Abstract
Plant architecture directly affects biomass in higher plants, especially grain yields in agricultural crops. In this study, we characterized a recessive mutant, plant architecture determinant (pad), derived from the Oryza sativa ssp. indica cultivar MH86. The mutant exhibited severe dwarf phenotypes, including shorter and stunted leaves, fewer secondary branches during both the vegetative and reproductive growth stages. Cytological studies revealed that pad mutant growth defects are primarily due to the inhibition of cell expansion. The PAD gene was isolated using a map-based cloning strategy. It encodes a plasma membrane protein OsMCA1 and a SNP responsible for a single amino acid change was found in the mutant. PAD was universally expressed in rice tissues from the vegetative to reproductive growth stages, especially in seedlings, nodes and rachillae. Quantitative real-time PCR analysis revealed that the most of the genes responding to gibberellin (GA) metabolism were up-regulated in pad mutant internodes. The endogenous GA content measurement revealed that the levels of GA1 were significantly decreased in the third internode of pad mutants. Moreover, a GA response assay suggested that OsMCA1/PAD might be involved in the regulation of GA metabolism and signal transduction. Our results revealed the pad is a loss-of-function mutant of the OsMCA1/PAD, leading to upregulation of genes related to GA deactivation, which decreased bioactive GA levels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant architecture directly affects grain yields. Rice plant architecture, which has recently been extensively investigated (Sakamoto and Matsuoka 2004; Springer 2010; Wang and Li 2005, 2008; Xing and Zhang 2010; Yang and Hwa 2008; Zhang et al. 2008), is a complicated agronomic trait that is mainly determined by tiller number, tiller angle, leaf arrangement, plant height and panicle morphology. In previous studies, several genes involved in controlling the above-mentioned components, such as TEOSINTE BRANCHED 1 (OsTB1), MONOCULM 1 (MOC1), LAZY1 (LA1), Tiller Angle Control 1 (TAC1), PROSTRATE GROWTH 1 (PROG1), YABBY1 (YAB1), OsDWARF4, Loose Plant Architecture1 (LPA1), Semi-Dwarf1 (SD1) and SOUAMOSA PROMOTER BINDING PROTEIN-LIKE 14 (OsSPL14), were identified based on natural and artificially generated mutants of various rice varieties (Dai et al. 2007; Jiao et al. 2010; Jin et al. 2008; Li et al. 2007; 2003; Miura et al. 2010; Sakamoto et al. 2006; Sasaki et al. 2002; Takeda et al. 2003; Tan et al. 2008; Wu et al. 2013; Yoshihara and Iino 2007; Yu et al. 2007). Plant height, an important trait that affects plant architecture, is determined by the number and lengths of internodes as well as environmental conditions (Wang and Li 2005). Plant height is primarily regulated by autologous phytohormones, including gibberellins (GAs) and brassinosteroids (BRs; Yang and Hwa 2008).
The GA plant hormones, whose major active forms include GA1 and GA4, are involved in plant height determination. The GAs is consisted of a large family of tetracyclic diterpenoid carboxylic acids and functions in diverse aspects of plant growth and development, especially internode elongation (Sakamoto 2006; Sakamoto and Matsuoka 2004). In recent years, the mechanisms of GA metabolism have been elucidated (Olszewski et al. 2002; Thomas et al. 2005; Yamaguchi 2008). Bioactive GAs are first synthesized from geranylgeranyl diphosphate (GGDP), which is a common diterpene precursor. GGDP is then converted to GA12 via a series of intermediates by four enzymes, ent-copalyl diphosphate (CDP) synthase (CPS), ent-kaurene synthase (KS), ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO). As an important checkpoint in the GA biosynthesis pathway, GA12 is converted to GA53 by undergoing 13-hydroxylation by GA13-oxidase (GA13ox; Magome et al. 2013). Then, GA53/GA12 is converted to GA20/GA9 by 2-oxoglutarate–dependent dioxygenase (ODD) GA20ox (GA20-oxidase) through two parallel pathways in a three- or four-step process. In the final step of bioactive GA synthesis, GA20 and GA9 are converted to GA1 and GA4, respectively, by ODD GA3-oxidase (GA3ox). In addition, GA1/GA4 and GA20/GA9 are deactivated by ODD GA2-oxidases (GA2oxs) and a cytochrome P450 monooxygenase (CYP714D1/EUI; Zhu et al. 2006). These genes have also been identified based on the corresponding GA-deficient mutants in rice (Sakamoto et al. 2004).
GA homoeostasis is controlled through negative feedback regulation of GA biosynthetic enzymes and positive feed-forward regulation of GA catabolic enzymes meditated by the GA signaling pathway (Hedden and Thomas 2012; Yamaguchi 2008). In plants harboring related gain-of-function mutations in repressors or loss-of-function mutations in components of positive regulation, the levels of bioactive GAs or GA biosynthetic gene expression are highly elevated. Loss-of-function mutations in repressors result in lower levels of bioactive GAs or GA20ox and GA3ox mRNAs compared with those of wild type (WT; Olszewski et al. 2002).
In the present study, a natural mutant of the rice indica variety Minghui 86 (MH86), pad, exhibited a visibly abnormal phenotype that consisted of reduced plant height, shorter and stunted leaves, fewer secondary branches, lower seed set and higher tiller numbers compared with WT (MH86). We cloned the pad gene using a map-based cloning strategy and found that it is encoded by OsMCA1, a homolog of AtMCA1 in Arabidopsis. A nucleotide substitution in the kinase domain was identified in the mutant. The OsMCA1/PAD gene is constitutively expressed throughout the whole plant and is highly expressed in seedling, leaf, young leaf, internode and young panicle tissues. Further studies indicated that the OsMCA1/PAD gene is involved in rice plant architecture and substantially affects rice plant height by regulating GA metabolism.
