT5 were down-regulated during drought and salinity treatments (Ibraheem et al. 2011) and the mRNA levels of the phloem Suc transporter PmSUC2 from broadleaf plantain (Plantago major) decreased after 24 h of salt stress (Pommerrenig et al. 2007). In addition, several reports showed that abiotic stress treatments resulted in activation of sucrose phosphate synthase (SPS) and sucrose synthase (SS), which led to the accumulation of Suc, in various plants (Basu et al. 2007; Dong et al. 2011; Hu et al. 2013; Kaur et al. 2003). In salt-stressed quinoa cotyledons (Chenopodium quinoa Willd.), higher SPS activity and higher Suc content were observed, suggesting that source-sink relations are changed in order to supply soluble sugars for the osmotic adjustment (Rosa et al. 2009). Compared with the well-watered CK, tall fescue seedlings had a higher level of Suc after deficit irrigation treatments, which enhanced tolerance to drought stress (Fu et al. 2010). The low levels of Glc observed in artichoke tubers in our study implied that the enzyme activities of INV or 1-SST, which catabolize Suc, were generally low. Though salinity or drought treatment induce 1-FEH transcription and increase its enzyme activity in vitro (Fig. 7a), the Suc content in vivo was probably enhanced due to the decreased Suc transport and the increased local Suc biosynthesis by SPS and SS under NaCl or PEG treatments. These elevated levels of Suc may inhibit FEH enzyme activities and delay the hydrolysis of high DP fructans in the tubers under salinity and drought stress. Why the tubers accumulated Suc instead of Fuc is not clear. Suc accumulation may facilitate instant osmotic adjustment in tubers subjected to abiotic stresses. In this way, Suc-mediated inhibition of enzyme activity may play a more prominent role than transcriptional or translational regulation in modulating 1-FEH activity under preliminary salinity or drought stress.
However, several reports have implicated fructans in protecting plants from cold or drought stress and in sustaining sucrose levels in the plant (Amiard et al. 2003; De Roover et al. 2000; Hincha et al. 2002; Ji et al. 2010; Konstantinova et al. 2002; Pilon-Smits et al. 1995; Pollock et al. 2003). Adjustments in the degree of polymerization of fructans may be as important as changes in abundance, and the balance between fructan biosynthesis and breakdown may be more important than the individual rates of biosynthesis or breakdown under abiotic stresses (Ji et al. 2007).
In summary, two 1-FEHs from Jerusalem artichoke were cloned and their enzymes properties were demonstrated to correspond to their native enzymes. Transcripts of both 1-FEHs were responded to salinity and drought stress. Under abiotic stress, 1-FEH enzyme activities may be inhibited by accumulated Suc in the tubers.
Abbreviations
- 1-FFT:
-
Fructan:fructan 1-fructosyltransferase
- 1F-F:
-
1F-Fructofuranosylnystose
- 1-SST:
-
Sucrose:sucrose 1-fructosyltransferase
- 1-K:
-
1-Kestose
- 6G-FFT:
-
Fructan:fructan 6G-fructosyltransferase
- 6-SFT:
-
Sucrose:fructan 6-fructosyltransferase
- CK:
-
Control
- DP:
-
Degree of polymerization
- DW:
-
Dry weight
- FEH:
-
Fructan exohydrolase
- FW:
-
Fresh weight
- Fru:
-
Fructose
- Glc:
-
Glucose
- HPLC:
-
High performance liquid chromatography
- INV:
-
Invertase
- LC–MS:
-
Liquid chromatography–mass spectrometry
- Nys:
-
Nystose
- qPCR:
-
Quantitative PCR
- RT-PCR:
-
