Abstract
The CCCH type zinc finger proteins are a super family involved in many aspects of plant growth and development. In this study, we investigated the response of one CCCH type zinc finger protein AtZFP1 (At2g25900) to salt stress in Arabidopsis. The expression of AtZFP1 was upregulated by salt stress. Compared to transgenic strains, the germination rate, emerging rate of cotyledons and root length of wild plants were significantly lower under NaCl treatments, while the inhibitory effect was significantly severe in T-DNA insertion mutant strains. At germination stage, it was mainly osmotic stress when treated with NaCl. Relative to wild plants, overexpression strains maintained a higher K+, K+/Na+, chlorophyll and proline content, and lower Na+ and MDA content. Quantitative real-time PCR analysis revealed that the expression of stress related marker genes KIN1, RD29B and RD22 increased more significantly in transgenic strains by salt stress. Overexpression of AtZFP1 also enhanced oxidative and osmotic stress tolerance which was determined by measuring the expression of a set of antioxidant genes, osmotic stress genes and ion transport protein genes such as SOS1, AtP5CS1 and AtGSTU5. Overall, our results suggest that overexpression of AtZFP1 enhanced salt tolerance by maintaining ionic balance and limiting oxidative and osmotic stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abiotic stresses such as cold, water deficit and salt stress severely reduce crop productivity. Soil salinization is a major problem worldwide (Boyer 1982) and limits production of various crops all over the world (Munns and Tester 2008). At present, there are approximately 10 billion hectares of saline soil in the world, which is more than 7 % of the total cultivated area (Tester and Davenport 2003). Salt stress has become one of the most important abiotic stress factors that limit plant growth. Soil salinization areas are rapidly expanding as is global land desertification, which is expected to reach half of the global arable land by the year 2050 (Vinocur and Altman 2005). For a long time, improving crop resistance has been regarded as an important goal for future agricultural development. Throughout evolution, plants have developed many mechanisms to adjust to stress resistance (such as salt-induced stress), such as causing a series of physiological and biochemical changes, and inducing the expression of functional and regulatory genes. The mechanisms linking salt stress to plant survival and resistance is complex, and requires further exploration.
Salt stress-induced genes are mainly divided into two categories (Shinozaki and Yamaguchi-Shinozaki 1997): the first class of functional genes encode proteins involved in plant stress tolerance, including aquaporin genes, membrane transporter protein genes, key enzyme synthesizing osmoprotectant genes (soluble sugar, proline and glycine betaine), membrane protection protein genes (LEA proteins, osmotic adjustment protein, anti-freeze protein and molecular chaperone protein), enzyme-modifying protein genes (thiol protease, Clp protease and ubiquitin protease) and genes related to the detoxification of reactive oxygen species (glutathione S-transferase enzyme, soluble epoxide hydrolase, catalase, superoxide dismutase and ascorbate peroxidase). The second class involves genes concerned with signal transduction, which can regulate the expression of target genes. This class includes protein kinase genes (including MAP kinase, CDP protein kinase, receptor protein kinase, ribosomal protein kinases and transcription regulatory protein kinase), transcription factors (including bZIP, MYC, MYB, NAC and DREB) and the PLC protein. A typical transcription factor generally contains a DNA binding domain, a dimerization site, a transcriptional regulatory domain and a nuclear localization signal (Liu et al. 1999). In plants, the typical DNA binding domain of plant transcription factors has bZIP (leucine zipper) domains, a Zinc Finger domain and a MYC (helix-loop-helix) domain. Previous work has identified transcription factors in various types of plant involved in tackling environmental stress and regulation of growth.
Plant hormones such as abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) are important components in stress-related signaling pathways (Finkelstein et al. 2002; Fujita et al. 2006; Torres and Dangl 2005; Wasilewska et al. 2008). Among them, ABA has been considered as a universal stress hormone in higher plants (Christmann et al. 2006; Fujita et al. 2006; Wasilewska et al. 2008). Exogenous application of ABA can induce the expression of a number of stress-related genes (Shinozaki et al. 2003; Zhu 2002). However, several other genes are not responsive to exogenous ABA treatment, although they can be induced by dehydration and cold stress (Shinozaki et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2005, 2006; Zhu 2002). For this reason, at least two pathways exist for how plants respond to abiotic stresses: ABA-dependent and ABA-independent pathways (Shinozaki et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2005). There is also some degree of crosstalk that exists between these two pathways (Yamaguchi-Shinozaki and Shinozaki 2005, 2006). In Arabidopsis, many transcription factors and their cognate cis-regulatory elements that function in these two pathways have been identified (Yamaguchi-Shinozaki and Shinozaki 2005, 2006). DREBs, AREB1, RD22BP1 and MYC/MYB are known to regulate ABA-responsive gene expression by interacting with their corresponding cis-acting elements, such as DRE/CRT, ABRE and MYCRS/MYBRS (Yamaguchi-Shinozaki and Shinozaki 2005). ABRE serves as the major cis-acting element in ABA-responsive gene expression, whereas DRE works as the essential cis-element in regulating the ABA-independent response to dehydration and cold stress (Yamaguchi-Shinozaki and Shinozaki 2005, 2006). Salt and drought stresses, as well as ABA signaling pathways, constitute a complex network and there is much overlap between their signaling pathways (Knight and Knight 2001; Shinozaki et al. 2003; Zhu 2002).
Salt stress-mediated signal transduction includes both ionic signaling pathways and pathways involved in osmotic homeostasis (Xiong et al. 2002; Zhu 2001, 2002, 2003). So far, our knowledge regarding the high salt-induced osmotic stress signaling pathway, especially the ABA-independent pathway, is still largely restricted to individual genes, and the unifying picture remains hidden. For the ionic aspect of salt stress, however, a signaling pathway based on the SOS (salt overly sensitive) genes has been established by using a conventional genetic screen, with well-defined signal transducers and signal output. SOS3, a calcineurin B-like protein, senses the rise in Ca2+ concentration caused by salt stress and interacts with SOS2, a protein kinase, which in turn activates SOS1, a plasma membrane Na+/H+ antiporter (Halfter et al. 2000; Liu et al. 2000; Liu and Zhu 1998; Qiu et al. 2002; Shi et al. 2000; Zhu 2003). In addition, SOS3–SOS2 regulates the expression of some salt-responsive genes and activates or suppresses the activities of other transporters involved in Na+ homeostasis (Zhu 2002, 2003). This specialized system restores ion homeostasis of cells under salt stress. Due to the existence of complex cross-talk and overlap between salt stress and other abiotic stress (as well as the ABA pathway), this type of conventional genetic screen has provided only a limited understanding of the high salt-induced osmotic stress signaling pathway (Xiong et al. 2002; Yamaguchi-Shinozaki and Shinozaki 2006; Zhu 2002).