Materials and methods
Plant materials
The rice mutant plant architecture determinant, pad, was identified from the O. sativa ssp. indica cultivar Minghui 86. All rice plants were planted in a field in Wuhan during the natural growing season.
Histological analysis
For microscopy studies, leaves and the uppermost internodes at the appropriate developmental stage were harvested, fixed in fixation solution (4 % paraformaldehyde (Sigma, Ronkonkoma, NY, USA) in 1× PBS, 0.02 % KCl, 0.8 % NaCl, 0.178 % Na2HPO4·2H2O, 0.024 % KH2PO4 and 0.1 % Triton X-100) overnight and sequentially dehydrated in 25, 50, 70, 80, 90, 95 and 100 % ethanol for 5 min each. Then, the dehydrated material was embedded in Steedman’s wax (polyethylene glycol 400 distearate: 1-hexadecanol; 9:1) and sectioned using a microtome to a thickness of 8 μm. The sections were adhered to slides coated with poly-l-lysine and then sequentially de-waxed in 100, 95, 80, 50 and 30 % ethanol for 5 min each. Finally, the sections were visualized and photographed on an inverted phase contrast microscope (IX51; Olympus).
Genetic analysis and map-based cloning
An F2 population derived from the pad mutant crossed with Nongken58 (japonica) was used for genetic linkage analysis and fine mapping. In total, 200 polymorphic SSR markers were identified from 600 SSR primer pairs used to screen the parents. Bulk Segregant Analysis (BSA) was used to assess SSR marker linkage to pad and roughly map the pad locus to chromosome 3. SNP markers were obtained by sequencing the region of interest. Finally, the pad locus was narrowed down to an 8.7-kb region between two SNP markers, S2 and S3, through 477 and 3,869 homozygous recessive individuals from the F2 population of the pad mutant × NK58, respectively. The candidate gene was identified by sequencing the genomic DNA in this region from the pad mutant and WT plants and generating pairwise sequence alignments using MacVector.
Plasmid construction
The coding region of the candidate gene was confirmed by comparing the sequence of the genomic DNA fragment with information in the GenBank, UniProtKB and Gramene databases. A 2,945-bp fragment containing 1,688 bp of upstream sequence and the 1,257 bp CDS was synthesized by GeneScript (Nanjing, China) and cloned into the binary vector pCAMBIA1300. The construct was then transformed into Agrobacterium strain EHA105 and designated pOsPMP618.
For the construction of the GFP-OsMCA1 fusion plasmid, the full-length OsMCA1 coding sequence was fused to the C terminus of the GFP coding sequence without a termination codon to create an in-frame fusion. The fragment was then cloned into a transient expression vector (pOsPMP550) under the control of the CaMV 35S promoter. The resulting plasmid was designated pOsPMP622.
For the construction of the OsMCA1-GUS fusion vector, an approximately 1.69 kb promoter region from the OsMCA1 gene was amplified and cut with HindIII and NaeI. The sequence was then inserted upstream of the 5′ end of the uidA gene encoding β-glucuronidase (GUS) in the binary vector pCAMBIA1300 to create the resulting plasmid, which was designated pOsPMP617.
Plant transformation
For the rescue of the pad mutant, the pOsPMP618 construct was introduced into calli derived from the pad mutant by Agrobacterium-mediated transformation. All transgenic rice plants were identified by PCR using the primer pair 618-1-F and 618-1-R (Supplemental Table S1). All transgenic plants were grown in a greenhouse at the Wuhan University Campus. For all Agrobacterium-mediated transformations, the plasmids were introduced into the Agrobacterium strain EHA105.
Tissue-specific analysis
For the tissue-specific expression of OsMCA1, pOsPMP617 was introduced into the japonica rice variety Nongken58, and positive transgenic plants were identified by PCR using the primer pair GUS-F and GUS-R (Supplemental Table S1). GUS activities were analyzed in situ according to a method described previously (Jefferson 1989). Briefly, tissues from the transgenic plants were washed in staining buffer containing 50 mM NaPO4 buffer (pH 7.0), 2 mM K3Fe(CN)6, 2 mM K4Fe(CN)6 and 0.2 % Triton X-100 three times on ice and then incubated in staining buffer supplemented with an X-Gluc stock solution (1 mM) at 37 °C overnight. GUS-stained tissues were observed on a stereo microscope and photographed with a camera (EOS 400D; Canon).
Transient assay on epidermal cells
The GFP alone (pOsPMP623) and GFP-OsMCA1 (pOsPMP622) fusion constructs were introduced into onion epidermal cells by particle bombardment using a PDS-1000/He (BIO-RAD). After incubation for 18–24 h at 26 °C, GFP fluorescence was observed on an inverted phase contrast microscope (IX51; Olympus).
Phylogenetic analysis
The putative homologs of OsMCA1 were identified using the Basic Local Alignment Search Tool from the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A phylogenetic tree of OsMCA1 homologs was constructed using the neighbor-joining (NJ) function of MEGA 5.2. The number of bootstrap replicates was estimated at 1,000, and all sites with alignment gaps/missing sequences in pairwise sequence comparisons were deleted.
RNA extraction and quantitative RT-PCR
All rice tissues were harvested, immediately frozen in liquid nitrogen and stored at −80 °C until use. Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and then was treated with RNase-free DNase I (New England Biolabs, Hitchin, UK) to remove DNA contamination prior to cDNA synthesis. First-strand cDNA synthesis was performed using M-MLV reverse transcriptase (Promega, Madison, USA) using 2–3 μg of RNA template and an oligo (dT) primer.
