Abstract
SymRK-interacting protein 1 (SIP1) has previously been shown to interact with the symbiosis receptor kinase, SymRK, in Lotus japonicus. A longer variant of the SIP1 transcript, SIP1L, was isolated and characterized. SIP1L contains an additional 17 amino acids that make its C-terminus a complete heat shock protein 20 (Hsp20)-like domain. In contrast to SIP1S, the longer splicing variant SIP1L could not interact with SymRK. Both SIP1L and SIP1S transcripts could be detected in developing nodules and other plant tissues, although the former was always more abundant than the latter. SIP1L and SIP1S formed heteromeric protein complexes, which were co-localized in the plasma membrane, cytoplasm and nuclei. Expression of SIP1-RNAi in transgenic hairy roots resulted in impairment in the nodule and arbuscular mycorrhizal development, suggesting an important role of SIP1 in the common symbiosis pathway. Overexpression of either SIP1L or SIP1S increased the number of nodules formed on transgenic hairy roots, indicating a positive role of SIP1 in nodulation. The SIP1S-like transcript was not detected in other higher plants tested, and the SIP1L-like proteins of these plants were capable of interacting with the SymRK orthologs. It is proposed that the loss of the ability of SIP1L to interact with SymRK in Lotus is compensated by the expression of a shorter splicing variant, SIP1S, which binds SymRK and may play a role in relaying the symbiosis signals to downstream cellular events.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Successful interactions between legume roots and specific rhizobia result in the formation of a highly specialized plant organ, the root nodule. The host cells provide organic carbon compounds to the microbe in exchange for the nitrogen nutrient derived from nitrogen fixation. Nodulation in legumes is initially activated in response to rhizobial signaling molecules, the nodulation factors (NFs) (Lerouge et al. 1990). Treatments with NFs at nano- to picomolar concentrations can induce a series of early host plant symbiotic responses, including root hair deformation, membrane potential depolarization and early nodulin gene expression (D’Haeze and Holsters 2002; Wais et al. 2002). Perceptive root systems allow the rhizobia to enter epidermal cells through the development of infection threads (ITs). Subsequently, rhizobia are released from the end of ITs into the continuously dividing host cortical cells. Inside the infected cells, rhizobia are enclosed in a unique symbiosome membrane, which is derived from the host plasma membrane. Within the symbiosomes, rhizobia reduce atmospheric nitrogen into ammonia as the root nodules become mature. In addition to the root nodule symbiosis (RNS), legumes are able to establish endosymbiosis with arbuscular mycorrhizal (AM) fungi Like rhizobia, AM fungi synthesize and secret Myc factors, which stimulate the formation of AM symbiosis (Maillet et al. 2011; Gough and Cullimore 2011). Rhizopheric AM fungi enter plant roots via hyphopodia, i.e. the intracellular penetration structures of epidermal cells. The fungal hyphae eventually extend into the root cortex, leading to the systematic colonization of host roots and intracellular arbuscule formation (Genre et al. 2005, 2008).
Recent molecular genetic studies have identified a series of host genes required for the establishment of symbioses with rhizobia and AM fungi in two model legumes, Lotus japonicus and Medicago truncatula. These genes play important roles in symbiotic signal perception and signal transduction (Oldroyd and Downie 2008). Legume root epidermal cells perceive the NF signals by two Lysin-motif (LysM) receptor-like kinases, NFR1 and NFR5 (Madsen et al. 2003; Radutoiu et al. 2003, 2007). These receptor kinases have been localized to the plasma membrane and may form a heteromeric protein complex capable of initiating signaling events to downstream components (Madsen et al. 2011). Acting downstream of the NF perception, at least eight genes participate and carry out conserved functions in a common symbiosis pathway (CSP), which is shared by RNS and AM symbiosis. These eight genes include SymRK/DMI2 (Endre et al. 2002; Stracke et al. 2002), CASTOR and POLLUX/DMI1(Ane et al. 2004; Imaizumi-Anraku et al. 2005; Charpentier et al. 2008), NUCLEOPORIN85 (NUP85), NUP133 (Saito et al. 2007; Kanamori et al. 2006), CCaMK/DMI3 (Levy et al. 2004; Tirichine et al. 2006), CYCLOPS/IPD3 (Messinese et al. 2007; Yano et al. 2008) and NENA (Groth et al. 2010). Recently, two more genes, Vapyrin and NSP2 from M. truncatula, have been shown to be required for AM symbiosis and may also be part of the common symbiosis pathway (Pumplin et al. 2010; Murray et al. 2011; Maillet et al. 2011).
The expression pattern and protein function of legume specific transcription factors (TFs) are known to play important roles in establishing the legume-specific traits (Libault et al. 2009). Besides the genes involved in the NF signal perception and the common symbiosis signaling pathway, several TF genes have been shown to participate in nodulation process. LjNIN, required for nodule inception, is the first gene identified to be essential for the formation of ITs and nodule primordia (Schauser et al. 1999). Two GRAS-type proteins, Nodulation Signaling Pathway 1 (NSP1) and NSP2, are essential for NF-induced gene expression, root hair infection and cortical cell division (CCD) (Oldroyd and Long 2003; Kalo et al. 2005; Smit et al. 2005; Heckmann et al. 2006). LjASTRAY encodes a bZIP TF, and the astray mutant exhibits the enhanced nodulation (Nishimura et al. 2002). Three closely related AP2/ERF TFs from Medicago truncatula, ERN1, ERN2 and ERN3, have been identified to bind to the ENOD11 promoter (Andriankaja et al. 2007). Additionally, the regulation of meristem specific MtHAP2.1 TF has been shown to contribute to the differentiation of nodule primordial cells (Combier et al. 2006).
Our previous work have identified a novel and ARID-containing TF, SIP1 for SymRK interacting protein 1, as an interactor of the SymRK protein kinase domain (SymRK-PK) and demonstrated that SIP1 binds the promoter of the NIN gene (Zhu et al. 2008). Here, we isolated a splicing variant of SIP1, which is longer than the original SIP1. The two variants, SIP1S and SIP1L, were derived from the same SIP1 gene through alternative splicing of the SIP1 pre-RNA. Our results suggested that SIP1L and SIP1S may form homo- and hetero-oligomers, and are required for nodule organogenesis and establishment of AM symbiosis.
Materials and methods
Plant growth
Plants of Lotus japonicus MG-20 Miyakojima were grown in a mixed soil medium containing sand and vermiculite at a 1:1 volume ratio supplied with a half-strength B&D medium (Broughton and Dilworth 1971) in growth cabinets with a 16/8 h light/dark cycle at 23 ± 1 °C. Plants of Nicotiana benthamiana were grown in growth chambers with a 16 h/8 h light/dark cycle at 26 °C.