Zinc finger proteins act as a type of transcription factor, typically by acting on a series of different types of zinc finger domains in series. These zinc-finger domains are formed by a combination zinc ions, cysteine and histidine. These zinc finger domains constitute the protein active sites, which can combine with RNA, DNA or protein (Brown 2005; Hall 2005; Lai and Blackshear 2001; Lai et al. 2000, 2002), and are requied for expression of several eukaryotic genes (Carballo et al. 1998).
There are a lot of zinc finger proteins in plants and many of them have been cloned from mode plants such as Arabidopsis thaliana, Oryza sativa and Gossypium hirsutum. The related zinc functional protein genes for these proteins have also been studied. From available reports, studies mainly focus on C2H2 zinc finger proteins. CCCH-type zinc finger proteins are a new class of zinc finger proteins that have been discovered in recent years (Gao et al. 2002; Hendriks et al. 2001; te Kronnie et al. 1999; Thompson et al. 1996). The typical amino acid sequence of these proteins is CX6–14CX4–5CX3H, but different classes of CCCH zinc finger proteins contain different numbers of CX6–14CX4–5CX3H zinc finger domains. Research into CCCH-type zinc finger proteins are mainly focused on animals (Lai et al. 2006; Liang et al. 2008; Paterou et al. 2006; Tenlen et al. 2006). Recently, CCCH-type zinc finger proteins in plants have also been reported. Li and Thomas isolated the CCCH-type zinc finger protein gene PEI1 of Arabidopsis, which was similar to the zinc finger proteins of animals and fungi. This investigation verified that PEI1 is a process-specific transcription factor that plays an important role in the formation of the Arabidopsis embryo (Li and Thomas 1998). Li et al. (2001) separated the CCCH-type zinc finger protein gene HUA1 from Arabidopsis. This protein has six typical CCCH domains, similar to Yeast, Caenorhabditis elegans, Drosophila melanogaster and mammals. Like the CCCH-type zinc finger protein of animals, the protein binds to RNA but not to DNA, and plays an important role in the regulation of the Arabidopsis flower developmental mechanism (Li et al. 2001). Schmitz found that FES1 can interact with FRI and FLC in Arabidopsis (Schmitz et al. 2005). Kong et al. (2006) found a CCCH-type zinc finger protein, OsDOS, which is localized to the nucleus and delays leaf senescence. Wang et al. (2008) studied the rice 68 and 67 CCCH-type zinc finger proteins in Arabidopsis and analyzed the phylogenetic trees, protein models and chromosomal gene locations. By analyzing the expression patterns of these genes, the proteins were divided into 11 and 8 subfamilies, respectively. The genes encoding these proteins are regulated by biotic and abiotic stress factors (Wang et al. 2008).
The zinc finger superfamily is involved in all stages of growth and development of Arabidopsis. Researchers have not only done a lot of work in analyzing CCCH-type zinc finger protein function under salt, drought, oxidation and other stresses within Arabidopsis, but also pointed out which areas of research require more exploration. The mechanisms of how plants adapt to salt stress are very complex, and the zinc finger protein family has diverse regulatory functions; therefore, understanding the effect of stress on CCCH-type zinc finger protein function is important for understanding the mechanism of how Arabidopsis adapts to high salt conditions.
Materials and methods
Plant materials and growth conditions
Arabidopsis Col-0 was selected as the wild-type control. Arabidopsis mutants SALK_112158 and SALK_024606 were ordered from the Arabidopsis Biological Resource Center and the homozygous mutant with a T-DNA insertion within AtZFP1 (AT2G25900) was verified by PCR. Arabidopsis seeds were sterilized by a 1 min 70 % ethanol treatment followed by 1 % NaClO incubation for 15 min and washed four times with distilled water. Seeds were sown on Murashige and Skoog (MS) medium plus 0, 80, 100, 120 mM NaCl or MS medium, 80 mM NaCl, 8 mM LiCl, 144 mM mannitol and then stratified for 3 d at 4 °C before being transferred to the culture room at 22 °C day/18 °C night under a 16/8 light/dark cycle 7 days. For adult stage experiments, the seeds of the T3 transgenic strains and the WT stain were germinated and grown in basal Hoagland nutrient solution, 15-day-old Arabidopsis plants were treated with Hoagland + 100 mM NaCl solution for 14 days at 22 °C day/18 °C night under a 16/8 light/dark cycle (150 μmol m−2 s−1 and 70 % relative humidity).
AtZFP1 bioinformatics analysis
The nucleic acid sequences of the AtZFP1 gene can be obtained from TAIR website. BLASTp online and software such as DNAstar, MegAlign and DNAman were used to translate nucleic acid sequences into protein sequences, homologous sequence alignment, homology analysis, and phylogenetic tree construction; the SMART online software was used to predict functional domains and functional classification.
The subcellular localization of AtZFP1
The encoded region of AtZFP1 without the stop codon was amplified using forward (5-CCCAAGCTTGGGCTCTCTCTCTTCTAGACTCAC-3) and reverse (5-CGCGGATCCGCGTGAGTTCTCTCTACTGAGTC-3) primers. The PCR fragment was fused in frame to the pJIT163–hGFP vector via HindIII and BamHI sites for sequence confirmation. The pJIT163–hGFP–AtZFP1 transient expression vector was successfully constructed and was transformed into Agrobacterium tumefaciens EHA105. Onion epidermal cells were then infected, pJIT163–hGFP–AtZFP1 expressed in the onion epidermal, and fluorescence microscopy was used to determine the localization of the CCCH-type zinc finger protein gene AtZFP1 (Sun et al. 2007b).
Expression pattern of the AtZFP1 gene
Plants were treated with salt by the submersion of 10-day-old Arabidopsis seedlings in a solution containing 100 mM NaCl. Samples were obtained after 0, 0.5, 1, 2, 3, and 6 h of salt stress (in each case, the samples were promptly frozen in liquid nitrogen and stored at −80 °C) (Lim et al. 2010). For quantitative real-time PCR, amplification was performed with oligonucleotides specific for the ATZFP1 gene (forward primer: 5-ACGGAGGTGGTTGTTCGTGGAGT-3, reverse primer: 5-CCGGAGCTTCTTCGATCTCAAACT-3). Amplification of ACTIN2 gene (forward primer: 5-AAGCTGGGGTTTTATGAATGG-3, reverse primer 5-TTGTCACACACAAGTGCATCAT-3) was used as an internal control (Wunderlich et al. 2003).
Generation of transgenic Arabidopsis strains
The encoded region of AtZFP1 amplified using forward primers with digestion site XbaI (5-CCCAAGCTTGGGCTCTCTCTCTTCTAGACTCAC-3) and reverse primers with digestion site SalI (5-CGCGGATCCGCGTGAGTTCTCTCTACTGAGTC-3). The full-length ORF of AtZFP1 was inserted into the plant binary vector pBI121 to construct pBI121–AtZFP1. Then, the AtZFP1 gene under the control of the CaMV35S promoter was transformed into Arabidopsis using the Agrobacterium-mediated transformation method (Zhang et al. 2006).