Quantitative RT-PCR was performed on a StepOne System (ABI) using the following program: 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s and a melting curve from 60 °C to 95 °C in 0.7 °C increments. Relative gene expression levels were calculated via the ΔΔCT method, and the rice Actin1 gene was used as an internal control to normalize the data. The primers used for qRT-PCR analysis are listed in Supplemental Table S1. All analyses were performed in triplicate. The transcripts of EUI1 were detected by Semi quantitative RT-PCR as described previously (Zhu et al. 2006). The rice Actin1 gene was used as an internal control with the prime pair as described previously (Huang et al. 2010).
Exogenous GA effects and GA content analysis
Exogenous GA treatment of rice seedlings was performed as described in (Ikeda et al. 2001) with minor modifications. Briefly, 15 seeds from WT and pad plants were surface sterilized for 30 min in a 2 % NaClO solution and washed four times with sterile distilled water. The sterilized seeds were soaked in sterile distilled water with or without 6.85 μM uniconazol for 36 h. The pretreated seeds were immersed in sterile distilled water for 24 h. Finally, the seeds were placed on 1 % agar plates supplemented with GA3 at different final concentrations ranging from 0.01 to 100 μM and grown at 26 °C under continuous light. After approximately 8 days, the length of the second leaf sheath was measured.
For GA content analysis, the individual internodes, including the uppermost internodes to the third internodes of WT plants and pad mutants, were harvested at the stage when the uppermost and second internodes were elongating and were immediately frozen in liquid nitrogen and stored at −80 °C until GA extraction. The endogenous GAs were detected by nano-LC–ESI-Q-TOF–MS analysis as described previously (Chen et al. 2012). All measurement were conducted three times.
Results
A pad mutant exhibits plant architecture defects
We identified a natural mutant of the O. sativa ssp. indica variety MH86 with plant architecture defects. This mutant exhibits a visibly abnormal phenotype during the vegetative and reproductive growth stages compared with WT. During growth and developmental stages, the mutant showed shorter root, smaller shoot and leaf sizes and reduced height (Fig. 1a–c). These mutated phenotypes, especially, became more apparent in the stem elongation stage to the seed setting stage (Fig. 1d). Compared with WT plants, the leaf length and plant height of the mutants were decreased by 59.16 % (17.90 ± 0.52 cm in the mutant vs. 43.83 ± 2.02 cm in WT; P < 0.001) and 46.87 % (67.1 ± 3.8 cm in the mutant vs. 126.3 ± 6.4 cm in WT; P < 0.001), respectively (Fig. 1e, f; Table 1). Moreover, the leaves of the mutant were curly and wrinkled with abnormal rachis from the base of the leaf (Fig. 1g).
Morphological characterization of the pad mutant. a Seed germination of WT (left) and pad (right) after 2 days. b WT (left) and pad (right) seedlings 6 days after germination. c WT (left) and pad (right) rice plants before heading. d WT (left) and pad (right) rice plants after grain filling. e Plant height of WT (left) and pad (right). f Leaf morphology of WT (left) and pad (right). g Morphology of the leaf base of WT (left) and pad (right). h Panicle of WT (left) and pad (right). i Grains per main panicle from WT (top) and pad (bottom). j Comparison of grain lengths between WT (top) and pad (bottom). k 1,000-grain weight and l grain yield per plant from WT and pad. The data are given as the mean ± SD (n = 8–15) in (j). The mean ± SD was calculated from three biological replicates and values were determined by the Student’s t test in k and l. Bars 1 cm in a; 2 cm in b and f–h; 10 cm in c–e; 3 mm in j
Furthermore, panicle size, spikelet number per panicle, grain length, 1,000-grain weight and seed set were obviously affected in the mutant (Fig. 1h–k; Table 1). The number of primary branches was significantly decreased by 33.73 % in the mutant compared with the WT (7.1 ± 0.7 in the mutant vs. 10.7 ± 0.5 in WT; P < 0.001), and the secondary branch number was also lower in the pad mutant (1.3 ± 1.0 in the mutant vs. 44.88 ± 4.8 in WT; P < 0.001; Table 1). In addition, the mutant exhibited a significant decrease in grain number per plant and grain size compared with WT (Table 1). Grain length and thickness were significantly reduced in the mutant (Fig. 1j; Table 1), whereas grain width was not affected. Compared with WT, 1,000-grain weight and grain yield per plant in the mutant were significantly decreased by 39.79 % (20.28 ± 0.24 g in the mutant vs. 28.35 ± 0.09 g in the WT; P = 1.47E−08) and 72.55 % (7.00 ± 1.38 g in the mutant vs. 25.50 ± 3.03 g in the WT; P = 1.12E−07), respectively (Fig. 1k, l). These results demonstrate that this mutation strongly affects rice plant architecture and in turn, grain yield. Therefore, we designated this mutant pad.
Cloning of the PAD gene through a map-based cloning approach
Genetic analysis of the pad mutant indicated that the phenotype of the F1 plants was WT. In the F2 population of MH86 × pad, 30 individuals exhibited a mutant phenotype out of 112 individuals (χ2(3:1) = 0.19, P = 0.663 > 0.5), fitting Mendel’s single gene segregation. These data indicate that pad mutant phenotypic traits are controlled by a single recessive locus.