Reverse transcriptase–polymerase chain reaction
- Suc:
-
Sucrose
References
Amiard V, Morvan-Bertrand A, Billard JP, Huault C, Keller F, Prud’homme MP (2003) Fructans, but not the sucrosyl-galactosides, raffinose and loliose, are affected by drought stress in perennial ryegrass. Plant Physiol 132:2218–2229
Asega AF, do Nascimento JRO, Schroeven L, Van den Ende W, Carvalho MAM (2008) Cloning, characterization and functional analysis of a 1-FEH cDNA from Vernonia herbacea (Vell.) Rusby. Plant Cell Physiol 49:1185–1195
Asega AF, do Nascimento TRO, Carvalho MAM (2011) Increased expression of fructan 1-exohydrolase in rhizophores of Vernonia herbacea during sprouting and exposure to low temperature. J Plant Physiol 168:558–565
Bancal P, Carpita NC, Gaudillere JP (1992) Differences in fructan accumulated in induced and field-grown wheat plants: an elongation-trimming pathway for their synthesis. New Phytol 120:313–321
Basu PS, Ali M, Chaturvedi SK (2007) Osmotic adjustment increases water uptake, remobilization of assimilates and maintains photosynthesis in chickpea under drought. Indian J Exp Biol 45:261–267
Chalmers J, Lidgett A, Cummings N, Cao Y, Forster J, Spangenberg G (2005) Molecular genetics of fructan metabolism in perennial ryegrass. Plant Biotechnol J 3:459–474
Claessens G, Van Laere A, De Proft M (1990) Purification and properties of an inulinase from chicory roots (Cichorium intybus L.). J Plant Physiol 136:35–39
Darwen CW, John P (1989) Localization of the enzymes of fructan metabolism in vacuoles isolated by a mechanical method from tubers of Jerusalem artichoke (Helianthus tuberosus L.). Plant Physiol 89:658–663
De Coninck B, Le Rov K, Francis I, Clerens S, Vergauwen R (2005) Arabidopsis AtcwINV3 and 6 are not invertases but are fructan exohydrolases (FEHs) with different substrate specificities. Plant, Cell Environ 28:432–443
de Mattos Paula, Arêas A, Oliveira ML, Romero Ramos CR, Sbrogio-Almeida ME, Raw I, Ho PL (2002) Synthesis of cholera toxin B subunit gene: cloning and expression of a functional 6 × His-tagged protein in Escherichia coli. Protein Expr Purif 25:481–487
De Roover J, Van Laere A, De Winter M, Van den Ende W (1999) Purification and properties of a second fructan exohydrolase from the roots of Cichorium intybus. Physiol Plant 106:28–34
De Roover J, Van den Branden K, Van Laere A, Van den Ende W (2000) Drought induces fructan synthesis and 1-SST (sucrose:sucrose fructosyltransferase) in roots and leaves of chicory seedlings (Cichorium intybus L.). Planta 210:808–814
Dong CJ, Wang XL, Shang QM (2011) Salicylic acid regulates sugar metabolism that confers tolerance to salinity stress in cucumber seedlings. Sci Hortic 129:629–636
Edelman J, Jefford T (1964) The metabolism of fructose polymers in plants. 4. β-Fructofuranosidases of tubers of Helianthus tuberosus L. Biochem J 93:148
Edelman J, Jefford T (1968) The mechanisim of fructosan metabolism in higher plants as exemplified in Helianthus tuberosus. New Phytol 67:517–531
Fu J, Huang B, Fry J (2010) Osmotic potential, sucrose level, and activity of sucrose metabolic enzymes in tall fescue in response to deficit irrigation. J Am Soc Hortic Sci 135:506–510
Gao K, Zhu T, Han G (2013) Water and nitrogen interactively increased the biomass production of Jerusalem artichoke (Helianthus tuberosus L.) in semi-arid area. Afr J Biotechnol 10:6466–6472