Isolation of the ATZFP1 T-DNA insertional mutants
In order to select plants homozygous for the T-DNA insertion, we utilized specific primers for SALK_112158, LP: 5-AGCAGTTTATATCACAGACACACACCTC-3, RP: 5-TCACACGAATCACCTCTTCTACAAC-3, and specific primers for SALK_024606, LP: 5-AGGTGTGTGTCTGTGATATAAACTG-3, RP: 5-ACGGCTGTTACTTTTGAACTC-3. Plants generating no polymerase chain reaction (PCR) products with the gene-specific primers were subsequently evaluated for the presence of the T-DNA insertion using the gene-specific forward primer in combination with the T-DNA left border specific primer LBb1: 5-GCGTGGACCGCTTGCTGCAACT-3.
Transcriptional activation analysis in yeast cells
The encoded region of AtZFP1 amplified using forward primers with digestion site EcoRI (5-GCGGAATTCGCAGTTTATATCACAGACACACA-3) and reverse primers with digestion site BamHI (5-CGGGATCCTCAACCGAGTGAGTTCTCTCTA-3). DNA fragments containing the whole ORF of AtZFP1 was inserted into the EcoRI/BamHI sites of the pGBKT7 vector to create the pGBKT7–AtZFP1 construct. According to the manufacturer’s protocol (Stratagene, USA), the negative control pGBKT7 plasmids were used to transform the AH109 yeast strain. The transformed strains were dripped onto SD/–Trp or SD/–Trp–His plates, and the trans-activation activity of each protein was evaluated according to their growth status and the activity of β-galactosidase.
Determination of seed germination of Arabidopsis
Emergence of radicles from the seed coat was used as the standard of seed germination and germination rate was measured after 24 h. Germination rate (%) = number of germinated seeds/total number of seeds × 100 %.
Determination of the cotyledon growth rate
Cotyledon growth was measured as the germ breaking through the seed coat and cotyledon grow rate was recorded. Cotyledon growth rate was measured on the fourth day and the cotyledon grow rate (%) = number of seeds grown cotyledon/total number of seeds × 100 %.
Determination of root length
The root length of the different strains was measured by ImageJ after 7 days (Ward et al. 2008).
Determination of Na+ and K+ content
Na+ and K+ contents were measured according to the method described by Wang and Zhao (1995). Arabidopsis leaves found at the same position and with different NaCl treatments were weighed (0.2 g), 10 mL of ddH2O added, sealed with parafilm, boiled for 48 h, filtered, the volume topped up to 25 mL, the 100 mM NaCl treatment group diluted fourfold with ddH2O and the control group without dilution, and finally, 4 mL samples were taken to measure Na+ and K+ contents using a flame photometer 410 flame photometer.
Determination of chlorophyll content
Chlorophyll content was determined using the method described by Li et al. (2003). Arabidopsis leaves were washed in distilled water and were extracted with 80 % acetone for 48 h at darkness. The absorbency at 663, 645 and 652 nm were determined using the TU-1810 UV-spectrophotometer. Chlorophyll content was calculated as follows: Ca (mg/l) = 12.7A663 − 2.69A64; Cb (mg/l) = 22.9A645 − 4.68A663; Ct (mg/l) = 8.02A663 + 20.21A645 or A652 × 1,000/34.5.
Determination of MDA content
Leaves of 0.5 g were ground in 5 mL of 10 % three trichloroacetic acid (TCA), and homogenate was centrifuged at 3,000g for 10 min. The absorbance of the supernatant was measured at 450, 532 and 600 nm. MDA content was calculated as following: MDA levels (mM/g FW) = △A × V/155 × W.
Determination of soluble sugar content
The determination of soluble sugar content was done using the method introduced by Marschner et al. (1981). Arabidopsis leaves given different treatments were taken at the same region, the extraction was measured using the TU-1810 UV spectrophotometer OD630. Soluble sugar content was calculated as: the content of soluble sugars obtained/lessons test solution volume × total volume of test solution diluted multiples/(sample fresh weight × 106) × 100 %.
Determination of soluble proline content
Determination of proline content was done using the ninhydrin method. Proline content (μg g−1 FW) = X × Vt/W × Vs; X was quantified from the standard curve; Vt is the volume of the extract; W is the mass of the sample; Vs is the sample volume used.
Quantitative real-time PCR of stress-related genes
To evaluate the effects of AtZFP1 expression, we investigated the expression profiles of several stress-responsive genes in the wild type and the AtZFP1 overexpression lines grown under salt stress conditions. 10-day-old seedlings grown on MS agar plates were sprayed with 100 mM NaCl solution. The seedlings were harvested after 0 and 2 h for extraction of total RNA (Jiang and Deyholos 2009; Li et al. 2013). Samples were flash frozen in liquid nitrogen and kept at −80 °C until used. For quantitative real-time PCR, amplification of KIN1 gene (forward primer: 5-AAGAATGCCTTCCAAGCCGGTCAG-3, reverse primer: 5-TACACTCTTTCCCGCCTGTTGTGC-3), RD29B gene (forward primer: 5-AGAAGGAATGGTGGGGAAAG-3, reverse primer: 5-CAACTCACTTCCACCGGAAT-3), RD22 gene (forward primer: 5-ATAATCTTTTGACTTTCGATTTTACCG-3, reverse primer: 5-CTTGGACGTTGGTACTTTTCTCG-3), AtP5CS1 gene (forward primer: 5-TAGCACCCGAAGAGCCCCAT-3, reverse primer: 5-TTTCAGTTCCAACGCCAGTAGA-3), AtGSTU5 gene (forward primer: 5-ATGGCTGAGAAAGAAGAAGTGAAGC-3, reverse primer: 5-TTAAGAAGATCTCACTCTCTCTGCC-3), SOS1 gene (forward primer: 5-TTCATCATCCTCACAATGGCTCTAA-3, reverse primer: 5-CCCTCATCAAGCATCTCCCAGTA-3) (Cheng et al. 2013; Huang et al. 2012; Lin et al. 2011). Amplification of ACTIN2 gene (forward primer: 5-AAGCTGGGGTTTTATGAATGG-3, reverse primer 5-TTGTCACACACAAGTGCATCAT-3) was used as an internal control (Wunderlich et al. 2003).
Statistical analysis
Each graphical plot represents the results from three independent experiments, and the values are mean ± SD. Statistical significance was determined by Student’s t tests, and p values = 0.05 or 0.01 were considered statistically significant.