To genetically map the pad gene, an F2 population of pad × Nongken 58 was subjected to a map-based cloning approach. A total of 80 polymorphic simple sequence repeat (SSR) markers distributed across 12 chromosomes were available for genetic analysis. The pad locus was mapped to the long arm of chromosome 3 by BSA (Fig. 2a). To fine map the pad locus, a large F2 population of 2,000 plants and nine polymorphic SSR markers flanking the region of the pad locus were used. A total of 25 homozygous recessive plants with mutant phenotypes were screened and used for further analysis. The pad locus was flanked by two SSR markers, RM22 and RM14491 (Fig. 2a). To further delimit the locus, a total of 470 homozygous recessive plants were screened and analyzed using four polymorphic SSR markers, RM 6301, RM14390, RM14400 and RM6883. The pad locus was mapped to the region between the SSR markers RM14390 and RM6883 with a physical distance of 387.79 kb based on the recombination frequency (Fig. 2b). To further localize the pad gene, another four polymorphic SSR markers, RM14400, RM14407, RM14408 and RM6883, were used to screen 3,869 homozygous recessive individuals, delimiting the pad locus to a region between RM14407 and RM6883 with a physical distance of 52 kb (Fig. 2c). To accurately determine the causative gene of the pad phenotype from this 52 kb region, we identified three SNPs within this region, S1, S2 and S3, based on sequences available in rice genomic databases. Finally, the pad locus was determined to fall within an 8.7 kb region flanked by the markers S2 and S3 located on a bacterial artificial chromosome contig, OSJNBa0011L14 (Fig. 2c). Only one open reading frame (ORF), Os03g0157300, was found in this region. Sequencing of the genomic DNA in this region revealed that the mutant allele harbored a G to C substitution at nucleotide position 131 in the predicted exon 2 of Os03g0157300 (Fig. 2d). Further sequencing of the different recombinant individuals verified that this SNP was completely linked to the pad locus; therefore, this gene was considered the candidate gene in the pad mutant. Further comparisons found that the single nucleotide substitution produced the amino acid change R44P (Fig. 2d, e; Supplemental Fig. S1). Os03g0157300 is predicted to encode Mid1-complementing activity1, OsMCA1, which is a homolog of AtMCA1 and AtMCA2 in Arabidopsis (Kurusu et al. 2012; Nakagawa et al. 2007).
Map-based cloning of the PAD gene. a The pad locus was mapped to chromosome 3. b The pad locus was further delimited to the region between two SSR markers, RM14390 and RM6883. The number under each marker represents the number of recombination events. c The pad locus was finally determined to fall within an 8.7-kb region flanked by two SNP markers, S2 and S3. d Schematic of the pad gene structure. Exons and introns are indicated by black boxes and lines, respectively. White boxes represent the 5′ and 3′ untranslated regions. The position of the mutation in pad is indicated by an arrow. e Schematic representation of the predicted domains of OsMCA1/PAD and the position of the mutation in the pad mutant. The black box indicates the SYTKc domain. The light grey box indicates the coiled–coil domain. The dark grey box indicates the PLAC8 domain. The bars indicate the positions of the potential transmembrane segments (TMs)
Bioinformatic analysis using the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/smart/job_status.pl?jobid=58191266854831383877959oJYXLFqQqK) indicated that the OsMCA1/PAD protein comprises a STYKc domain at the N terminus, a coiled-coil region in the middle and a PLAC8 motif at the C terminus (Fig. 2e; Supplemental Fig. S2). Two putative transmembrane segments (TM1 and TM2) are predicted by the SOSUI system (http://bp.nuap.nagoya-u.ac.jp/sosui/cgi-bin/adv_sosui.cgi). TM1 lies in the STYKc domain, and TM2 locates in the PLAC8 motif (Fig. 2e; Supplemental Fig. S2).
Genetic complementation for the pad phenotype
To confirm that OsMCA1 corresponds to the pad locus, we artificially synthesized the OsMCA1 based on the cDNA sequence of the annotated OsMCA1 polypeptide from the Gramene database (http://ensembl.gramene.org/Oryza_sativa/Info/Index). We amplified the native promoter region and the 5′ untranslated region (UTR), from WT genome and fused into synthetic gene of OsMCA1, and then cloned into the binary vector pCAMBIA1300. A total of 27 independent transformants were obtained via Agrobacterium-mediated transformation into the mutant background. As shown in Fig. 3, all T1 transgenic lines exhibited normal plant architecture (Fig. 3a–d). The genetic complementation experiment demonstrated that the expression of OsMCA1 gene in mutant background corrects the mutant phenotypes.
Genetic complementation of the pad mutant. Transgenic rice morphology at the vegetative stage (a) and the mature stage (b). c Grain size of transgenic rice. d Panicle morphology at the mature stage. WT (left), the pad mutant (middle) and a genetic complementation line (right) are shown. Bars 10 cm in (a) and (b); 3 cm in (c); 2 cm in (d)
The OsMCA1/PAD gene is constitutively expressed in rice plants
To investigate the tissue-specific expression profile of the OsMCA1/PAD gene during rice development, we performed qRT-PCR analyses on total RNA extracted from various tissues from WT (MH86) and the mutant. In accordance with the expression pattern observed previously, OsMCA1/PAD was constitutively expressed in all tissues (Kurusu et al. 2012). In addition, the expression of OsMCA1/PAD was significantly decreased in mature, young leaf and seedling tissues in the pad mutant compared with WT (Fig. 4a). Conversely, higher expression levels in stems were observed in the pad mutant compared with WT, suggesting that OsMCA1/PAD may play different roles in leaves and stems. The decrease in OsMCA1/PAD gene expression in leaves and the increase in stems are consistent with the curly, wrinkled leaves and reduced plant height in the mutant.