Protein–protein interaction in yeast cells
The full-length SIP1L was amplified by PCR using SIP1L-yF and SIP1L-yR primers (Supplementary Table 2). The amplified SIP1L cDNA was cloned into the NdeI/SalI and NdeI/XhoI sites of pGBKT7 and pGADT7, generating pSIP1L-BD and pSIP1L-AD, respectively. The C-terminus of SIP1L, or SIP1Lc, was amplified using SIP1Lc-yF and SIP1L-yR primers and cloned to the NdeI/SalI and NdeI/XhoI sites of pGBKT7 and pGADT7, yielding pSIP1Lc-BD and pSIP1Lc-AD, respectively. pSymRK-PK-BD, pSymRK-PK-AD, pSIP1S-BD, pSIP1Sc-BD, pSIP1S-AD, pSIP1Sc-AD were described previously (Zhu et al. 2008). The MtDMI2-PK cDNA (GenBank acc. No. CAD10809.1) was amplified using MtDMI2-PK-yF and MtDMI2-PK-yR primers, and the OsSymRK-PK cDNA (BAC83202.1) were amplified using OsSymRK-PK-yF and OsSymRK-PK-yR primers (Supplementary Table 2). The amplified fragments were cloned into the NdeI/SalI and NdeI/XhoI sites of pGBKT7, respectively, yielding pMtDMI2-PK-BD and pOsSymRK-PK-BD. The full-length MtSIP1LL1 cDNA (Medtr3g116200.1) was amplified using MtSIP1LL1-yF and MtSIP1LL1-yR primers, and OsSIP1LL1 (LOC_Os06g41730.1) was obtained using OsSIP1LL1-yF and OsSIP1LL1-yR primers. The resulting PCR fragments were cloned into the NdeI/EcoRI and NdeI/XhoI of pGADT7, respectively, generating pMtSIP1LL-AD and pOsSIP1LL1. Assays for protein–protein interactions and Western blot analysis were performed according to the procedures described in the yeast-two hybrid handbook (Clontech, USA).
In vitro protein–protein interaction
For in vitro protein–protein pull-down assays, the SIP1L cDNA was amplified using SIP1L-eF and SIP1L-eR primers and cloned into SalI/NotI site of pGEX-6p1 to generate pGEX-6p1-SIP1L. The SIP1S cDNA was amplified using SIP1S-eF and SIP1S-eR primers and cloned at the XbaI/HindIII site of pGEX-KG, yielding pGEX-KG-SIP1S. GST and GST-tagged recombinant proteins were expressed in E. coli cells as described previously (Zhu et al. 2008), and purified using glutathione-Sepharose 4B beads (GE Healthcare). The glutathione beads containing a bound recombinant protein were incubated with a purified and soluble His-tagged protein. After incubation, proteins absorbed to the glutathione beads were eluted in SDS-sample loading buffer and resolved on SDS-PAGE. Subsequent immunoblotting of pull-down proteins was carried out as described previously (Zhu et al. 2008).
Gene expression profiling
Total RNA was isolated from fresh plant tissues using TRIZOL reagent (Invitrogen) and treated with DNase Ι (Takara) to eliminate potential genomic DNA contamination. Reverse transcription products were used for subsequent PCR reactions under the condition of 5 min at 95 °C, and 27–30 cycles of 30 s at 94 °C, 30 s at 57 °C and 25 s at 72 °C, followed by 6 min at 72 °C. The amplified products were separated on 2.5 % agarose gels. Primers of SIP1-rF and SIP1-rR were used for RT-PCR. These primers flanked the alternative RNA splicing region of SIP1, and were able to amplify both of SIP1L and SIP1S mRNA, producing two amplified DNA bands after separation on agarose gels. For identification splicing variants of the SIP1 genes in other plant species, the following primers were synthesized: MtSIP1LL1-rF, MtSIP1LL1-rR, OsSIP1LL1-rF, OsSIP1LL1-rR, AtSIP1LL1-rF and AtSIP1LL1-rR (Supplementary Table 2). These primers were designed according to the corresponding sites of SIP1-rF/R that were able to produce approximate 400-bp bands if alternative splicing did not occur in these plant species. For real-time PCR, total RNA of transgenic hairy roots was isolated and the first-strand cDNA was prepared as described above. Quantitative real-time PCR (qRT-PCR) was performed using One-Step SYBR PrimeScript qRT-PCR Mix (Toyobo). The ATP synthase (AW719841) and ubiquitin (AW720576) genes were amplified using ATPase-rtF and ATPase-rfR, and Ub-rtF and Ub-rtR primers and used as reference controls.
Subcellular localization in tobacco epidermal cells
The SIP1L/S coding sequences were amplified using SIP1L/S-coloc-F and SIP1L/S-coloc-R primers (Supplementary Table 2) and cloned as translational fusion with the GFP and DsRED coding sequences in pC1302 and pC1302GUS-DsRED at the NcoI/SpeI sites, generating pC1302-35S:SIP1L-GFP and pC1302GUS-35S:SIP1S-DsRED, respectively. pC1302GUS-DsRED was a modified plasmid in such a way that the GFP and Hygromycin-resistance coding sequences of pC1302 were replaced with the DsRED and GUS genes. These constructs were transferred into Agrobacterium tumefaciens strain EHA105. The Agrobacterium cells were harvested by centrifugation and resuspended in infiltration buffer (10 mM MES-KOH, pH 5.8, 10 mM MgCl2, 150 μM acetosyringone). The strains were mixed to a final OD600 of about 0.5 for each Agrobacterium strain. The cells were incubated on bench for 2–4 h at room temperature. The top leaves of N. benthamiana were used for infiltration with a 2-ml syringe. Fluorescence signals were observed in live leaf cells using Zeiss LSM510 laser scanning microscope two to four days after infiltration with Agrobacterium cells.
RNAi and overexpression constructs
Two SIP1 DNA fragments were amplified by PCR using L. japonicus genomic DNA and SIP1L cDNA as template with primers listed in Supplementary Table 2. The forward primers contained PstI-SmalI sites, and the reverse primers had XbaI-BamHI sites. Amplified products were digested by SmalI/BamHI or PstI/XbaI and ligated into the SmaI/BamHI site and then the PstI/XbaI site of pCAMBIA1301-35S-int-T7. The resulting construct contained a sense and an antisense SIP1 DNA sequence interrupted by an intron of the Actin gene. The SIP1L and SIP1S DNA fragments were amplified by PCR with SIP1L/S overexpression primers (Supplementary Table 2). The amplified products were digested by BspHI/BstEII and ligated into the NcoI/BstEII sites of pC1302GUS. pC1302GUS is a modified vector in such a way that the hygromycin-resistance gene of pC1302 is replaced by the GUS coding sequence, which serves as a marker in hairy root transformation. The resulting plasmids were transferred into A. rhizogenes LBA1334 strain for generation of transgenic hairy roots in L. japonicus. Transgenic hairy roots generated using the empty vector, pCAMBIA1301-35S-int-T7 and pC1302GUS, served as controls. The knock-down or the overexpression effect of SIP1 was assessed by quantitative RT-PCR analysis or semi-quantitative RT-PCR.
Hairy root transformation
Generation of transgenic hairy roots of L. japonicus was performed as described previously (Kumagai and Kouchi 2003). Briefly, Agrobacterium rhizogenes strain LBA1334 cells carrying a plasmid were used to infect the hypocotyls of Lotus seedlings. Hairy roots emerged from the hypocotyls were labeled individually. A root tip of 2–3 mm was excised for GUS staining. If a root tip was GUS-positive, the hairy root was preserved on the hypocotyl. One transgenic hairy root was preserved on each plant, and the main root and all GUS-negative hairy roots were removed. Plants with transgenic hairy roots were maintained in pots filled with sand/vermiculite (1/1 volume ratio) supplied with a half-strength B&D medium. Six days after plantation, Mesorhizobium loti was used to inoculate the transgenic hairy roots to induce nodulation.
Phenotyping of rhizobial infection
Mesorhizobium loti strain MAFF303099 harboring a plasmid pPN28 constitutively expresses a β-galactosidase (lacZ) gene (Scott and Ronson 1982) and was used to visualize the positions of ITs in hairy roots after staining with X-gal. Hairy roots were harvested and pretreated in 2.5 % glutaraldehyde in 1 × PBS buffer (pH7.0) for 2 h at room temperature for endogenous β-galactosidase inactivation. The hairy roots were then stained in X-gal solution (0.8 mg/ml of X-Gal, 2.5 mM each of potassium ferricyanide and potassium ferrocyanide in 0.2 × PBS, pH7.0) overnight at 30 °C. After washing, the roots were observed under a light microscope.