Results
Sequence analysis of AtZFP1
The AtZFP1 gene contains a complete ORF of 948 bp and the protein product of AtZFP1 is comprised of 315 amino acids with a molecular mass of 35 kDa. The AtZFP1 protein contains two CCCH-type zinc fingers. The two zinc fingers are between 130–156 and 165–188 amino acids, respectively (Fig. 1a). To investigate the evolutionary relationship among plant CCCH-type zinc finger proteins, a phylogenetic tree of the full-length amino acid sequences was constructed using the Neighbor–Joining method and compared homologous amino acid sequences of Arabidopsis, Oryza, Populus, Zea mays, Sorghum and Vitis. High homology was found at amino acids 100–200 (the red box) between the regions containing two typical CCCH zinc finger structures (Fig. 1b). By constructing phylogenetic relationships of different ZFP amino acid sequences of different plants, we found that ZFP in Arabidopsis, sorghum, maize, rice, poplar and grape have a relatively distant kinship (Fig. 1c).
The AtZFP1 protein CCCH domains (a). Multiple alignment of amino acid sequences of AtZFP1 and other plant ZFPs (b). Phylogenetic relationships between different plant ZFP family members (c). The DNAman software service was used for analysis of phylogenetic relationships. Arabidopsis (At2g25900) sequences were obtained from the TAIR database. Populus (XP_002440755.1), Vitis (XP_002275926.1), Oryza (Os01g0192000), Zea may (NP_001144558.1), Sorghum (XP_002440755.1) sequences were obtained from the NCBI database
Subcellular localization of AtZFP1
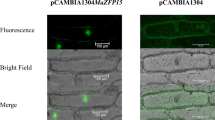
To examine the subcellular localization of AtZFP1, the recombinant constructs of the AtZFP1–pJIT163–hGFP fusion vector and the pJIT163–hGFP vector alone were transformed into onion epidermal cells via infection with Agrobacterium. As a result, the AtZFP1–pJIT163–hGFP fusion protein accumulated solely in the nucleus (Fig. 2a), whereas GFP alone was present throughout the whole cell (Fig. 2b), suggesting that AtZFP1 is a nuclear localized protein (Fig. 2).
Subcellular localiztion of AtZFP1 in onion epidermal cells. GFP alone (a) and the AtZFP1–GFP fusion proteins (b) that were driven by the CaMV 35S promoter transiently expressed in onion epidermal cells and were observed under a differential interference contrast microscope. Dark field green fluorescence A and B were obtained
Salt induces the expression of the AtZFP1 gene
To determine whether AtZFP1 expression could be induced by abiotic stress, accumulation of AtZFP1 mRNA in wild type Arabidopsis seedlings was assessed during salt treatment using quantitative Real-time PCR (qPCR). As shown, in NaCl treated samples, the AtZFP1 transcript reached its maximum level at about 2 h, but slightly declined thereafter (Fig. 3).
Expression patterns of AtZFP1. Total RNA was isolated from leaves of seedlings grown in culture. The expression levels were normalized to Arabidopsis actin. Wild-type Col-0 without treatment as control; seedling treated with 100 mM NaCl for 0, 0.5, 1, 2, 3 and 6 h as salt experimental treatment. Data were expressed as mean ± SD (n = 3 experiments)
Identification of AtZFP1 overexpression strains
To understand the role of AtZFP1 in the plant abiotic stress response, we overexpressed AtZFP1 in Arabidopsis under the control of the CaMV 35S promoter, and generated three T3 homozygous transgenic strains (S23, S28 and S32) that exhibit relatively high expression level of AtZFP1 (Fig. 4).
Identification of different Arabidopsis transgenic strains. Antibiotic (Kana) screening in overexpressing strains (a). Genomic DNA PCR of overexpression strains, lane 1 blank control, with ddH2O as template; lane 2 negative control wild type, with wild type DNA as template; lane 3–11 different overexpression strains (b). The transgenic plants were tested by real time PCR, the Arabidopsis actin gene was used as a reference gene. S15–S104 indicated the different transgenic Arabidopsis strains while WT indicated the wild type Arabidopsis (c)
Identification of T-DNA insertion mutants of AtZFP1
To locate the T-DNA inserts at the At2g25900 genomic locus, we performed PCR analysis and sequenced the amplified fragments by PCR (Fig. 5a, b). These investigations revealed that in SALK_112158 and SALK_024606, AtZFP1 was expressed at extremely low levels (Fig. 5c). The schematic diagrams of the T-DNA insertions in SALK_112158 and SALK_024606 are shown in Fig. 5d with the filled boxes representing coding regions and dashed lines denoting the untranscribed regions.
Identification of T-DNA insertion mutants of AtZFP1 strains. T-DNA insertion was confirmed using PCR with the indicated primer sets (a, b). AtZFP1 transcripts were determined by qPCR (c). Schematic representation of the AtZFP1 (At2g25900) genomic fragments. T-DNA insertion positions are illustrated by triangles (d)
Transcriptional activity of the AtZFP1 protein in yeast
To provide more evidence on the potential role of AtZFP1 as a transcription factor, we performed a transcription assay for AtZFP1 using a yeast system. The coding region of the gene was fused to the GAL4 DNA-binding domain to generate the pBD-AtZFP1 fusion plasmid. All transformants could grow well only on medium lacking tryptophan. On selection medium without tryptophan and histidine, the transformants harboring pBD-AtZFP1 grew well while the transformants containing pBD alone could not grow on the same medium. These results indicated that AtZFP1 has transactivation capacity (Fig. 6).
Transcription activity of the AtZFP1 protein in yeast. The coding sequence of AtZFP1 was cloned into pGBKT7 vectors to express the pBD-AtZFP1 fusion protein. The construct pBD-AtZFP1 and the negative control pGBKT7 vector alone were transformed into the yeast host. The transformants were dropped on SD–Trp and SD–Trp–His-mediums, respectively, and incubated at 30 °C for 2 days
Overexpressing AtZFP1 enhances salt tolerance in transgenic Arabidopsis
As shown in Fig. 7, there is no difference in phenotype of wild-type and AtZFP1 overexpressing Arabidopsis strains in conditions lacking NaCl. When the salt concentration was equal or greater than 100 mM NaCl, the inhibition effect was significantly greater in wild type plants compared to transgenic plants. When NaCl concentration was 80 mM, the seed germination rate of wild-type Arabidopsis was significantly inhibited compared to the overexpressing strains; When the NaCl concentration was 100 mM, the germination rate of overexpression strains began to be inhibited; when the NaCl concentration was 120 mM, overexpression strains and wild type plants were significantly inhibited, but the germination rate of overexpression strains was significantly higher than that of wild type plants. When the NaCl concentration was 120 mM, the cotyledon growth rate and the root length in transgenic strains and wild type plants were significantly inhibited, but the cotyledons growth rate and the root length of overexpression strains were significantly higher than that of wild type. When NaCl concentration was 80 mM, the root length of wild type and three overexpression strains of S23, S28 and S32 decreased 56.7, 36.4, 26.0 and 26.7 %, respectively; When NaCl concentration was 100 mM, the root length of wild type and three overexpression strains decreased 76.9, 51.4, 51.6 and 45.9 %, respectively; when NaCl concentration was 120 mM, the reduction percentage of root length of wild type and three overexpression strains of S23, S28 and S32 were 82.3, 62.9, 49.8 and 47.6 %, respectively. These results indicated that overexpressing AtZFP1 increased seed germination, cotyledons growth rate and root length under salt treatment (Fig. 7).