Goetz M, Roitsch T (1999) The different pH optima and substrate specificities of extracellular and vacuolar invertases from plants are determined by a single amino-acid substitution. Plant J 20:707–711
Guo M, Hang H, Zhu T, Zhuang Y, Chu J, Zhang S (2008) Effect of glycosylation on biochemical characterization of recombinant phytase expressed in Pichia pastoris. Enzym Microb Technol 42:340–345
Hendry GA (1993) Evolutionary origins and natural functions of fructans—a climatological, biogeographic and mechanistic appraisal. New Phytol 123:3–14
Hincha DK, Zuther E, Hellwege EM, Heyer AG (2002) Specific effects of fructo- and gluco-oligosaccharides in the preservation of liposomes during drying. Glycobiology 12:103–110
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Circ Calif Agric Exp Stn 347:1–32
Hu T, Hu LX, Zhang XZ, Zhang PP, Zhao ZJ, Fu JM (2013) Differential responses of CO2 assimilation, carbohydrate allocation and gene expression to NaCl stress in perennial ryegrass with different salt tolerance. PLoS ONE 8:e66090
Huang Z, Zhao L, Chen D, Liang M, Liu Z, Shao H, Long X (2013) Salt stress encourages proline accumulation by regulating proline biosynthesis and degradation in Jerusalem artichoke plantlets. PLoS ONE 8:e62085
Ibraheem O, Dealtry G, Roux S, Bradley G (2011) The effect of drought and salinity on the expressional levels of sucrose transporters in rice (Oryza sativa Nipponbare) cultivar plants. Plant Omics 4:68–74
Ji X, Van den Ende W, Schroeven L, Clerens S, Geuten K, Cheng S, Bennett J (2007) The rice genome encodes two vacuolar invertases with fructan exohydrolase activity but lacks the related fructan biosynthesis genes of the Pooideae. New Phytol 173:50–62
Ji X, Shiran B, Wan J, Lewis DC, Jenkins CL, Condon AG, Dolferus R (2010) Importance of pre-anthesis anther sink strength for maintenance of grain number during reproductive stage water stress in wheat. Plant, Cell Environ 33:926–942
Joudi M, Ahmadi A, Mohamadi V, Abbasi A, Vergauwen R, Mohammadi H, Van den Ende W (2012) Comparison of fructan dynamics in two wheat cultivars with different capacities of accumulation and remobilization under drought stress. Physiol Plant 144:1–12
Kaur S, Gupta AK, Kaur N (2003) Effect of kinetin on starch and sucrose metabolising enzymes in salt stressed chickpea seedlings. Biol Plant 46:67–72
Kawakami A, Yoshida M, Van den Ende W (2005) Molecular cloning and functional analysis of a novel 6&1-FEH from wheat (Triticum aestivum L.) preferentially degrading small graminans like bifurcose. Gene 358:93–101
Kerepesi I, Galiba G, Bányai É (1998) Osmotic and salt stresses induced differential alteration in water-soluble carbohydrate content in wheat seedlings. J Agric Food Chem 46:5347–5354
K-I Tamura, Sanada Y, Tase K, Komatsu T, Yoshida M (2011) Pp6-FEH1 encodes an enzyme for degradation of highly polymerized levan and is transcriptionally induced by defoliation in timothy (Phleum pratense L.). J Exp Bot 62:3421–3431
Kim J, Mahé A, Brangeon J, Prioul J (2000) Maize vacuolar invertase, IVR2, is induced by water stress. Organ/tissue specificity and diurnal modulation of expression. Plant Physiol 124:71–84
Konstantinova T, Parvanova D, Atanassov A, Djilianov D (2002) Freezing tolerant tobacco, transformed to accumulate osmoprotectants. Plant Sci 163:157–164
Lammens W, Le Roy K, Schroeven L, Van Laere A, Rabijns A, Van den Ende W (2009) Structural insights into glycoside hydrolase family 32 and 68 enzymes: functional implications. J Exp Bot 60:727–740