Spatial expression of the GUS reporter driven by the SIP1 promoter
A genomic fragment of ~ 2.4 kb upstream of the SIP1 start codon (ATG) containing the putative SIP1 promoter was prepared by PCR according to the genomic DNA sequence of the SIP1 locus (Chr1.CM0113.1720.r2.m) using SIP1-pF and SIP1-pR primers. The amplified DNA fragment was inserted into the HindIII/SalI site of pCAMBIA1391Z, generating pC1391Z-SIP1p::GUS. Two additional constructs, pC1391Z-SymRKp::GUS and pC1391Z-NINp::GUS were made for comparison of the gene expression patterns of SymRK and NIN with SIP1 during nodule development. They were amplified using SymRK-pF and SymRK-pR, and NIN-pF and NIN-pR primers, and inserted into the HindIII/SalI site and PstI/BamHI sites of pCAMBIA1391Z, respectively. Transgenic hairy roots produced by the plasmids was inoculated with Mesorhizobium loti and examined for GUS expression pattern.
Mycorrhizal infection assay
Transgenic hairy roots were grown in pots containing sand/vermiculite (1/1 volume ratio) and inoculated with Glomus intraradices. Three weeks after inoculation with the mycorrhizal fungus, roots were stained with trypan blue (Kumar et al. 2008) for quantification of mycorrhizal infection events under microscope.
Results
Molecular characterization of SIP1L and SIP1S
SIP1 has been described previously as an interacting protein of the protein kinase domain of SymRK, or SymRK-PK (Zhu et al. 2008) and is referred to as SIP1S in order to distinguish it from the longer splicing variant, SIP1L. The two cDNAs were identical in nucleotide sequence except that SIP1L contained an additional 51 bp sequence at the nucleotide position between 1,030 and 1,080 (Supplementary Fig. 1). Both cDNAs were derived from a single gene locus (Chr1.CM0113.1720.r2.m of Lotus japonicus assembly Build 2.5, http://www.kazusa.or.jp/lotus/) by alternative splicing of the SIP1 pre-mRNA. The presence of SIP1L and SIP1S variants in Lotus roots was confirmed by amplification and DNA sequencing of their cDNAs from different RNA samples of L. japonicus roots. The SIP1 gene has 14 exons (Fig. 1a). When all of the 14 exons were linked together, the splicing would generate the SIP1L mRNA. The SIP1S mRNA was produced without exon 12. Exon 12 was 51 bp in length and encoded an additional 17 amino acid residues in SIP1L. The additional 17 amino acids matched the missing portion of the conserved Hsp20 domain in SIP1S. Thus, SIP1L contained two conserved domains, ARID and Hsp20, while SIP1S had the ARID domain and a partial Hsp20 domain missing the β2 strand (Fig. 1b, c). The Hsp20 domain (333–419 residues in SIP1L) consists of seven β-strands (Fig. 1b) arranged in two β-sheets (Fig. 1c). The 17 amino acid sequence constitutes β2 strand and the spacer between β2 and β3 (Fig. 1b). Hsp20 domain is also referred to as the alpha-crystallin domain (ACD), which is found in alpha-crystallin-type small heat-shock proteins (HSPs), or p23-like domain, which is found in p23 and p23-like proteins. Because small HSP chaperones are involved in protein–protein interactions (Basha et al. 2004; Studer and Narberhaus 2000), the Hsp20 domain of SIP1L may be responsible for interaction with other proteins. During evolution, the truncated version of the Hsp20 domain in SIP1S may have evolved to interact with SymRK in legumes. The sequence information of SIP1L has been submitted to GenBank (accession number JN602367).
Splicing variants of the SIP1 mRNA. a SIP1 contains 14 exons (solid boxes). Exon 12 (red) is present in the SIP1L mRNA, but absent in SIP1S. The AT-rich interaction domain (ARID) and Hsp20-like domain are indicated. b Deduced peptide sequence of SIP1L. ARID and Hsp20-like domains are highlighted in light blue and orange, respectively. The 17 amino acid residues encoded by exon 12 in SIP1L are shaded in red. Seven predicted β-strands are boxed. Note that β2 strand is part of the sequence encoded by exon 12. c Homology modeling of the Hsp20-like domain of SIP1L (right) and the alpha crystallin domain (ACD; PCD:2Wj5a) from rat Hsp20 protein (left). The 3D structure of the Hsp20-like domain (PCD:1shs) of the small heat shock protein from Methanocaldodoccus jannaschii was used as the template. The 3D images were produced by Modeller at http://ps2.life.nctu.edu.tw/. The absence of the 17 amino acids in SIP1S would eliminate the β2 strand and disrupt the 3D structures of the highly conserved β2 sheets of the Hsp20-like domain
Inducible and specific expression of SIP1L and SIP1S
The expression of the SIP1 gene is elevated in L. japonicus roots after rhizobial inoculation. We designed a pair of primers that flanked the alternative splicing site in order to amplify the cDNA regions corresponding to both SIP1L and SIP1S mRNAs (Fig. 2a). The quantity of amplified products in agarose gels were estimated by ImageJ software (Fig. 2b). The results showed that the SIP1L transcript (upper band) was more abundant than SIP1S (lower band), and the two transcripts had a similar pattern of expression during nodule development. The expression of both splicing variants were induced by rhizobial inoculation and reached the highest level at 2–8 days post rhizobial inoculation. To identify whether the splice variants of SIP1L and SIP1S were present in other tissues of L. japonicus in addition to the root nodules, the same primers were used to PCR amplification using cDNAs reverse transcribed from total RNAs isolated from L. japonicus leaves, flowers and pods. We also designed three pairs of primers for amplification of the corresponding cDNA regions of MtSIP1LL1, OsSIP1LL1 and AtSIP1LL1. We found that isoforms of SIP1L/S were detected in every tissue of L. japonicus. However, the shorter variant, SIP1S, was not detected in non-legume plants O. sativa and A. thaliana and the legume plant M. truncatula (Fig. 2c, d). The DNA bands (Fig. 2d) of the O. sativa roots and leaves marked by arrows indicate the nonspecific PCR products corresponding to the EST sequences, FG962411.1 (upper) and FG958933.1 (lower).