Expression pattern of OsMCA1/PAD. a qRT-PCR analysis of OsMCA1/PAD in seedling leaf, whole seedling, young leaf (at the tillering stage), old leaf (at the heading stage), stem, young panicle, flower and root tissues. The mean ± SD were calculated from three biological replicates and values were determined by the Student’s t test. b–j GUS analysis of OsMCA1/PAD expression; b seedlings 2 days after germination; c shoot apex of the seedlings 2 days after germination; d seedling 4 days after germination; e leaf morphology at the vegetative stage; f stem at the booting stage; g inflorescence; h young florets; i rachilla; j grain husk. Bars 2 mm in (b); 100 µm in (c) and (i); 3 mm in (d) and (f–h); 5 mm in (e); 200 µm in (j)
To further understand the tissue specificity of OsMCA1/PAD expression in rice, a GUS reporter construct driven by a native promoter was generated and transformed into the rice variety NK58 via Agrobacterium-mediated transformation. GUS expression was detected in all tissues throughout the developmental stages (Fig. 4b–j), which was consistent with the previous results and showed the similar expression pattern of AtMCA2 in Arabidopsis (Kurusu et al. 2012; Yamanaka et al. 2010). However, strong expression was found in nodes, inflorescences, young florets and rachillae (Fig. 4f–i), implying that OsMCA1/PAD could function in the elongation of nodes and the cell development necessary for plant height and smaller grains, which is consistent with the plant architecture defect in the pad mutant.
To determine the subcellular distribution of OsMCA1/PAD, a transient assay was performed. The fluorescent signal from the GFP-OsMCA1 fusion was only detected in the plasma membrane (Fig. 5d–f), whereas GFP alone exhibited no obvious subcellular distribution (Fig. 5a–c). This result is consistent with OsMCA1 encoding a membrane protein (Kurusu et al. 2012).
OsMCA1/PAD affects cell elongation in stems and roots
Based on the expression pattern of OsMCA1 and the pad mutant phenotype, OsMCA1/PAD may affect plant architecture. To further investigate whether OsMCA1/PAD regulates cell elongation or division, we examined cell morphology in the leaves, the roots of 7-day-old seedling and the uppermost internode at the early stage of heading. Cytological studies revealed that the cell sizes in the leaves and roots of the pad mutant were smaller than in those of WT (Fig. 6a, b). In particular, the uppermost internode was significantly longer in WT than that in the mutant (Fig. 6c), which is consistent with the reduced plant height in the mutant (Fig. 1e; Table 1). Taken together, these findings suggest that cell expansion is severely disrupted in the mutant.
OsMCA1/PAD is involved in the regulation of GA metabolism
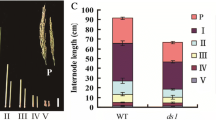
To categorize the internode elongation pattern of pad mutant, we measured the length of each internode in WT and the pad mutant. We found that internode elongation in the mutant was greatly restrained. As shown in Fig. 7a, the length from the uppermost to sixth internodes in the pad mutant was significantly reduced compared with WT. Furthermore, we found the length of each internode was uniformly reduced in the pad mutant. The internode elongation pattern of pad mutant presented as the same pattern as WT (Supplemental Fig. S3). This result suggested that the pad is a typical dn-type dwarf mutant (Takeda 1979). Moreover, the pattern is also similar to that of some GA-related dwarf mutants (Itoh et al. 2004), implying pad could be related to GA metabolism. These results were consistent with cytological studies and indicate that the decreased internode length in the pad mutant occurred mainly due to restrained cell elongation in the internodes (Fig. 6c). It simultaneously excludes the pad mutant belongs to BR-related dwarf mutant. According to the internode elongation pattern and the dwarf phenotype in the pad mutant, which are similar to some rice dwarf mutants that exhibit altered GA responses (Guo et al. 2013; Sakamoto et al. 2004; Ueguchi-Tanaka et al. 2000). Therefore, we hypothesize that the pad mutant could involve in GA metabolism. To explore whether GA plays a role in the dwarf phenotype of the pad mutant, we first compared the effects of GA on shoot elongation in the mutant and WT. Sterilized WT and mutant seeds were pretreated with or without 6.85 μM uniconazol, an inhibitor of GA biosynthesis, and then placed on 1 % agar plates supplemented with GA3 at concentrations ranging from 0.01 to 100 µM. By analyzing the length of the second leaf sheath after 8 days of treatment, we found that the pad mutant exhibited less sensitivity to exogenous GA3 compared with WT irrespective of uniconazol pretreatment (Fig. 7b–d). Although the pad mutant exhibited insensitive to exogenous GA3, it did respond to GA in a concentration-dependent manner. Furthermore, the difference in the length of the second sheath became more obvious with increasing GA3 concentrations (Fig. 7d). Therefore, OsMCA1/PAD could be involved in regulating GA metabolism.