Lasseur B, Lothier J, Djoumad A, De Coninck B, Smeekens S, Van Laere A, Van den Ende W (2006) Molecular and functional characterization of a cDNA encoding fructan:fructan 6G-fructosyltransferase (6G-FFT)/fructan:fructan 1-fructosyltransferase (1-FFT) from perennial ryegrass (Lolium perenne L.). J Exp Bot 57:2719–2734
Lasseur B, Schroeven L, Lammens W, Le Roy K, Spangenberg G, Manduzio H, Vergauwen R, Van den Ende W (2009) Transforming a fructan:fructan 6G-fructosyltransferase from perennial ryegrass into a sucrose:sucrose 1-fructosyltransferase. Plant Physiol 149:327–339
Le Roy K, Lammens W, Verhaest M, De Coninck B, Rabijns A, Van Laere A, Van den Ende W (2007a) Unraveling the difference between invertases and fructan exohydrolases: a single amino acid (Asp-239) substitution transforms Arabidopsis cell wall invertase 1 into a fructan 1-exohydrolase. Plant Physiol 145:616–625
Le Roy K, Vergauwen R, Cammaer V, Yoshida M, Kawakami A, Van Laere A, Van den Ende W (2007b) Fructan 1-exohydrolase is associated with flower opening in Campanula rapunculoides. Funct Plant Biol 34:972–983
Li H, Xu H, Zhao G, Liang M (2014) The effect of deformation on dry matter and sugar accumulation and their distribution profiles in Jerusalem artichoke. Acta Pratacult Sin 23:149–157
Liang M, Hole D, Wu J, Blake T, Wu Y (2012) Expression and functional analysis of NUCLEAR FACTOR-Y, subunit B genes in barley. Planta 235:779–791
Liang M, Yin X, Lin Z, Zheng Q, Liu G, Zhao G (2014) Identification and characterization of NF-Y transcription factor families in Canola (Brassica napus L.). Planta 239:107–126
Liu D, Li J, Zhao S, Zhang R, Wang M, Miao Y, Shen Y (2013) Secretome diversity and quantitative analysis of cellulolytic Aspergillus fumigatus Z5 in the presence of different carbon sources. Biotechnol Biofuels 6:149
Livingston DP, Henson CA (1998) Apoplastic sugars, fructans, fructan exohydrolase, and invertase in winter oat: responses to second-phase cold hardening. Plant Physiol 116:403–408
Livingston DP III, Hincha DK, Heyer AG (2009) Fructan and its relationship to abiotic stress tolerance in plants. Cell Mol Life Sci 66:2007–2023
Lothier J, Lasseur B, Le Roy K, Van Laere A, Barre P, Van den Ende W (2007) Cloning, gene mapping, and functional analysis of a fructan 1-exohydrolase (1-FEH) from Lolium perenne implicated in fructan synthesis rather than in fructan mobilization. J Exp Bot 58:1969–1983
Marx SP, Nosberger J, Frehner M (1997) Seasonal variation of fructan-β-fructosidase (FEH) activity and characterization of a β-(2-1)-linkage specific FEH from tubers of Jerusalem artichoke (Helianthus tuberosus). New Phytol 135:267–277
Nguyen G, Hailstones D, Wilkes M, Sutton B (2010) DROUGHT STRESS: role of carbohydrate metabolism in drought-induced male sterility in rice anthers. J Agron Crop Sci 196:346–357
Oliveira V, Silva E, Zaidan L, Carvalho M (2013) Effects of elevated CO2 concentration and water deficit on fructan metabolism in Viguiera discolor Baker. Plant Biol 15:471–482
Pilon-Smits EA, Ebskamp MJ, Paul MJ, Jeuken MJ, Weisbeek PJ, Smeekens SC (1995) Improved performance of transgenic fructan-accumulating tobacco under drought stress. Plant Physiol 107:125–130
Pollock C, Farrar J, Tomos D, Gallagher J, Lu C, Koroleva O (2003) Balancing supply and demand: the spatial regulation of carbon metabolism in grass and cereal leaves. J Exp Bot 54:489–494