Expression of SIP1L and SIP1S variants during nodule development and subcellular co-localization of the two proteins. a Semi-quantitative RT-PCR was used to assess the expression pattern of SIP1L and SIP1S. Total RNA was isolated from L. japonicus roots 30 min post inoculation (mpi) with M. loti or the indicated days post inoculation (dpi). A pair of primers flanking exon 12 of SIP1 were used to amplify the cDNA fragments of SIP1L (400 bp) and SIP1S (350 bp) by PCR at 31 or 27 cycles. The expression of the polyubiquitin (UBI) gene was used as the internal control. b The density of the amplified PCR products (a) was estimated quantitatively by ImageJ software. c Expression of both SIP1L and SIP1S splicing variants in roots (R), leaves (L), flowers (F) and pods (P) of L. japonicus (Lj) plants. d Absence of the SIP1S splicing variant in the seedlings (S), roots (R), leaves (L) of O. sativa (Os), Arabidopsis (At) and M. truncatula (Mt). Arrows indicate nonspecific PCR bands corresponding to the rice EST sequences, FG962411.1 (upper) and FG958933.1 (lower), as determined by DNA sequencing. e Co-expression of SIP1S::DsRED and SIP1L::GFP in tobacco epidermal cells. The red and green fluorescent signals detected using a confocal microscope were superimposed on the bright field image of the same cells (Overlay). Both SIP1L and SIP1S were co-localized to the nuclei (N), plasma membrane (PM) and cytoplasm. Bar 50 μm
Subcellular co-localization of SIP1L and SIP1S
Our previous study has shown that SIP1S is localized to the nuclei in onion epidermal cells (Zhu et al. 2008). Because the N-terminal DsRED-tagged SIP1S (Zhu et al. 2008) was found to be unstable, we constructed a new plasmid, SIP1S::DsRED, which expressed C-terminal DsRED-tagged SIP1S. We also constructed another plasmid expressing SIP1L::GFP. The two recombinant proteins were co-expressed under the CaMV 35S promoter in tobacco epidermis cells through infiltration of a mixture of two A. tumefaciens strains carrying the two plasmids. The results showed that the GFP and DsRED fluorescence signals were overlapped in the nuclei, plasma membrane (PM) and cytoplasm (Fig. 2e). The nuclei, PM and cytoplasm localization of SIP1L/S made it possible that SIP1S interact with PM receptor SymRK and transmit signals from the PM to the nucleus.
No interaction between SIP1L and SymRK
Previously, we have demonstrated that the C-terminal 184 amino acid residues of SIP1S are critical for the interaction with SymRK-PK (Zhu et al. 2008). The extra 17 amino acids of SIP1L were found to insert in this C-terminal region. To test if this insertion affected the interaction, we constructed a series of plasmids that expressed recombinant SymRK, SIP1L and SIP1S fused with the Gal4 DNA-binding domain (BD) or activation domain AD (Fig. 3a). Western blot analysis showed that all recombinant proteins were expressed properly in yeast cells (Supplementary Fig. 2). In contrast to SIP1S that was identified through interaction with SymRK (Zhu et al. 2008), SIP1L failed to interact with SymRK-PK in either of the BD or AD fusion directions (Fig. 3b), suggesting that the 17 amino acid insertion in SIP1L appears to alter the protein structure that prevents the interaction with SymRK-PK. Thus, during nodule development, the SIP1 gene is transcribed into pre-mRNA that is processed, via alternative splicing, into two mRNAs, the minor one encoding a SymRK-interacting isoform (SIP1S), whereas the major one encoding SIP1L that does not interact with SymRK and may play a different role during nodule development.
Interaction of SIP1L with SymRK, SIP1L and SIP1S. a Deletion constructs of SIP1L and SIP1S. b Interaction of SIP1L with SymRK. The indicated constructs were expressed as fusion proteins with the Gal4 DNA binding domain (BD) in pGBKT7, or activation domain (AD) in pGADT7. Yeast cells containing both plasmids were grown on SD/-Trp/-Leu containing X-gal (SD-TL + X-gal) medium and selected for protein–protein interaction on SD/-Trp/-Leu/-His/-Ade containing X-gal (SD-TLHA + X-gal) plates. Cell extracts were used for measurements of β-galactosidase activity (Miller units). The combinations of P53/SV40, Lam/SV40 (Clontech) and SymRK-PK/SIP1S were used as controls. c Self-interactions of SIP1L. d Interaction of SIP1L with SIP1S. e Pull-down assay of protein–protein interaction between SIP1L and SIP1S. The positions of His-SIP1L, GST-SIP1 (SIP1L and SIP1S) and GST are indicated. Asterisks (Lane 3) indicate degrade proteins of GST-SIP1S. f Interactions between orthologs of SymRK and SIP1L in M. truncatula and O. sativa
To investigate if SIP1L-like proteins of M. truncatula and nonlegume O. sativa and A. thaliana interact with SymRK orthologs, we performed protein–protein interaction test in the yeast two-hybrid system. For this, plasmids pBD-MtDMI2-PK and pAD-MtSIP1LL1-1, as well as pBD-OsSymRK-PK and pAD-OsSIP1LL1, were transformed into yeast cells. The results showed that OsSymRK interacted with OsSIP1LL1, and the interaction between MtDMI2 and MtSIP1LL1 was weak, yet readily detectable (Fig. 3e). Thus, the interactions between orthologs of SymRK and SIP1L are conserved in legumes and nonlegume plants, and may play important roles in vascular development, which is required for not only for nodule formation, but also for leaf, stem and flower functions. In L. japonicus, LjSIP1L could not interact with LjSymRK. To compensate this, a shorter variant, SIP1S, was produced, which interacted with LjSymRK.
Formation of oligomeric protein complex
SIP1S has previously been shown to dimerize with itself (Zhu et al. 2008). We asked if SIP1L retained the dimerization property and if it dimerized with SIP1S. We constructed a set of plasmids that expressed recombinant proteins fused with the BD or AD domain (Fig. 3a). These BD/AD fusion proteins were properly expressed in yeast cells as indicated by Western blot analysis (Supplementary Fig. 3). The result showed that SIP1L could interact with itself and with SIP1S, and the C-terminal region is critical for these interactions (Fig. 3c, d). The interaction between SIP1Lc-BD and SIP1S-AD was very strong. However, the interactions of SIP1L/SIP1S, SIP1L/SIP1Sc and SIP1Lc/SIP1Sc combinations were relatively weak, as assessed on the basis of the poor cell growth on SD/-Trp/-Leu/-His/-Ade selective plates (Fig. 3d). Proper protein conformation of the C-terminus of SIP1 may thus be critical for protein–protein interactions, and may play a role in relaying symbiotic signals from the SymRK receptor in legume roots. Similar observations have also been reported for yeast cells expressing human WASP and C5a receptor proteins (Tardif et al. 2003) or expressing Lotus CIP73 and CCaMK (Kang et al. 2011).
Interaction between SIP1L and SIP1S in vitro
We performed in vitro pull-down assays to test if the interaction between SIP1L and SIP1S took place in a yeast-free system. SIP1L and SIP1S were expressed as GST-tag fusion proteins and immobilized on glutathione-beads. Purified and soluble His-tag SIP1L protein was incubated with the GST-SIP1L and GST-SIP1S proteins absorbed on glutathione-beads. GST alone and glutathione-beads were used as negative controls. After washing, proteins that were pulled-down with the beads were resolved on SDS-PAGE and detected using anti-His antibody or anti-GST antibody. The results clearly showed that SIP1L could interact with itself and with SIP1S (Fig. 3e), forming homomeric and heteromeric oligo-protein complexes in vitro.
A family of plant-specific transcription factors
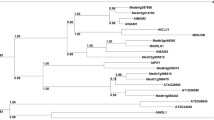
Analysis of plant protein databases revealed that there are a large number of SIP1L-like proteins (with an identity of 50–75 %) in the plant kingdom. In contrast, there are only a few proteins that contained either the ARID-like or the Hsp20-like domain, and none containing both domains in the animal and microbe protein databases. This suggests that SIP1L-like proteins are a family of plant-specific TFs that may play roles in cellular and developmental processes unique to plants, such as nodulation and flower formation. The plant SIP1L-like (SIP1LL) proteins share a high sequence similarity among themselves (Fig. 4b). By losing the 17 amino acid residues in the Hsp20-like domain (Fig. 1a), SIP1S appeared to have gained the ability to interact with SymRK. Phylogenetic tree analysis suggested that SIP1L-like proteins in dicot and monocot plants may have evolved from different ancestors (Fig. 4a). In legumes, LjSIP1L and LjSIP1S, along with GmSIP1LL1, GmSIP1LL2 and MtSIP1LL1, proceeded in a highly conserved phylogenetic clade (Fig. 4a), suggesting a possible legume-specific function of these proteins. It is interesting to note that there is a conserved tripeptide GAV motif at the C-termini of the legume SIP1L orthologs (Fig. 4a), the function of which remains unknown.