Alterations in GA metabolism in pad. a Lengths of each internode in WT and pad plants. b Seedling phenotypes of WT (1 and 2) and the pad mutant (3 and 4) 8 days after germination. The seedlings were pretreated with (2 and 4) or without (1 and 3) 6.85 μM uniconazol on a 1 % agar plate without GA3. c Seedling phenotypes of WT (1 and 2) and the pad mutant (3 and 4) 8 days after germination. The seedlings were pretreated with (2 and 4) or without (1 and 3) 6.85 μM uniconazol on a 1 % agar plate supplemented with 1 µM GA3. d Second leaf sheath comparison between WT (circle) and pad (square) in response to GA treatment in seedlings pretreated with (closed circles and closed squares) or without (open circles and open squares) 6.85 μM uniconazol. e qRT-PCR analysis of OsGA20ox2 and OsMCA1/PAD in the uppermost to third internodes of WT and pad. f qRT-PCR analysis of the genes involved in GA metabolism in the third internode of WT and pad. The data are given as the mean ± SD (n = 10) in (a) and (d). The mean ± SD were calculated from three biological replicates in (e) and (f). Values were determined by the Student’s t test in (a) and (e–f)
To determine whether there was a difference in GA content between WT and the mutant, we monitored endogenous GA levels in the early 13-hydroxylation pathway of GA metabolism in the third internode. In the pad mutant, the levels of the precursors of GA1, GA53, GA44 and GA19, were reduced by approximately one-third to one-half compared with WT. In the pad mutant, bioactive GA (GA1) contents significantly decreased to less than one-third of WT levels (Table 2). To determine whether GA metabolism was regulated by OsMCA1/PAD, we analyzed the expression levels of genes involved in GA metabolism. The expression pattern of OsGA20ox2, the key gene involved in GA biosynthesis in rice, was similar to that of OsMCA1/PAD (Fig. 7e) and was significantly increased from the uppermost internode to the third internode in the mutant compared with WT. Moreover, other genes involved in GA biosynthesis, including OsCPS1, OsKS1, OsKO2, OsKAO and OsGA3ox2, were also up-regulated in the third internode of the pad mutant. Interestingly, the expression of GA deactivation genes OsGA2ox1 and OsGA2ox3 also significantly increased in the third internode of the pad mutant (Fig. 7f). These results suggest that OsMCA1/PAD may regulate GA biosynthesis and catabolism.
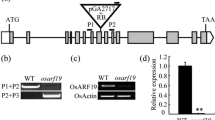
Furthermore, similar expression patterns of the genes involved in GA metabolism were also observed in the uppermost internode and second internode (Supplemental Fig. S4). Moreover, significant difference in the transcription levels of Eui1 was also found in the uppermost internode and second internode between pad and the WT, while no significant difference between pad and the WT in the third internode (Fig. 8).
Taken together, these results indicated that the loss-of-function mutation in the OsMCA1/PAD led to upregulation of the expression of the genes related to GA deactivation, which decreased bioactive GA levels. As a consequence, the lower active GA contents could induce to constitutively upregulate the expression of the genes for GA biosynthesis by feedback regulation cycles, consequently, affecting plant architecture by inhibiting cell elongation.
Discussion
MCA1 genes are highly conserved in higher plants
Two MCA1 homologs, Mid1-Complementing Activity 1 (AtMCA1) and Mid1-Complementing Activity 2 (AtMCA2), which encode plasma membrane proteins with mechanosensitive (MS) channel activity corresponding to Ca2+ uptake, have been reported in Arabidopsis (Nakagawa et al. 2007; Yamanaka et al. 2010). Increased AtMCA1 expression can promote Ca2+ uptake in roots and elevate cytoplasmic free Ca2+ concentrations upon hypo-osmotic shock. Furthermore, the N terminus of both proteins contain EF hand-like domains and are indispensable for Ca2+ uptake (Nakano et al. 2011). In addition, OsMCA1, a rice homolog of MCA in Arabidopsis, also plays a critical role in Ca2+-permeable mechanosensitive channel activity and participates in the regulation of reactive oxygen species (ROS) generation induced by hypo-osmotic stress in cultured rice cells (Kurusu et al. 2012). Phylogenetic tree analysis revealed that the OsMCA1/PAD homologous proteins group into two clades, one belonging to monocots and the other belonging to dicots. A putative uncharacterized protein encoded on chromosome 6, EEE65313.1 (O. sativa), shares 87 % sequence identity with OsMCA1/PAD (Supplemental Fig. S5). Even so, OsMCA1/PAD was described as a sole homolog of AtMCAs in the rice genome based on the genome data search (Kurusu et al. 2012). These data indicate that the OsMCA1/PAD protein may exhibit a highly conserved biological function in higher plants.
OsMCA1 is involved in the modulation of rice plant architecture
Previous studies have reported that the MCA1 gene involved in calcium signaling in Arabidopsis and rice (Nakagawa et al. 2007; Nakano et al. 2011; Yamanaka et al. 2010) and fruit size in tomato (Frary et al. 2000). No reports have shown that MCA1 affects plant architecture via regulating GA metabolism. In the present study, we isolated a natural pad mutant with defective plant architecture, including reduced plant height, a shorter root, stunted leaves, smaller panicles, fewer secondary branches, an altered number of spikelets per panicle and a lower seed rating compared with WT. Using qRT-PCR analysis and a GUS activity assay, we found that the OsMCA1/PAD gene is expressed in the majority of tissues during the vegetative and reproductive growth stages, which is consistent with the abnormal mutant phenotype. As a result, the combined mutant phenotypic characteristics could cause a reduction in grain yield.
Using a map-based cloning strategy and rescue assay, we confirmed that a point mutation in a single gene, OsMCA1, caused defects throughout the rice plant developmental stages. The genetic complementation experiment confirmed the pad is a loss-of-function mutation. This SNP was found in the kinase domain and converted a hydrophilic amino acid to a hydrophobic amino acid. A single amino acid change in the kinase domain led to major plant architecture defects, revealing that the OsMCA1/PAD gene involves in the regulation of plant architecture, which has not been reported previously. Further investigation demonstrated that mutated OsMCA1/PAD affects cell size and cell elongation in leaves, stems and internodes. This result is inconsistent with results for the OsMCA1/PAD orthologs fw2.2 and cell number regulators (CNRs) in tomato and maize, which control plant organ size by regulating cell proliferation but not cell expansion (Cong et al. 2002; Frary et al. 2000; Guo et al. 2010). Our data show that a single SNP (R44P) might endow OsMCA1/PAD with a novel function in rice.