Overexpression of AtZFP1 improves salt tolerance. The phenotype of wild type and transgenic Arabidopsis plants under different NaCl concentrations for 7 days (a); effect of different NaCl treatments on the seed germination rate of wild type and overexpressing Arabidopsis plants (b); effect of different NaCl treatment on cotyledon rate of wild type and overexpressing Arabidopsis plants (c); effect of different NaCl treatment on root length of wild type and overexpressing Arabidopsis plants (d). Data represent the means of 6 replicates ± SD. For each column, different letters a, b, c, d, e, f, g, h indicate significant differences at P = 0.05
The Li+ toxicity is 10 times more poisonous than Na+ and the mass fraction of 10 % LiCl does not produce osmotic stress. The mass fraction of 10 % of LiCl (8 mM LiCl), 80 mM NaCl and 144 mM mannitol solution, which were of the same osmotic potential, were added to the MS medium. As shown in Fig. 8, under 80 mM NaCl, 8 mM LiCl and 144 mM mannitol treatment conditions, overexpression strains and wild type plants had different degrees of inhibition, but the inhibition degree of the wild type strain was significantly greater than the overexpression strains. By measuring the germination rate, cotyledon growth rate and root lengths, we found that NaCl stress had the maximum inhibitory effect and LiCl stress had the lowest inhibitory effect, which revealed that at the seed germination stage, the inhibitory effect of osmotic stress is higher than that of ionic stress (Fig. 8).
The phenotype of wild type and overexpressing Arabidopsis strains under different treatment (a); effect of different treatments on seed germination rate (b), cotyledon growth rate (c) and root length (d) of wild type and overexpressing Arabidopsis plants. Data are represented as means of 6 replicates ± SD. For each column, different letters a, b, c, d, e, f, g, h, indicate significant differences at P = 0.01
At the Arabidopsis germination stage, the NaCl suppression effect was mainly due to osmotic stress while ionic stress effects were minor. Compared to the wild type strain, overexpression strains had strong salt resistance capacity. We then asked whether at the adult stage overexpression strains still have the ability to resist saline stress.
As shown in Fig. 9, using the Hoagland nutrient solution, the overexpression and wild type Arabidopsis strains grew well overall. In three overexpression strains leaves appeared greener. One overexpression strain, S28, exhibited obvious developmental delay: the strain had less wheel seat, tillering and pods relative to wild type plants. After treatment with 100 mM NaCl for 14 days, growth of wild type and overexpression strains was inhibited. Overexpression strains, however, have longer roots, thicker stems, much larger leaf area, and less withered yellow leaves relative to that of wild type. These results suggest that overexpression strains had also a stronger capability of resisting salinity compared to wild type at the adult stage (Fig. 9a).
Phenotype of NaCl stress on the seedling stage of Arabidopsis (a); effect of NaCl treatment on the chlorophyll a, b, a + b contents of wild type and overexpression Arabidopsis strains (b); effect of NaCl treatment on Na+, K+ contents and Na+/K+ of wild type and overexpression Arabidopsis strains (c); effect of NaCl treatment on MDA (d), soluble sugar (e) and proline (f) content of wild type and overexpression Arabidopsis strains. Data are presented as the average of 3 replicates ± SD. For each column, different letters a, b indicate significant differences at P = 0.05
Under growth conditions without salt treatment, we found that overexpression strains had slightly elevated chlorophyll content compared to wild type. With the 100 mM NaCl treatment, chlorophyll content of all strains declined but chlorophyll a, chlorophyll b and chlorophyll a + b contents of overexpression strains were significantly higher than wild type, suggesting under salt stress, compared with the wild type, overexpression strains had a smaller degree of chlorophyll degradation (Fig. 9b).
After treatment with 100 mM NaCl for 14 days, leaves taken from the same position in overexpression strains and wild type Arabidopsis plants were taken to measure Na+, K+ content and the Na+/K+ ratio. Using the Hoagland nutrient solution treatment, the Na+, K+ content and Na+/K+ ratio of three overexpression strains and wild type plants showed no significant differences. After treatment with 100 mM NaCl, however, the Na+ content and Na+/K+ ratio were elevated both in the wild type and overexpression strains, but increased more rapidly in wild type plants. The K+ content decreased more significantly in the wild type. In summary, relative to wild type Arabidopsis, the overexpression strains can maintain more intracellular K+, lower level Na+ content and lower Na+/K+ ratio under salt stress, which may result in better maintenance of ionic balance and reduce harmful ion accumulation in plant cells while reducing ionic stress (Fig. 9c).
Salinity is often accompanied by oxidative stress. MDA is an important indicator of membrane lipid peroxidation. Under NaCl treatment, MDA levels increased in all strains but the increased content was more evident in wild type Arabidopsis plants (Fig. 9d).
Soluble sugar and proline act as important osmolytes and play an important role in the resistance process. As shown in Fig. 9e, under salt stress conditions, soluble sugar increased more significantly in overexpression strains than in wild type Arabidopsis. As shown in Fig. 9f, under salt stress conditions, proline increased more significantly in overexpression lines compared to the wild type Arabidopsis strain.
T-DNA mutants are sensitive to salt tolerance
As shown in Fig. 10, there is no difference in the phenotype of wild type and transgenic Arabidopsis strains in conditions lacking NaCl. At 100 mM NaCl, the inhibitory effect by T-DNA mutants is significantly larger than wild type plants. Seed germination rate, the cotyledon growth rate and the root length of T-DNA mutants were inhibited significantly compared to wild type. The reduction percentage of root length of wild type and two mutants strains SALK_112158 and SALK_024606 were 73.2, 85.7 and 82.1 %, respectively (Fig. 10).