Phylogenetic analysis of plant SIP1L-like (SIP1LL) proteins. a SIP1LL sequences were aligned using Clustal X and the alignment was used to generate the phylogenetic tree using the neighbor-joining method of MAGA4.0. Numbers at branch nodes indicate bootstrap values (%) obtained from 1,000 trials. Included are SIP1LL sequences (GenBank acc. Numbers in supplementary Table 1) from L. japonicus (Lj), M. truncatula (Mt), Glycine max (Gm), Ricinus communis (Rc), Populus trichocarpa (Pt), Vitis vinifera (Vv), A. thaliana (At), Sorghum bicolor (Sb), Zea mays (Zm), Hordeum vulgare (Hv) and O. sativa (Os). They all contain both the ARID and Hsp20-lke domains except SIP1S, which lacks a complete Hsp20-like domain. LjSIP1 belongs to a conserved clade of legume-specific SIP1LLs. Three other clades are designated as monocot-specific 1 and 2, and dicot-specific 2 SIP1LLs. b Alignment of the Hsp20-like domains of LjSIP1L (LjL), LjSIP1S (LjS), GmSIP1LL1 (Gm), MtSIP1LL1 (Mt), AtSIP1LL1 (At), OsSIP1LL1 (Os), ZmSIP1LL1 (Zm) and PtSIP1LL1 (Pt). The seven conserved β-strands and positions of amino acid residues in the proteins are indicated
Fewer nodules in hairy roots expressing SIP1-RNAi
We examined the effect of the suppression of SIP1 expression on the nodulation phenotype using A. rhizogenes-mediated RNAi approach (Limpens et al. 2004; Kumagai and Kouchi 2003). Because the difference between the two variants was only the insertion of 51 bp nucleotides, we could not differentiate SIP1L and SIP1S using the RNAi approach, and have to silence them both together. We constructed two RNAi expression plasmids targeting the 3′-untranslated region (RNAi-1) and the middle portions (RNAi-2) of the coding region, (Fig. 5b, supplementary Fig. 1). These plasmids would produce double-strand hairpin RNAs that would degrade both SIP1L and SIP1S mRNAs. We searched the L. japonicus genome database and found no homolog sequence that would be targeted by these RNAi constructs. Transgenic hairy roots expressing RNAi constructs and the empty vector control (pCAMBIA1301-35S-int-T7) were inoculated with M. loti and grown in the absence of nitrogen fertilizer in order to induce nodulation. As shown in Fig. 5c, an average of 0.7 and 0.6 nodules per plant was found in hairy roots expressing RNAi-1, RNAi-2, respectively, as compared to the control (3.2 nodules per plant) 10 days after inoculation. We also counted nodules 20 days after inoculation, and the results showed that the number of nodules was reduced from 8.5 in the control to an average value of 3.0 nodules per plant for the two RNAi hairy root groups (Fig. 5a, c). In addition, we found that nearly half of RNAi hairy roots had no nodule, while almost all control plants produced nodules 20 days after rhizobial inoculation. In order to confirm that the SIP1 mRNA level was knocked down by the RNAi expression, we extracted total RNA from the transgenic hairy roots and measured the SIP1 mRNA level by semi-quantitative RT-PCR (data not shown) and quantitative RT-PCR (Supplementary Fig. 8). Our data showed that the SIP1 mRNA was repressed effectively in hairy root samples tested (Supplementary Fig. 8). We also tested if the RNAi expression affected other growth phenotype of the hairy roots. Our results showed no significant difference in the length and density of hairy roots between RNAi and the control (Supplementary Fig. 4). Taken together, the expression of SIP1-RNAi in transgenic hairy roots resulted in significant reduction in the number of nodules per plant as compared to that in the control hairy roots expressing the empty vector. The RNAi expression did not affect root growth except nodule development. Thus, SIP1 appears to play an important role in nodule formation.
Nodulation phenotypes of transgenic hairy roots. a Transgenic hairy roots were generated using Agrobacterium rhizogenes containing SIP1-RNAi constructs (1–2) or empty vector pCAMBIA1301-35S-int-T7 (Control). Ten and Twenty days after inoculation with M. loti, hairy roots were harvested and photographed. Note the reduced nodule numbers in RNAi hairy roots as compared to the control. Approximately 50 % of the RNAi hairy roots developed no nodule at all. Arrowheads indicate nodules. Bar 1 cm. b SIP1-RNAi construct. The cDNA sequences used for SIP1-RNAi-1, RNAi-2 (see Supplementary Fig. 1) were cloned in pCAMBIA1301-35S-int-T7 so that a sequence was inserted in both forward and inverted directions separated by an Actin intron sequence. The RNAi construct, a GUS reporter, and a hygromycin B-resistance marker (HygR) were flanked by T-DNA left and right boarders (LB and RB). The GUS reporter was used for identification of transgenic hairy roots. c Nodulation phenotypes of SIP1-RNAi hairy roots. Nodules were scored 10 and 20 days post-inoculation (DPI) with Rhizobium. The numbers of independent hairy root systems scored are indicated by (n). The statistical significance levels of observed differences as determined by Student’s t test are indicated by p values. Nodulation ratio is the value of the numbers of transgenic hairy root systems with nodules versus total transgenic hairy root systems. d-e Nodulation phenotypes of hairy roots overexpressing SIP1S and SIP1L. More nodules were formed on the SIP1-overexpressing hairy roots than on the control hairy roots (Ctrl). Stars indicate statistical significance below a statistical p value of 0.05. Bar 1 cm in (a, d)
No effect of SIP1-RNAi on rhizobial infection
We asked if SIP1-RNAi had any effect on root hair responses to rhizobial infection and on the formation and extension of ITs. We examined 10 hairy root systems from the control and 10 from the SIP1-RNAi-1 group. We scored the rhizobial infection phenotypes including the shepherd’s hook of root hair curling (Fig. 6a), different stages of IT formation (Fig. 6b, c, e), nodule primordial formation (Fig. 6d) and mature nodules (Fig. 6e). As shown in the results (Fig. 6f), there was no significant difference in root hair curling and IT formation between the control and the SIP1- RNAi hairy roots. The formation of nodule primordia was also not affected by the expression of the SIP1-RNAi (Fig. 6d). Therefore, the reduced number of mature nodules observed in the SIP1- RNAi hairy roots was likely due to the abortion of nodule organogenesis at a stage of nodule maturation. Thus, SIP1 may play a role in the early development of nodule organogenesis in L. japonicus.