mOsMCA1 may have gained a novel biological function
Bioinformatics and subcellular localization analyses indicated that OsMCA1/PAD encodes a transmembrane protein and contains a putative STYKc domain, a conserved coiled–coil region and a PLAC8 motif. In addition, an EF hand-like domain adjacent to the ARPK domain of MCA1 has been predicated in Arabidopsis (Nakagawa et al. 2007; Nakano et al. 2011). The STYKc domain was described as a possible dual-specificity Ser/Thr/Tyr kinase domain based on the annotation in the SMART protein database. The specificity of this class of kinases cannot be predicted, and its function is poorly understood in rice. In Arabidopsis, the STYKc domain may be involved in mediating morphological alterations (Uchida et al. 2011), intercellular communication during meristem maintenance regulation and differentiation (Hattan et al. 2004) and the formation of normal epidermal surfaces during embryogenesis (Tsuwamoto et al. 2008). There is also some information regarding STYKc function in animals. As a novel effector caspase, the characteristic kinase domain STYKc may play a role in innate immunity in sponges (Wiens et al. 2007). Cyclin-dependent kinase 5 (cdk5)/p53-regulated kinase (cprk), which was characterized as a novel neuronal kinase, harbors a STYKc domain that has kinase activity and functions in the brain (Kesavapany et al. 2003). These researches have suggested that R44P in OsMCA1/PAD could affect the kinase activity of MCA1.
OsMCA1 involves in regulating GA deactivation
Previous studies have reported that MCA1 is involved in calcium signaling and fruit size (Kurusu et al. 2012; Nakagawa et al. 2007; Yamanaka et al. 2010). However, our data show that OsMCA1/PAD affects plant architecture via regulating GA metabolism. Cytological studies indicated that internode elongation is regulated by the suppression of cell elongation. Compared with another classical GA-deficient rice mutant sd1, the pad was less responsive to exogenous GA3 treatment than WT and the sheath length of pad was not completely recovered to that of the WT at 100 µM of GA3 in seedling stage. While the sheath length of sd1 was fully rescued under exogenous GA3 treatment (Ashikari et al. 2002), implying that OsMCA1/PAD gene is distinct functions from SD1. Further analyses revealed that the key genes for GA biosynthesis, OsGA20ox2 and OsGA3ox2, were upregulated in the uppermost to third internodes of pad mutant. The expression of OsGA20ox2 in the pad mutant was significantly increased in the second and third internodes compared with WT, whereas higher expression was detected in the first internode of the pad mutant in contrast to the d1 mutant (Ueguchi-Tanaka et al. 2000). In addition, endogenous GA levels generally decreased in the third internode of the pad mutant compared with WT. In contrast, higher GA levels accumulated in the third internode of the d1 mutant. Moreover, we surprisingly observed that the expression of other genes involved in the GA biosynthesis and deactivation were upregulated. The expression profiles of those genes are highly correlated to the upregulation of OsMCA1 expression in pad mutant. As shown in Supplemental Fig. S6, Osmca1/pad upregulated the expression of the genes for GA deactivation, such as OsGA2ox1, OsGA2ox3 and Eui1, leading to the significant decrease of bioactive GA contents in the third internode. As a consequence, the reduction of bioactive GA contents could induce to constitutively upregulate the expression of the genes for GA biosynthesis by feedback regulation cycles.
In summary, our results show that OsMCA1/PAD gene is responsible for the plant architecture defect. A SNP leads to an amino acid change R44P at a STYKc domain on N terminus of OsMCA1 and inhibits cell elongation. The loss-of-function of the OsMCA1/PAD leads to upregulation of gene expression related to GA deactivation and subsequent downregulation of bioactive GA levels to finally cause the plant dwarf phenotype. Therefore, OsMCA1/PAD may function as a positive regulator involved in plant growth and development. This finding will provide insight to further understanding of the mechanism of GA metabolism regulation and the function of kind of OsMCA1/PAD genes in higher plants.
References
Ashikari M, Sasaki A, Ueguchi-Tanaka M, Itoh H, Nishimura A, Datta S, Ishiyama K, Saito T, Kobayashi M, Khush GS (2002) Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice ‘green revolution’. Breed Sci 52:143–150
Chen ML, Fu XM, Liu JQ, Ye TT, Hou SY, Huang YQ, Yuan BF, Wu Y, Feng YQ (2012) Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J Chromatogr B Analyt Technol Biomed Life Sci 905:67–74
Cong B, Liu J, Tanksley SD (2002) Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc Natl Acad Sci USA 99:13606–13611
Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou DX (2007) The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol 144:121–133
Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289:85–88
Guo M, Rupe MA, Dieter JA, Zou J, Spielbauer D, Duncan KE, Howard RJ, Hou Z, Simmons CR (2010) Cell Number Regulator1 affects plant and organ size in maize: implications for crop yield enhancement and heterosis. Plant Cell 22:1057–1073
Guo X, Hou X, Fang J, Wei P, Xu B, Chen M, Feng Y, Chu C (2013) The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J 75:403–416
Hattan J, Kanamoto H, Takemura M, Yokota A, Kohchi T (2004) Molecular characterization of the cytoplasmic interacting protein of the receptor kinase IRK expressed in the inflorescence and root apices of Arabidopsis. Biosci Biotechnol Biochem 68:2598–2606
Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444:11–25
Huang J, Tang D, Shen Y, Qin B, Hong L, You A, Li M, Wang X, Yu H, Gu M, Cheng Z (2010) Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J Genet Genomics 37:23–36
Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13:999–1010
Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M (2004) A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol 54:533–547
Jefferson RA (1989) The GUS reporter gene system. Nature 342:837–838
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544
Jin J, Huang W, Gao JP, Yang J, Shi M, Zhu MZ, Luo D, Lin HX (2008) Genetic control of rice plant architecture under domestication. Nat Genet 40:1365–1369
Kesavapany S, Lau KF, Ackerley S, Banner SJ, Shemilt SJ, Cooper JD, Leigh PN, Shaw CE, McLoughlin DM, Miller CC (2003) Identification of a novel, membrane-associated neuronal kinase, cyclin-dependent kinase 5/p35-regulated kinase. J Neurosci 23:4975–4983
Kurusu T, Nishikawa D, Yamazaki Y, Gotoh M, Nakano M, Hamada H, Yamanaka T, Iida K, Nakagawa Y, Saji H, Shinozaki K, Iida H, Kuchitsu K (2012) Plasma membrane protein OsMCA1 is involved in regulation of hypo-osmotic shock-induced Ca2 + influx and modulates generation of reactive oxygen species in cultured rice cells. BMC Plant Biol 12:11
Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F, Yuan M, Luo D, Han B, Li J (2003) Control of tillering in rice. Nature 422:618–621
Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, Li B, Wang X, Li J (2007) LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res 17:402–410
Magome H, Nomura T, Hanada A, Takeda-Kamiya N, Ohnishi T, Shinma Y, Katsumata T, Kawaide H, Kamiya Y, Yamaguchi S (2013) CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci USA 110:1947–1952
Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42:545–549
Nakagawa Y, Katagiri T, Shinozaki K, Qi Z, Tatsumi H, Furuichi T, Kishigami A, Sokabe M, Kojima I, Sato S, Kato T, Tabata S, Iida K, Terashima A, Nakano M, Ikeda M, Yamanaka T, Iida H (2007) Arabidopsis plasma membrane protein crucial for Ca2 + influx and touch sensing in roots. Proc Natl Acad Sci USA 104:3639–3644
Nakano M, Iida K, Nyunoya H, Iida H (2011) Determination of structural regions important for Ca(2+) uptake activity in Arabidopsis MCA1 and MCA2 expressed in yeast. Plant Cell Physiol 52:1915–1930
Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14(Suppl):S61–S80
Sakamoto T (2006) Phytohormones and rice crop yield: strategies and opportunities for genetic improvement. Transgenic Res 15:399–404
Sakamoto T, Matsuoka M (2004) Generating high-yielding varieties by genetic manipulation of plant architecture. Curr Opin Biotechnol 15:144–147
Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, Miyao A, Hirochika H, Kitano H, Ashikari M, Matsuoka M (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134:1642–1653
Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, Tanaka H, Kitano H, Matsuoka M (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24:105–109
Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416:701–702
Springer N (2010) Shaping a better rice plant. Nat Genet 42:475–476
Takeda K (1979) Internode elongation and dwarfism in some gramineous plants. Gamma field symp 16:1–18
Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33:513–520
Tan L, Li X, Liu F, Sun X, Li C, Zhu Z, Fu Y, Cai H, Wang X, Xie D, Sun C (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet 40:1360–1364
Thomas SG, Rieu I, Steber CM (2005) Gibberellin metabolism and signaling. Vitam Horm 72:289–338
Tsuwamoto R, Fukuoka H, Takahata Y (2008) GASSHO1 and GASSHO2 encoding a putative leucine-rich repeat transmembrane-type receptor kinase are essential for the normal development of the epidermal surface in Arabidopsis embryos. Plant J 54:30–42
Uchida N, Igari K, Bogenschutz NL, Torii KU, Tasaka M (2011) Arabidopsis ERECTA-family receptor kinases mediate morphological alterations stimulated by activation of NB-LRR-type UNI proteins. Plant Cell Physiol 52:804–814
Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M (2000) Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA 97:11638–11643
Wang Y, Li J (2005) The plant architecture of rice (Oryza sativa). Plant Mol Biol 59:75–84
Wang Y, Li J (2008) Rice, rising. Nat Genet 40:1273–1275
Wiens M, Korzhev M, Perovic-Ottstadt S, Luthringer B, Brandt D, Klein S, Muller WE (2007) Toll-like receptors are part of the innate immune defense system of sponges (demospongiae: porifera). Mol Biol Evol 24:792–804
Wu X, Tang D, Li M, Wang K, Cheng Z (2013) Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol 161:317–329
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251
Yamanaka T, Nakagawa Y, Mori K, Nakano M, Imamura T, Kataoka H, Terashima A, Iida K, Kojima I, Katagiri T, Shinozaki K, Iida H (2010) MCA1 and MCA2 that mediate Ca2 + uptake have distinct and overlapping roles in Arabidopsis. Plant Physiol 152:1284–1296
Yang XC, Hwa CM (2008) Genetic modification of plant architecture and variety improvement in rice. Heredity 101:396–404
Yoshihara T, Iino M (2007) Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol 48:678–688
Yu B, Lin Z, Li H, Li X, Li J, Wang Y, Zhang X, Zhu Z, Zhai W, Wang X, Xie D, Sun C (2007) TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J 52:891–898
Zhang Q, Li J, Xue Y, Han B, Deng XW (2008) Rice 2020: a call for an international coordinated effort in rice functional genomics. Mol Plant 1:715–719
Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, Zhu X, Mander LN, Kamiya Y, Yamaguchi S, He Z (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18:442–456
Acknowledgments
We thank Prof. Yu-Qi Feng for technical assistance of GA content analysis. This work was supported by the National Natural Science Foundation of China (No.3087496).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, Z., Cheng, Q., Sun, Y. et al. A SNP in OsMCA1 responding for a plant architecture defect by deactivation of bioactive GA in rice. Plant Mol Biol 87, 17–30 (2015). https://doi.org/10.1007/s11103-014-0257-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-014-0257-y