The phenotype of wild type and T-DNA mutant Arabidopsis strains under different treatment (a); effect of different treatments on seed germination rate (b), cotyledon growth rate (c) and root length (d) of wild type and T-DNA mutant Arabidopsis plants. Data are presented as means of 6 replicates ± SD. For each column, different letters a, b, c indicate significant differences at P = 0.05
Expression of stress-related genes in AtZFP1 transgenic Arabidopsis plants
To elucidate the possible role of AtZFP1 in response to salt stress further, the expression of several stress-related marker genes of KIN1, RD29B and RD22 in AtZFP1 transgenic lines and wild type were analyzed. In order to elucidate the functional gene changes associated with stress, we evaluated expression of the ion transport related gene SOS1, oxidation related gene AtGSTU5 and osmotic substance synthesis gene AtP5CS1, encoding pyrroline-5-carboxylate synthetase, all of which are induced by various environmental stresses (Rizhsky et al. 2004). Expression levels of all these genes were induced by salt treatment both in WT and transgenic plants (Fig. 11). Under salt treatment, the expression levels of all three stress-related genes in AtZFP1 overexpressing plants were increased more than those in WT plants. This data is in agreement with other reports that suggest AtZFP1 is an upstream regulator of these genes and plays an important role in regulating the expression of stress-responsive genes (Cheng et al. 2013; Huang et al. 2011; Li et al. 2013).
Induction studies of salt-responsive marker genes and function genes in wild type and AtZFP1 overexpressing plants under salt stress. Transcript levels of marker genes of RD29B, RD22, KIN1 (a) and function genes of AtP5CS1, AtGSTU5 and SOS1 (b) were determined by real-time qRT-PCR analysis of 10-day-old seedlings treated with 100 mM NaCl for 2 h. Data is represented as the mean of 3 replicates ± SD. Mean values were normalized to the transcript levels of an internal actin control
Discussion
The cDNA of AtZFP1 (At2g25900) is 948 bp and one single open reading frame (ORF) encodes a protein of 315 amino acids. The deduced amino acid sequence of the cDNA showed that it encoded a polypeptide of approximately 35 kDa, which was a member of the Arabidopsis CCCH TZF protein family and was identified as an environmental NaCl-inducible gene in the genome wide analysis of the CCCH-type ZF family (Wang et al. 2008). Some Arabidopsis zinc finger genes have been investigated, such as Arabidopsis salt-inducible zinc finger 1 (AtSZF1) and zinc finger 2 (AtSZF2) involved in the tolerance of Arabidopsis to salt stress (Sun et al. 2007a), PEI1 involved in embryogenesis regulation (Li and Thomas 1998) and SOMNUS involved in the phytochrome signal transduction pathway (Kim et al. 2008). Lin’s results showed that expression of AtTZF1 (AtCTH/AtC3H23) mRNA is repressed by a hexokinase-dependent sugar signaling pathway. AtTZF1 acts as a positive regulator of ABA/sugar responses and a negative regulator of GA responses, at least in part by modulating gene expression. Overexpressing AtTZF1 plants showed late flowering and enhanced stress tolerance (Lin et al. 2011). In the present study, we analyzed the phenotypes of wild type, AtTZF1-overexpressing strains and loss of function mutants in the presence of salt. Our results indicated that AtZFP1 positively regulated plant responses to salt treatment (Fig. 3).
We have determined that AtZFP1 localizes to the onion nucleus (Fig. 1). Pomeranz found that AtTZF1 can traffic between the nucleus and cytoplasm, but localizes to the nucleus with higher frequency. These cytoplasmic foci vary in size, shape, intensity, and number, depending on the conditions of individual cells. The subcellular localization patterns of zinc finger proteins from different organisms are similar in Arabidopsis, maize, and rice protoplasts (Pomeranz et al. 2010).
By observing the germination stage phenotype of Arabidopsis, germination, cotyledon growth rate and root length of overexpression strains and wild type plants under 100 and 120 mM NaCl treatment, we found that these traits were inhibited significantly in transgenic strains. The suppression ability of overexpression strains was significantly lower than the wild type plants. It was also shown that when the salt-induced OSISAP1 zinc finger protein gene was transformed into Arabidopsis, the seed germination rate of transgenic plants was significantly higher than that of wild type plants under salt stress (Mukhopadhyay et al. 2004). Huang isolated and cloned a salt-induced CCCH-type zinc finger protein gene, AtOZF2, which localized to the cell membrane. The cotyledon growth rate was inhibited more strongly in the overexpression strains compared to the wild type and mutant strains at 150 mM NaCl, and the degree of inhibition in mutant strains was the largest (Huang et al. 2012). Sun found that the root growth of the atszf1-1/atszf2-1 double mutant was more inhibited than that of the wild type or single mutant plants (Sun et al. 2007a). In the present study, it was shown that overexpression of AtZFP1 increased salt tolerance of plants to a certain extent at the seed germination stage. We also found that salt stress had both ionic and osmotic effects at the germination stage. Additionally, we found that the inhibitory effect of LiCl was significantly less than that of isotonic NaCl and mannitol, suggesting that the inhibitory effect of NaCl was mainly due to osmotic stress and that the effect of the ion stress was of second-most importance at the germination stage.
Chlorophyll is an important part of the chloroplast thylakoid membrane pigment protein. Chlorophyll content reflects the photosynthesis strength of plants. Judging from the experimental data, salt treatment increased chlorophyll content in overexpression strains of Arabidopsis compared to wild type Arabidopsis plants. Kong found that the CCCH-type zinc finger protein gene OsDOS regulates rice leaf senescence and gene silencing experiments revealed that the chlorophyll content of leaves decreased from the beginning of the booting stage, followed by rapid leaf yellowing and senescence (Kong et al. 2006). Jan found that the overexpression strains of a salt-induced CCCH-type zinc finger gene, OsTZF1, in rice contains higher chlorophyll content than wild type rice. This effect was presumably due to a delay in the aging process and due to a higher maximum photochemical efficiency (Fv/Fm) of PSII (Jan et al. 2013). In summary, higher chlorophyll content in overexpression strains under salt stress preserves photosynthetic activity.
In plants, salt stress often disrupts membrane permeability. Na+ pours into cells and is accompanied by K+ leakage, so that Na+/K+ ratio decreases and the ionic balance of cells is destroyed. Under different salt treatment, Arabidopsis leaves of overexpression strains keep less Na+ and have more K+ content, maintaining a smaller Na+/K+ ratio and developing a better ability to adapt to salt stress. This observation may be explained by the fact that expression of AtZFP1 is involved in increasing Na+ efflux, maintaining K+ content to adapt salt stress in some way (Shabala et al. 2014). A salt-related Cys2/His2 type zinc finger protein gene, STZ, from the Arabidopsis cDNA library also plays an important role in directly or indirectly regulating the expression of Na+ and Li+ via the ENA efflux gene (Lippuner et al. 1996). HAL1, HAL2 and HAL3 are involved in the regulation of ion transport systems, and the expression of HAL1 was induced by NaCl and abscisic acid (Ferrando et al. 1995). To further elucidate the above mechanism, the effects of salt stress on the expression of genes responsible for ion efflux, such as SOS1, was investigated. The expression levels of SOS1 in the transgenic Arabidopsis plants increased after salt stress for 2 h and were higher in transgenic plants than that of WT plants (Fig. 9c). As a result, AtZFP1 may regulate ion transport associated with certain genes, such as certain ion channels, transporters, or proton pumps, but the specific mechanism of action requires further study.