Rhizobial infection assay of transgenic hairy roots expressing SIP1-RNAi. The transgenic hairy roots were inoculated with M. loti strain MAFF303099 that constitutively expresses a lacZ marker. Rhizobial infection phenotypes were scored based the formation of typical shepherd’s hook (a), and the position of the IT tips at the root hairs (b), at epidermis (c), at the nodule primordia (d) and at the mature nodule (e). Arrows indicate the characteristic features used for scoring the phenotypes. The recorded data of each category are presented in (f). Bar 25 μm
More nodules in hairy roots overexpressing SIP1L or SIP1S
We also examined the effect of overexpression of SIP1L and SIP1S on nodule development. We overexpressed SIP1L and SIP1S driven by the CaMV 35S promoter in transgenic hairy roots. The elevated levels of the SIP1L and SIP1S mRNA in transgenic hairy roots were confirmed by RT-PCR (Supplementary Fig. 8). The number of nodules was recorded 15 days after inoculation with M. loti. As shown in Fig. 5d-e, the average number of nodules was increased from 5.61 to 8.20 per plant and from 5.61 to 8.21 per plant in hairy roots overexpressing SIP1S and SIP1L, respectively. Similar results were obtained by a repeated experiment, suggesting that SIP1S and SIP1L both play a positive role in nodulation, which is consistent with the conclusion drawn from the SIP1-RNAi experiments.
Spatial pattern of SIP1 expression during nodule organogenesis
A construct expressing the GUS reporter driven by the SIP1 promoter was made and expressed in transgenic hairy roots of L. japonicus. Analysis of the GUS staining results revealed that the SIP1 gene was expressed constitutively in the central vascular tissues, root cap and vascular junction at the base of lateral roots in the un-inoculated hairy roots (Supplementary Fig. 5). In hairy roots inoculated with rhizobia, the GUS reporter was expressed in the whole central infected area of nodule primordia and mature nodules (Fig. 7). There was overlap in gene expression pattern between SIP1 and SymRK and NIN genes (Fig. 7a, b, c, e, f). The SIP1 gene was also found to express in the vascular tissues and vascular junction at the base of nodules (Fig. 7d, e, f and Supplementary Fig. 5) in addition to the infected zone. This suggests that SIP1 may play a role in vascular tissue differentiation at the early stages of nodule development and in vascular transport in mature nodules (Brewin 1991; Lotocka 2008).
Spatial pattern of SIP1 gene expression in nodules. GUS reporter was expressed under the control of a 2.4 kb LjSIP1 promoter. Transgenic hairy roots expressing SIP1pro::GUS were inoculated with M. loti to induce nodulation. The expression of SIP1pro::GUS was observed in the central infected zone (c), and was elevated in the vascular tissues (arrowheads) of nodules (d-f). SymRKpro::GUS (a) and NINpro::GUS (b) were highly expressed in the whole nodules. Bar 0.5 mm
Impaired arbuscule formation in SIP1-RNAi hairy roots
SymRK has been proposed to be the starting point of the common symbiosis pathway. In the absence of SymRK, AM fungal infection is arrested in the epidermal step (Gherbi et al. 2008). The expression levels of SIP1 mRNA did not vary significantly after inoculation with AM fungi as evaluated by RT-PCR (Supplementary Fig. 6b). To assess if arbuscule formation was also affected by SIP1-RNAi, we chose transgenic hairy roots of RNAi-1 for evaluation of the ability to form AM symbiosis. The transcript level of SIP1 gene in SIP1 RNAi-1 transgenic hairy roots inoculated with Glomus intraradices was examined by real-time PCR, revealing SIP1 gene was efficiently suppressed (Supplementary Fig. 6c). In the control hairy roots expressing the empty vector (pCAMBIA1301-35S-int-T7), the arbuscular fungus Glomus intraradices developed typical AM fungal hyphae that penetrated through epidermal cells from the hyphopodia and grew intracellularly through the cortex, and formed arbuscules in the inner cortical cells (Fig. 8a, b). In the SIP1 RNAi-1 transgenic hairy roots, AM hyphae were able to penetrate through the epidermis (Fig. 8e) and grew intercellularly through the entire cortex without entering into the cortical cells (Fig. 8c). The extending fungal hyphae across the intercellular space resulted in a lattice-like infection (Fig. 8f), which was similar to the AM symbiosis phenotype observed in the MtVapyrin knockdown roots (Pumplin et al. 2010). Few AM fungal hyphae were able to enter into the cortical cells, and as a consequence, arbuscules were unable to develop in SIP1 RNAi-1 hairy roots (Fig. 8c, d), which was in contrast with the control hairy roots where arbuscules were developed everywhere (Fig. 8a). We performed more detailed characterization on the arbuscule phenotypes. We scored the presence and absence of AM fungal infection events and arbuscule formation in eight hairy root systems in the control and nine in the SIP1 RNAi-1 group. AM fungal infections that did not reach the cortex were ignored. Our results showed that the total infection events were of no significant difference between the RNAi and control roots (Fig. 8g). We also did not find any significant difference in plant size and shape between the RNAi and control groups (Supplementary Fig. 6a). However, the control hairy roots had much more fungal infections that led to arbuscule formation (117 per root system) than the RNAi hairy roots (34 per root; Fig. 8h). Moreover, 84 % of the infections in the control hairy roots led to arbuscular formation, whereas only 28 % of them did so in the RNAi hairy roots (Fig. 8i). Taken together, the knock-down expression of SIP1 by RNAi had no significant effects on AM fungal infection, but could block the entry of the fungal hyphae into the root cells and subsequent arbuscular formation. These observations demonstrate a role of SIP1 in the formation of arbuscules, but not in the fungal infection step, during the establishment of symbiosis with AM fungi.
Colonization of Glomus intraradices in SIP1-RNAi hairy roots. Hairy roots expressing the empty vector (Control) and SIP1 RNAi-1 were inoculated with Glomus intraradices and stained with trypan blue to detect the arbuscules. The fungal hyphae penetrated through the epidermal cells from the hyphopodia and grew intracellularly through the cortex to form arbuscules in the control hairy roots (a, b). In SIP1 RNAi-1 hairy roots, the fungal hyphae were able to penetrate through the epidermal cells, but failed to enter the cortical cells (c-e). Arbuscules (AR) were absent completely in RNAi roots (c, e), or were observed only occasionally (d). Arrows depict the penetrating sites. Bars 0.1 mm (a, c–f) and 25 μm in (b). Comparison of numbers of AM infection events per plant (g), number of arbuscules per plant (h) and percentage of the infection events that resulted in arbuscular development (i) in the control (Ctrl) and RNAi hairy roots. Infection events were recorded based on the presence of characteristic hyphopodia on the epidermis
Discussion
SIP1S is a nuclear protein that interacts with SymRK at the plasma membrane and binds to the NIN promoter in the nuclei (Zhu et al. 2008). In this report, we described the isolation and functional analysis of SIP1L, the more abundant splicing variant of the SIP1 gene. We presented evidence showing that the two proteins were co-expressed during nodule development and might form heteromeric complex in plant cells. We performed careful investigation on the subcellular co-localization of SIP1L and SIP1S in tobacco epidermal cells. These results reveal that SIP1L and SIP1S co-localize in the plasma membrane and cytoplasm (Fig. 2e), in addition to the nuclei as shown previously (Zhu et al. 2008). SIP1S appeared to gain the ability to interact with SymRK through losing a portion of the Hsp20-like domain at its C-terminus. We hypothesize that SIP1S may transfer the symbiosis signal from SymRK receptor kinase at the plasma membrane to the nuclei where it interacts with SIP1L, and regulates downstream gene expression during nodulation process.