Pommerrenig B, Papini-Terzi FS, Sauer N (2007) Differential regulation of sorbitol and sucrose loading into the phloem of Plantago major in response to salt stress. Plant Physiol 144:1029–1038
Resina D, Ac Serrano, Valero F, Ferrer P (2004) Expression of a Rhizopus oryzae lipase in Pichia pastoris under control of the nitrogen source-regulated formaldehyde dehydrogenase promoter. J Biotechnol 109:103–113
Ritsema T, Smeekens S (2003a) Fructans: beneficial for plants and humans. Curr Opin Plant Biol 6:223–230
Ritsema T, Smeekens S (2003b) Engineering fructan metabolism in plants. J Plant Physiol 160:811–820
Rosa M, Hilal M, Gonzalez JA, Prado FE (2009) Low-temperature effect on enzyme activities involved in sucrose-starch partitioning in salt-stressed and salt-acclimated cotyledons of quinoa (Chenopodium quinoa Willd.) seedlings. Plant Physiol Biochem 47:300–307
Schroeven L, Lammens W, Van Laere A, Van den Ende W (2008) Transforming wheat vacuolar invertase into a high affinity sucrose: sucrose 1-fructosyltransferase. New Phytol 180:822–831
Spollen WG, Nelson CJ (1994) Response of fructan to water deficit in growing leaves of tall fescue. Plant Physiol 106:329–336
Sprenger N, Bortlik K, Brandt A, Boller T, Wiemken A (1995) Purification, cloning, and functional expression of sucrose:fructan 6-fructosyltransferase, a key enzyme of fructan synthesis in barley. Proc Natl Acad Sci 92:11652–11656
Trouverie J, Thevenot C, Rocher J, Sotta B, Prioul J (2003) The role of abscisic acid in the response of a specific vacuolar invertase to water stress in the adult maize leaf. J Exp Bot 54:2177–2186
Ueno K, Ishiguro Y, Yoshida M, Onodera S, Shiomi N (2011) Cloning and functional characterization of a fructan 1-exohydrolase (1-FEH) in edible burdock (Arctium lappa L.). Chem Cent J 5:1–9
Valluru R, Van den Ende W (2008) Plant fructans in stress environments: emerging concepts and future prospects. J Exp Bot 59:2905–2916
Van den Ende W, Michiels A, De Roover J, Verhaert P, Van Laere A (2000) Cloning and functional analysis of chicory root fructan 1-exohydrolase I (1-FEH I): a vacuolar enzyme derivedfrom a cell-wall invertase ancestor? Mass fingerprint of the 1-FEH I enzyme. Plant J 24:447–456
Van den Ende W, Michiels A, Van Wonterghem D, Clerens SP, De Roover J, Van Laere AJ (2001) Defoliation induces fructan 1-exohydrolase II in witloof chicory roots. Cloning and purification of two isoforms, fructan 1-exohydrolase IIa and fructan 1-exohydrolase IIb. Mass fingerprint of the fructan 1-exohydrolase II enzymes. Plant Physiol 126:1186–1195
Van den Ende W, Clerens S, Vergauwen R, Van Riet L, Van Laere A, Yoshida M, Kawakami A (2003a) Fructan 1-exohydrolases. β-(2, 1)-trimmers during graminan biosynthesis in stems of wheat? Purification, characterization, mass mapping, and cloning of two fructan 1-exohydrolase isoforms. Plant Physiol 131:621–631
Van den Ende W, De Coninck B, Clerens S, Vergauwen R, Van Laere A (2003b) Unexpected presence of fructan 6-exohydrolases (6-FEHs) in non-fructan plants: characterization, cloning, mass mapping and functional analysis of a novel ‘cell-wall invertase-like’ specific 6-FEH from sugar beet (Beta vulgaris L.). Plant J 36:697–710
Van den Ende W, De Coninck B, Van Laere A (2004) Plant fructan exohydrolases: a role in signaling and defense? Trends Plant Sci 9:523–528
Van den Ende W, Yoshida M, Clerens S, Vergauwen R, Kawakami A (2005) Cloning, characterization and functional analysis of novel 6-kestose exohydrolases (6-KEHs) from wheat (Triticum aestivum). New Phytol 166:917–932