Apart from osmotic stress and ionic stress, NaCl stress also leads to increasing production of intracellular reactive oxygen species, causing oxidative damage to cells. In this process, malondialdehyde (MDA) is widely recognized as a marker for lipid peroxidation (Székely et al. 2008). The lower MDA content indirectly reflects stronger resistance ability of plants to salt stress. During salt treatment, MDA content of overexpression strains were significantly lower than that of wild type Arabidopsis (Fig. 9d). The transcript levels of antioxidant enzyme genes and increased antioxidant activity were observed with salt treatment. Results showed that the AtGSTU5 gene had the highest transcriptional level in overexpression strains (Fig. 11b). These findings are similar to those of Huang (Huang et al. 2012). We reveal that the expression of AtZFP1 plays a positive regulatory role in salt and peroxide stress and reduces damage by reactive oxygen species.
The accumulation of compatible osmolytes, including soluble sugars, maintains cell homeostasis under stress (Rosa et al. 2009). Proline has been proposed to act as an osmoprotective molecule because it participates in the maintenance of redox balance, ROS detoxification, and the protection of protein structures through its chaperone-like features (Székely et al. 2008). In this study, an accumulation of soluble sugars and free proline was found in the AtZFP1-overexpressing plants, which was significantly higher than that of wild type plants (Fig. 9f). To further characterize this mechanism, the expression level of AtP5CS1, responsible for proline biosynthesis, was investigated. The expression level of AtP5CS1 was much higher in transgenic plants compared to WT plants (Fig. 11b). Experiments by Cheng also proved that proline biosynthesis genes, AtP5CS1 (AB022784.2) and AtP5CS2 (Y09355.1), are associated with salt tolerance (Cheng et al. 2013).
To test whether AtZFP1 affects the expression of the known stress pathway genes, several marker genes of RD29B, RD22 and KIN1 were analyzed. These genes showed a hypersensitive salt response in transgenic plants. RD29B is a cold, high salt, and dessication-inducible gene with two ABA-responsive elements (ABREs) present in its promoter region (Nakashima et al. 2006). RD22 transcription is induced by salt and ABA treatment, but no ABRE has been identified in its promoter region (Yamaguchi-Shinozaki and Shinozaki 1993; Zhang et al. 2007). KIN1, which contains the C repeat/dehydration responsive element (CRT/DRE) motif in its promoter, can be induced by cold, or ABA and dehydration treatment (Knight et al. 1999). Figure 11 A showed that the transcript levels of Rd29B, RD22 and KIN1 displayed enhanced salt induction in AtZFP1-overexpressing plants compared to WT. These observations support the concept that AtZFP1 regulates the expression of these stress marker genes under salt-, dehydration- and ABA-induced stress conditions.
Many studies have shown that salt stress, drought stress, ABA and other signaling pathways normally constitute a complex regulatory network, and some regulation genes play a role in the process (Knight and Knight 2001; Shinozaki et al. 2003; Zhu 2002). Recent studies have proven that the CCCH-type zinc finger protein gene is an important regulatory factor in different biological processes, particularly by regulating the mRNA and protein level of some regulation genes. Finding the specific target genes for CCCH-type zinc finger proteins presents a significant challenge. Sun found that Arabidopsis CCCH-type zinc finger protein genes, AtSZF1 and AtSZF2, play an important role in Arabidopsis salt stress but the mechanism is unclear (Sun et al. 2007a). The study of cotton showed that the CCCH-type zinc finger protein gene GhZFP1 plays an important role in plants under certain stresses and during the emergency response (Guo et al. 2009). The enhanced cold and drought tolerance of AtTZF1-overexpressing plants were not associated with increased ABA accumulation, suggesting that it is mainly ABA responses that are affected (Lin et al. 2011).
Transcription factors regulate many genes within plants. AtZFP1 affects not only salt stress-related genes, but also the genes controlling plant growth and development, thus affecting the plant’s overall phenotype. At the seedling stage, we found that overexpression strains had stunting phenotypes on plant height, tillering and flowering both under salt treatment and non-salt treatment conditions, which was consistent with the results of Lin (Lin et al. 2011). It was also found that expression of the CCCH-type zinc finger protein gene MsZFN delays flowering time in transgenic Arabidopsis (Chao et al. 2014).
In conclusion, AtZFP1 transcription is activated by high salt and it confers stress tolerance. Results above suggest that AtZFP1 assists in regulating cellular ion balance, osmotic balance and ROS homeostasis. In this study, the expression of marker genes are upregulated by AtZFP1 expression or by salt stress-mediated ABA signaling, suggesting that AtZFP1 activates the transcription of functional genes via an ABA-dependent or ABA-independent signaling pathway.