SIP1L contains an ARID domain and a complete Hsp20-like domain, and represents a member of a widely distributed TF family in the plant kingdom (Fig. 4). None of the members of this family of proteins, except SIP1S and SIP1L, has been studied before. The fact that animals and microbes lack TFs that contain both ARID and Hsp20-like domains suggests that the SIP1L-like proteins may function as TFs regulating cellular and developmental processes that are unique to plants, such as nodulation, flower development and alkaloid biosynthesis. There are at least six SIP1L-like proteins containing both the ARID and Hsp20-like domains in Arabidopsis thaliana, none of which have been characterized with regard to their biological functions. In addition to Lotus japonicus, other legumes also express SIP1L-like genes (Fig. 4). Because these legume SIP1L-like proteins have evolved in a highly conserved phylogenetic clade, the pair of SIP1L and SIP1S might have been evolutionarily recruited from another development mechanism into the regulation of nodule development. To investigate whether or not a SIP1S-like variant is also present in other legumes and nonlegume plants, we performed RT-PCR tests (Fig. 2d), and our results reveal that the SIP1S-like variant was absent in M. truncatula and was also not present in O. sativa and A. thaliana. Our results further show that SymRK and SIP1L orthologs of M. truncatula and O. sativa are able to interact with each other (Fig. 3f). In contrast, LjSymRK is unable to interact with LjSIP1L. We propose that the splicing variant of LjSIP1S has evolved in L. japonicus during evolution in order to compensate the lack of interaction between LjSymRK and LjSIP1L. The conserved interactions between SymRK orthologs and SIP1LL proteins may imply the biological functions that are common in plants, such as vascular development that is required for not only for nodule organogenesis, but also for the functions of root, leaf and stem and flowers.
The ARID family of DNA binding proteins was first recognized in the studies of the mouse BRIGHT protein (Herrscher et al. 1995) and Drosophila DRI (Gregory et al. 1996). The animal ARID proteins have been implicated in the control of cell growth, differentiation and development (Wilsker et al. 2002; Miransari 2011; Dallas et al. 2000). Because the SIP1 gene is also expressed in non-symbiotic tissues, such as the lateral root primordia and central vascular tissues of roots and stem (Supplementary Fig. 5), SIP1L and SIP1S may also have other biological functions related to the control of cell growth and plant development. The major difference between the animal ARID proteins and the plant SIP1L-like TFs is that the animal ARID proteins do not contain the Hsp20-like domain. Studies on the animal proteins containing the Hsp20-like domain have provided evidence for the involvement of the Hsp20-like domain in protein–protein interactions. The Hsp20-like domain is required for the interaction between human Sip1 and Skp1 (Garcia-Ranea et al. 2002; Matsuzawa and Reed 2001). Size exclusion chromatography analyses and immunoprecipitation experiments of mouse BRIGHT protein reveal that it exists as a stable dimer (Nixon et al. 2004). However, when BRIGHT is bound to DNA, a higher order of protein complexes like a tetramer may be formed (Herrscher et al. 1995). In this report, we provided evidence for the interaction of SIP1L and SIP1S (Fig. 3). Additionally, like SIP1S, SIP1L could also bind to the NIN promoter (Supplementary Fig. 7). There is a possibility that SIP1L and SIPS may form a heterodimer or heterotetramer in the nuclei and bind to NIN promoter. How the formation of a heteromeric protein complex between SIP1L and SIP1S affects the ability and specificity of DNA binding is a topic of our ongoing investigation.
Our research has focused on the biological function of SIP1 gene in nodulation. Our data showed that the expression of SIP1 RNAi in transgenic hairy roots had no observable effect on the IT formation, but could severely impair nodule development, leading to drastic reduction in the number of nodules per plant (Figs. 5, 6). The inhibitory effect of SIP1 RNAi on nodulation could be attributed to the abortion of developing nodules. Additionally, the nodule number was increased in the hairy roots overexpressing SIP1L or SIP1S, suggesting that both variants play a positive role in nodulation formation. Because the SIP1 gene was highly expressed in the vascular tissue (Fig. 7), the inhibition of nodule formation by SIP1 RNAi might be achieved via disruption in vascular differentiation and/or vascular transport (Imaizumi-Anraku et al. 2000; Lotocka 2008), both of which would block the nutrition supply for nodule development. How the vascular formation is impaired by the SIP1-RNAi expression or a mutation in the SIP1 gene remains to be investigated in future. Interestingly, the SymRK gene is also known to be constitutively transcribed in the legume root including the vascular tissue, and is not restricted to the symbiotic processes. Recently, SymRK has been shown to be involved in the vascular bundle development in Phaseolus vulgaris nodules (Sanchez-Lopez et al. 2011), implying the possible biological significance of the interaction between SymRK and SIP1S in vascular tissue development. Because SIP1L-like proteins are TFs, the role of SIP1 in controlling the vascular systems may be exerted through regulation of gene expression. Identification of genes that are targeted by SIP1L and SIP1S may provide new insight into the regulation of nodule development. In this regard, NIN, MtHAP2.1, NSP1 and NSP2 that are involved in nodule development (Schauser et al. 1999; Combier et al. 2006, 2008) could be one of the possible targets of SIP1L/SIP1S TFs. Our qRT-PCR results showed that the transcript levels of the NIN gene in transgenic hairy roots was not affected significantly by SIP1-RNAi (Supplementary Fig. 8). It is possible that other TFs may compensate the reduction of SIP1 proteins in RNAi hairy roots, as reported before (Hirsch et al. 2009). More targets of SIP1L and SIP1S could be found through comparing the transcription profiles of the sip1 mutant and the wild type plant. SIP1L and SIP1S do not appear to contain either a transcription activation domain or a suppression domain. Therefore, they may have to interact with other transcription activators or suppressors in order to regulate downstream gene expression. Identification of these possible transcription co-factors will help elucidate the mechanism of SIP1-mediated transcription regulation during nodule development.
In this study, we also aimed at evaluating the possible effect of SIP1 RNAi on AM symbiosis. SymRK is known to be required for the establishment of arbuscular mycorrhization (Parniske 2008). Mutations in SymRK can nearly completely block the infection of AM fungi in legume roots. We observed that reduced SIP1 expression by RNAi did not affect the infection of the AM fungus Glomus intraradices in root cells (Fig. 8g), suggesting that SIP1 is not involved the fungal infection process, which is consistent with the observation that SIP1 RNAi had no effect on rhizobial infection (Fig. 6). The expression of SIP1 RNAi appeared to specifically block the entry step of Glomus intraradices into root cortical cells, resulting in a lattice-like infection across the cortex. This phenotype is similar to that of MtVapyrin RNAi roots (Pumplin et al. 2010) and nsp2-2 mutant (Maillet et al. 2011). The question of how SIP1 RNAi blocks the fungal entry into cortical cells remains open and warrants further experimental investigations.