Van den Ende W, Clerens S, Vergauwen R, Boogaerts D, Le Roy K, Arckens L, Van Laere A (2006) Cloning and functional analysis of a high DP fructan:fructan 1-fructosyl transferase from Echinops ritro (Asteraceae): comparison of the native and recombinant enzymes. J Exp Bot 57:775–789
Van den Ende W, Lammens W, Van Laere A, Schroeven L, Le Roy K (2009) Donor and acceptor substrate selectivity among plant glycoside hydrolase family 32 enzymes. FEBS J 276:5788–5798
Van Der Meer IM, Koops AJ, Hakkert JC, Van Tunen AJ (1998) Cloning of the fructan biosynthesis pathway of Jerusalem artichoke. Plant J 15:489–500
Van Laere A, Van den Ende W (2002) Inulin metabolism in dicots: chicory as a model system. Plant, Cell Environ 25:803–813
Vandoorne B, Mathieu A-S, Van den Ende W, Vergauwen R, Périlleux C, Javaux M, Lutts S (2012) Water stress drastically reduces root growth and inulin yield in Cichorium intybus (var. sativum) independently of photosynthesis. J Exp Bot 63:4359–4373
Verhaest M, Van den Ende W, Roy KL, De Ranter CJ, Van Laere A, Rabijns A (2005) X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus. Plant J 41:400–411
Verhaest M, Lammens W, Le Roy K, De Ranter CJ, Van Laere A, Rabijns A, Van den Ende W (2007) Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor. New Phytol 174:90–100
Vijn I, Smeekens S (1999) Fructan: more than a reserve carbohydrate? Plant Physiol 120:351–360
Wagner W, Wiemken A (1986) Properties and subcellular localization of fructan hydrolase in the leaves of barley (Hordeum vulgare L. cv Gerbel). J Plant Physiol 123:429–439
Wang L, Huang Y, Long X, Meng X, Liu Z (2011) Cloning of exoinulinase gene from Penicillium janthinellum strain B01 and its high-level expression in Pichia pastoris. J Appl Microbiol 111:1371–1380
Zhang J, Dell B, Conocono E, Waters I, Setter T, Appels R (2009) Water deficits in wheat: fructan exohydrolase (1-FEH) mRNA expression and relationship to soluble carbohydrate concentrations in two varieties. New Phytol 181:843–850
Zhao G, Liu Z, Chen M, Kou W (2006) Effect of saline aquaculture effluent on salt-tolerant Jerusalem Artichoke (Helianthus tuberosus L.) in a semi-arid coastal area of China. Pedosphere 16:762–769
Zhao G, Liu Z, Chen M (2008) Soil properties and yield of Jerusalem artichoke (Helianthus tuberosus L.) with seawater irrigation in North China Plain. Pedosphere 18:195–202
Acknowledgments
We thank Kathleen Farquharson for valuable comments on the manuscript revision. This research was supported by grants from the National Natural Science Foundation of China (31201842), the National High Technology Research and Development Program (“863”Program, 2011AA100209), the Doctoral Program of Higher Education of China (20120097120015), Fundamental Research Funds for the Central Universities (KYZ201206), the Priority Academic Program Development of Jiangsu Higher Education Institutions (RAPD program, 809001), and the Scientific Research Foundation of the State Human Resource Ministry.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, H., Liang, M., Xu, L. et al. Cloning and functional characterization of two abiotic stress-responsive Jerusalem artichoke (Helianthus tuberosus) fructan 1-exohydrolases (1-FEHs). Plant Mol Biol 87, 81–98 (2015). https://doi.org/10.1007/s11103-014-0262-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-014-0262-1