References
Boyer JS (1982) Plant productivity and environment. Science 218:443–448
Brown RS (2005) Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol 15:94–98
Carballo E, Lai WS, Blackshear PJ (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281:1001–1005
Chao Y, Zhang T, Yang Q, Kang J, Sun Y, Gruber MY, Qin Z (2014) Expression of the alfalfa CCCH-type zinc finger protein gene MsZFN delays flowering time in transgenic Arabidopsis thaliana. Plant Sci 215:92–99
Cheng L et al (2013) Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol Biochem 70:252–260
Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E (2006) Integration of abscisic acid signalling into plant responses. Plant Biol 8:314–325
Ferrando A, Kron SJ, Rios G, Fink GR, Serrano R (1995) Regulation of cation transport in Saccharomyces cerevisiae by the salt tolerance gene HAL3. Mol Cell Biol 15:5470–5481
Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell Online 14:S15–S45
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442
Gao G, Guo X, Goff SP (2002) Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703–1706
Guo YH, Yu YP, Wang D, Wu CA, Yang GD, Huang JG, Zheng CC (2009) GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol 183:62–75
Halfter U, Ishitani M, Zhu J-K (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci 97:3735–3740
Hall TMT (2005) Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 15:367–373
Hendriks EF, Robinson DR, Hinkins M, Matthews KR (2001) A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. EMBO J 20:6700–6711
Huang P, Chung M-S, Ju H-W, Na H-S, Lee DJ, Cheong H-S, Kim CS (2011) Physiological characterization of the Arabidopsis thaliana Oxidation-related Zinc Finger 1, a plasma membrane protein involved in oxidative stress. J Plant Res 124:699–705
Huang P, Ju H-W, Min J-H, Zhang X, Chung J-S, Cheong H-S, Kim CS (2012) Molecular and physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 2, a plasma membrane protein involved in ABA and salt stress response through the ABI2-mediated signaling pathway. Plant Cell Physiol 53:193–203
Jan A et al (2013) OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol 161:1202–1216
Jiang Y, Deyholos MK (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69:91–105
Kim DH et al (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell Online 20:1260–1277
Knight H, Knight MR (2001) Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci 6:262–267
Knight H, Veale EL, Warren GJ, Knight MR (1999) The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell Online 11:875–886
Kong Z, Li M, Yang W, Xu W, Xue Y (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141:1376–1388
Lai WS, Blackshear PJ (2001) Interactions of CCCH Zinc Finger Proteins with mRNA TRISTETRAPROLIN-MEDIATED AU-RICH ELEMENT-DEPENDENT mRNA DEGRADATION CAN OCCUR IN THE ABSENCE OF A POLY (A) TAIL. J Biol Chem 276:23144–23154
Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ (2000) Interactions of CCCH zinc finger proteins with mRNA binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem 275:17827–17837
Lai WS, Kennington EA, Blackshear PJ (2002) Interactions of CCCH zinc finger proteins with mRNA non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J Biol Chem 277:9606–9613
Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ (2006) Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol 26:9196–9208
Li Z, Thomas TL (1998) PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell Online 10:383–398
Li J, Jia D, Chen X (2001) HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell Online 13:2269–2281
Li XG, Meng QW, Jiang GQ, Zou Q (2003) The susceptibility of cucumber and sweet pepper to chilling under low irradiance is related to energy dissipation and water–water cycle. Photosynthetica 41:259–265
Li J, Han Y, Zhao Q, Li C, Xie Q, Chong K, Xu Y (2013) The E3 ligase AtRDUF1 positively regulates salt stress responses in Arabidopsis thaliana. PLoS ONE 8:e71078
Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE, Fu M (2008) A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem 283:6337–6346
Lim GH, Zhang X, Chung MS, Lee DJ, Woo YM, Cheong HS, Kim CS (2010) A putative novel transcription factor, AtSKIP, is involved in abscisic acid signalling and confers salt and osmotic tolerance in Arabidopsis. New Phytol 185:103–113
Lin PC et al (2011) The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA-and GA-mediated growth, stress and gene expression responses. Plant J 65:253–268
Lippuner V, Cyert MS, Gasser CS (1996) Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J Biol Chem 271:12859–12866
Liu J, Zhu J-K (1998) A calcium sensor homolog required for plant salt tolerance. Science 280:1943–1945
Liu L, White MJ, MacRae TH (1999) Transcription factors and their genes in higher plants. Eur J Biochem 262:247–257
Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci 97:3730–3734
Marschner H, Kuiper PJC, Kylin A (1981) Genotypic differences in the response of sugar beet plants to replacement of potassium by sodium. Physiol Plant 51:239–244
Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101:6309–6314
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Nakashima K et al (2006) Transcriptional regulation of ABI3-and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60:51–68
Paterou A, Walrad P, Craddy P, Fenn K, Matthews K (2006) Identification and stage-specific association with the translational apparatus of TbZFP3, a CCCH protein that promotes trypanosome life-cycle development. J Biol Chem 281:39002–39013
Pomeranz M, Lin P-C, Finer J, Jang J-C (2010) AtTZF gene family localizes to cytoplasmic foci. Plant Signal Behav 5:190–192
Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K (2002) Regulation of SOS1, a plasma membrane Na +/H + exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci 99:8436–8441
Rizhsky L, Davletova S, Liang H, Mittler R (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279:11736–11743
Rosa M, Prado C, Podazza G, Interdonato R, González JA, Hilal M, Prado FE (2009) Soluble sugars: metabolism, sensing and abiotic stress: a complex network in the life of plants. Plant Signal Behav 4:388–393
Schmitz RJ, Hong L, Michaels S, Amasino RM (2005) FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development 132:5471–5478
Shabala S, Shabala L, Barcelo J, Poschenrieder C (2014) Membrane transporters mediating root signalling and adaptive responses to oxygen deprivation and soil flooding. Plant Cell Environ. doi:10.1111/pce.12339
Shi H, Ishitani M, Kim C, Zhu J-K (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na +/H + antiporter. Proc Natl Acad Sci 97:6896–6901
Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115:327
Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6:410–417
Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, Li C (2007a) The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol 48:1148–1158
Sun W, Cao Z, Li Y, Zhao Y, Zhang H (2007b) A simple and effective method for protein subcellular localization using Agrobacterium-mediated transformation of onion epidermal cells. Biologia 62:529–532
Székely G et al (2008) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 53:11–28
te Kronnie G, Stroband H, Schipper H, Samallo J (1999) Zebrafish CTH1, a C3H zinc finger protein, is expressed in ovarian oocytes and embryos. Dev Genes Evol 209:443–446
Tenlen JR, Schisa JA, Diede SJ, Page BD (2006) Reduced dosage of pos-1 suppresses Mex mutants and reveals complex interactions among CCCH zinc-finger proteins during Caenorhabditis elegans embryogenesis. Genetics 174:1933–1945
Tester M, Davenport R (2003) Na + tolerance and Na + transport in higher plants. Ann Bot 91:503–527
Thompson MJ, Lai WS, Taylor GA, Blackshear PJ (1996) Cloning and characterization of two yeast genes encoding members of the CCCH class of zinc finger proteins: zinc finger-mediated impairment of cell growth. Gene 174:225–233
Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8:397–403
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16:123–132
Wang BS, Zhao KF (1995) Comparison of extractive methods of Na and K in wheat leaves. Plant Physiol Commun 31:50–52 (in Chinese)
Wang D, Guo Y, Wu C, Yang G, Li Y, Zheng C (2008) Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom 9:44
Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG (2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol 147:1181–1191
Wasilewska A et al (2008) An update on abscisic acid signaling in plants and more…. Mol Plant 1:198–217
Wunderlich M, Werr W, Schöffl F (2003) Generation of dominant-negative effects on the heat shock response in Arabidopsis thaliana by transgenic expression of a chimaeric HSF1 protein fusion construct. Plant J 35:442–451
Xiong L, Schumaker KS, Zhu J-K (2002) Cell signaling during cold, drought, and salt stress. Plant Cell Online 14:S165–S183
Yamaguchi-Shinozaki K, Shinozaki K (1993) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet MGG 238:17–25
Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci 10:88–94
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Zhang X, Henriques R, Lin S–S, Niu Q-W, Chua N-H (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641–646
Zhang Y et al (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell Online 19:1912–1929
Zhu J-K (2001) Cell signaling under salt, water and cold stresses. Curr Opin Plant Biol 4:401–406
Zhu J-K (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zhu J-K (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31300205, 31370420), Natural Science Research Foundation of Shandong (ZR2013CQ009) and the Program for Scientific research innovation team in Colleges and universities of Shandong Province.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Han, G., Wang, M., Yuan, F. et al. The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana . Plant Mol Biol 86, 237–253 (2014). https://doi.org/10.1007/s11103-014-0226-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-014-0226-5