References
Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F (2007) AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19:2866–2885
Ane JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Levy J, Debelle F, Baek JM, Kalo P, Rosenberg C, Roe BA, Long SR, Denarie J, Cook DR (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303:1364–1367
Basha E, Lee GJ, Demeler B, Vierling E (2004) Chaperone activity of cytosolic small heat shock proteins from wheat. Eur J Biochem 271:1426–1436
Brewin NJ (1991) Development of the legume root nodule. Annu Rev Cell Biol 7:191–226
Broughton WJ, Dilworth MJ (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125:1075–1080
Charpentier M, Bredemeier R, Wanner G, Takeda N, Schleiff E, Parniske M (2008) Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20:3467–3479
Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernie T, Ott T, Gamas P, Crespi M, Niebel A (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20:3084–3088
Combier JP, de Billy F, Gamas P, Niebel A, Rivas S (2008) Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes Dev 22:1549–1559
Dallas PB, Pacchione S, Wilsker D, Bowrin V, Kobayashi R, Moran E (2000) The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol Cell Biol 20:3137–3146
D’Haeze W, Holsters M (2002) Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12:79R–105R
Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417:962–966
Garcia-Ranea JA, Mirey G, Camonis J, Valencia A (2002) p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett 529:162–167
Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17:3489–3499
Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P (2008) Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20:1407–1420
Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, Auguy F, Peret B, Laplaze L, Franche C, Parniske M, Bogusz D (2008) SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc Natl Acad Sci USA 105:4928–4932
Gough C, Cullimore J (2011) Lipo-chitooligosaccharide signalling in endosymbiotic plant-microbe interactions. Mol Plant Microbe Interact 24:867–878
Gregory SL, Kortschak RD, Kalionis B, Saint R (1996) Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol Cell Biol 16:792–799
Groth M, Takeda N, Perry J, Uchida H, Draxl S, Brachmann A, Sato S, Tabata S, Kawaguchi M, Wang TL, Parniske M (2010) NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22:2509–2526
Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142:1739–1750
Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R, Tucker PW (1995) The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev 9:3067–3082
Hirsch S, Kim J, Munoz A, Heckmann AB, Downie JA, Oldroyd GE (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21:545–557
Imaizumi-Anraku H, Kouchi H, Syono K, Akao S, Kawaguchi M (2000) Analysis of ENOD40 expression in alb1, a symbiotic mutant of Lotus japonicus that forms empty nodules with incompletely developed nodule vascular bundles. Mol Gen Genet 264:402–410
Imaizumi-Anraku H, Takeda N, Charpentier M, Perry J, Miwa H, Umehara Y, Kouchi H, Murakami Y, Mulder L, Vickers K, Pike J, Downie JA, Wang T, Sato S, Asamizu E, Tabata S, Yoshikawa M, Murooka Y, Wu GJ, Kawaguchi M, Kawasaki S, Parniske M, Hayashi M (2005) Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433:527–531
Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, Kiss GB, Downie JA, Oldroyd GE (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308:1786–1789
Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, Jensen TH, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J (2006) A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA 103:359–364
Kang H, Zhu H, Chu X, Yang Z, Yuan S, Yu D, Wang C, Hong Z, Zhang Z (2011) A novel interaction between CCaMK and a protein containing the Scythe_N ubiquitin-like domain in Lotus japonicus. Plant Physiol 155:1312–1324
Kumagai H, Kouchi H (2003) Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol Plant Microbe Interact 16:663–668
Kumar T, Majumdar A, Das P, Sarafis V, Ghose M (2008) Trypan blue as a fluorochrome for confocal laser scanning microscopy of arbuscular mycorrhizae in three mangroves. Biotech Histochem 83:153–159
Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Prome JC, Denarie J (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344:781–784
Levy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ane JM, Lauber E, Bisseling T, Denarie J, Rosenberg C, Debelle F (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303:1361–1364
Libault M, Joshi T, Benedito VA, Xu D, Udvardi MK, Stacey G (2009) Legume transcription factor genes: what makes legumes so special? Plant Physiol 151:991–1001
Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55:983–992
Lotocka B (2008) Vascular system within developing root nodules of Lupinus luteus L. Part 1. Juvenile stage. Acta Biol Cracov Bot 50:79–88
Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425:637–640
Madsen EB, Antolin-Llovera M, Grossmann C, Ye JY, Vieweg S, Broghammer A, Krusell L, Radutoiu S, Jensen ON, Stougaard J, Parniske M (2011) Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J 65:404–417
Maillet F, Poinsot V, Andre O, Puech-Pages V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, Martinez EA, Driguez H, Becard G, Denarie J (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469:58–63
Matsuzawa SI, Reed JC (2001) Siah-1, SIP, and Ebi collaborate in a novel pathway for β-catenin degradation linked to p53 responses. Mol Cell 7:915–926
Messinese E, Mun JH, Yeun LH, Jayaraman D, Rouge P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, Ane JM (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20:912–921
Miransari M (2011) Interactions between arbuscular mycorrhizal fungi and soil bacteria. Appl Microbiol Biotechnol 89:917–930
Murray JD, Muni RR, Torres-Jerez I, Tang Y, Allen S, Andriankaja M, Li G, Laxmi A, Cheng X, Wen J, Vaughan D, Schultze M, Sun J, Charpentier M, Oldroyd G, Tadege M, Ratet P, Mysore KS, Chen R, Udvardi MK (2011) Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J 65:244–252
Nishimura R, Ohmori M, Kawaguchi M (2002) The novel symbiotic phenotype of enhanced-nodulating mutant of Lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol 43:853–859
Nixon JC, Rajaiya J, Webb CF (2004) Mutations in the DNA-binding domain of the transcription factor Bright act as dominant negative proteins and interfere with immunoglobulin transactivation. J Biol Chem 279:52465–52472
Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59:519–546
Oldroyd GE, Long SR (2003) Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol 131:1027–1032
Parniske M (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6:763–775
Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ (2010) Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J 61:482–494
Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425:585–592
Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26:3923–3935
Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi-Anraku H, Umehara Y, Kouchi H, Murooka Y, Szczyglowski K, Downie JA, Parniske M, Hayashi M, Kawaguchi M (2007) NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19:610–624
Sanchez-Lopez R, Jauregui D, Nava N, Alvarado-Affantranger X, Montiel J, Santana O, Sanchez F, Quinto C (2011) Down-regulation of SymRK correlates with a deficiency in vascular bundle development in Phaseolus vulgaris nodules. Plant, Cell Environ 34:2109–2121
Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402:191–195
Scott DB, Ronson CW (1982) Identification and mobilization by cointegrate formation of a nodulation plasmid in Rhizobium trifolii. J Bacteriol 151:36–43
Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308:1789–1791
Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, Parniske M (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417:959–962
Studer S, Narberhaus F (2000) Chaperone activity and homo- and hetero-oligomer formation of bacterial small heat shock proteins. J Biol Chem 275:37212–37218
Tardif M, Brouchon L, Rabiet MJ, Boulay F (2003) Direct binding of a fragment of the Wiskott-Aldrich syndrome protein to the C-terminal end of the anaphylatoxin C5a receptor. Biochem J 372:453–463
Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, Downie A, Sato S, Tabata S, Kouchi H, Parniske M, Kawasaki S, Stougaard J (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441:1153–1156
Wais RJ, Keating DH, Long SR (2002) Structure-function analysis of nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol 129:211–224
Wilsker D, Patsialou A, Dallas PB, Moran E (2002) ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ 13:95–106
Yano K, Yoshida S, Muller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, Asamizu E, Tabata S, Murooka Y, Perry J, Wang TL, Kawaguchi M, Imaizumi-Anraku H, Hayashi M, Parniske M (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105:20540–20545
Zhu H, Chen T, Zhu M, Fang Q, Kang H, Hong Z, Zhang Z (2008) A novel ARID DNA-binding protein interacts with SymRK and is expressed during early nodule development in Lotus japonicus. Plant Physiol 148:337–347
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program grant no. 2010CB126502), the National Natural Science Foundation of China (grant no. 31170224, 30900096), the Ministry of Agriculture of China (grant no. 2009ZX08009-116B), the Fok Ying-Tong Education Foundation, China (grant no. 122038), the Fundamental Research Funds for the Central Universities (grant no. 2011PY136), the Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (grant no.2011SC02) and the China National Fundamental Fund of Personnel Training (Grant no. J1103510).
Author information
Authors and Affiliations
Corresponding author
Additional information
Chao Wang and Hui Zhu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, C., Zhu, H., Jin, L. et al. Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus . Plant Mol Biol 82, 97–111 (2013). https://doi.org/10.1007/s11103-013-0042-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0042-3