Abstract
Microarray analysis of a salt-tolerant wheat mutant identified a gene of unknown function that was induced by exposure to high levels of salt and subsequently denoted TaSIP (Triticum aestivum salt-induced protein). Quantitative PCR analysis revealed that TaSIP expression was induced not only by salt, but also by drought, abscisic acid (ABA), and other environmental stress factors. Transgenic rice plants that expressed an RNA interference construct specific for a rice gene homologous to TaSIP was more susceptible to salt stress than wild-type rice plants. Subcellular localization studies showed that the TaSIP localized to the cell membrane. Under conditions of salt and drought stress, transgenic Arabidopsis plants that overexpressed TaSIP showed superior physiological properties compared with control plants, including lower Na+ content and upregulation of several stress resistance genes. Staining of transgenic tissues with β-glucuronidase (GUS) failed to indicate tissue-specific activity of the full-length TaSIP promoter. Quantitative analysis of GUS fluorescence in transgenic plants treated with ABA or salt stress revealed that the region 1,176–1,410 bp from the start codon contained an ABA-responsive element and that the region 579–1,176 bp from the start codon upstream of the exon contained a salt-stress-responsive element. Based on these results, we conclude that the key part of the TaSIP gene is the region of its promoter involved in salt tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil salinization poses a major challenge to modern agriculture. Salt stress affects many plant features, including yield, protein synthesis, photosynthesis, and energy metabolism (Parida and Das 2005). Exposure to high salinity conditions results in low intracellular osmotic pressure, which causes an imbalance of ions and disrupts the dynamic ion balance between the cell interior and the apoplastic space. The resultant ion toxicity greatly inhibits plant growth and development. The plant growth cycle, from seed germination and seedling growth to vegetative and reproductive growth, is thus suppressed under conditions of salt stress (Sairam and Tyagi 2004).
Salt tolerance enables plants to complete their life cycle in an environment with a high concentration of soluble salt. Studies on the biology of salt stress have been performed for over 20 years (Flowers et al. 1977; Greenway and Munns 1980; Hasegawa et al. 2000; Zhu 2002), but despite such extensive research effects and the accumulation of substantial knowledge on plant responses to salt stress, the functions of many stress-inducible genes remain unknown. This lack of information underscores the urgent need to discover more genes that confer salt tolerance and to better understand both salt tolerance mechanisms and the physiological function of proteins involved in salt tolerance.
Wheat is a major staple crop. Improvements in the tolerance of wheat cultivars to environmental stress could increase yields and provide a secure food source to those living on land that is marginally cultivable. Due to its hexaploid genomic structure and large genome, genetic studies on wheat are challenging, especially those related to mechanisms involved in tolerance to environmental stress. However, the cloning and functional validation of stress tolerance genes continue to provide a deeper understanding of the networks of signaling in response to stress in wheat and may eventually lead to the development of more stress-tolerant crops.
Materials and methods
Plant materials and growth conditions
The salt-tolerant wheat mutant RH8706-49 and salt-susceptible mutant RH8706-34 used in this study are near-isogenic lines (Zhou et al. 2009). The Arabidopsis thaliana ecotype ‘Columbia’ and Oryza sativa group Japonica were from our laboratory stock. Seeds from RH8706-49 were soaked in water until radicle emergence, after which seedlings were transferred to a hydroponic tank and grown at 23 °C under a 16/8-h light/dark photoperiod. Plants with two true leaves were subjected to stress treatment (Ge et al. 2007). Arabidopsis plants were cultured on 1× Murashige Skoog (MS) medium (pH 5.7) that contained 30 g/L sucrose and 10 g/L agar (Ishitani et al. 1997).
Analysis of TaSIP gene expression
Wheat plants at the two-leaf stage were subjected to treatment with 170 mM NaCl, 50 μmol/L abscisic acid (ABA), or 15 % polyethylene glycol (PEG) for 0, 1, 6, 12, 24, or 72 h. Total RNA extracts for each sample were subjected to reverse transcription. The resulting cDNA was analyzed by quantitative (q)PCR using a Rotor-Gene 3000 PCR system (Gene Co. Ltd., Hong Kong, PRC). A wheat β-actin gene (GenBank accession no. AB181991) was used as an endogenous housekeeping gene to normalize expression, with SYBR Green as the fluorescent reporter dye. The CT comparative quantification method was used to determine the transcript level. The RNA from three biological replicates of each treatment was extracted and assayed by qPCR. The primers used for the quantitative reverse transcription (qRT)-PCR are shown in Table 1. The cloned TaSIP cDNA sequence is shown in Electronic Supplementary Material (ESM) Fig. S1.
Generation of transgenic Arabidopsis plants overexpressing TaSIP
Total RNA was extracted from RH8706-49 plants at the two-leaf stage and reverse-transcribed into cDNA (Ge et al. 2007) for use as the template during RT-PCR amplification. The cloned TaSIP cDNA was subcloned behind the cauliflower mosaic virus (CaMV) 35S promoter in the expression vector pCAMBIA1300. The freeze–thaw method was used to transfer the vector into Agrobacterium tumefacieus strain GV3101. Positive clones were used to transform Arabidopsis (Clough and Bent 1998). The wild-type (WT) plant was used as control plants. Positive transgenic Arabidopsis plants were selected on MS medium containing hygromycin (25 mg/L), and their transgenic nature was confirmed using RT-PCR. The primers used for qRT-PCR are shown in Table 1.
Analysis of salt and drought tolerance in TaSIP-expressing Arabidopsis plants
Seeds from homozygous transgenic lines and a control were surface-sterilized and plated onto half-strength (½) MS plates. After 3 days, germinated seeds were transferred to fresh 1/2MS plates containing either 170 or 200 mM NaCl, which were then placed vertically to allow the roots of the seedlings to elongate along the surface of the agar. Root length was recorded after 6 days of culture. After 10 days, the seedlings were transferred to pots filled with vermiculite and grown under normal light at 22 °C; after a further 2 weeks, plants were watered with plain water, 180 mM NaCl, 200 mM NaCl, 15 % PEG, or 20 % PEG. Phenotypic changes were recorded for both treated and control plants.
Generation and analysis of rice overexpressing TaSIP
The amplified TaSIP gene was cloned into the pTCK303 overexpression vector for transformation into rice. An RNA interference (RNAi) knockdown construct was prepared using the rice TaSIP homologous sequence Os05g0199100, which was then cloned into the expression vector pTCK303 as a hairpin construct. Rice plants were transformed with these two constructs, and control plants were transformed with the empty vector pTCK303.
Seeds harvested from transformed plants and control plants were soaked in a solution containing 1 μg/mL hygromycin. After 7 days, total RNA was isolated from germinated and rooted plants and assayed by RT-PCR (to validate gene overexpression) and by qPCR (to validate gene knockdown). Positive transgenic plants were grown in either Hoagland nutrient solution (HNS) or in HNS supplemented with 170 mM NaCl for 7 days at 28 °C in an illuminated incubator under a 16/8-h (light/dark) photoperiod. Phenotypic characteristics were recorded after 7 days of treatment. The primers used for qRT-PCR are shown in Table 1.
Subcellular localization of TaSIP protein
Total RNA was extracted from RH8706-49 plants at the two-leaf stage and reverse-transcribed into cDNA (Ge et al. 2007) to be used as the template for RT-PCR amplification using the primers listed in Table 1. The cloned TaSIP gene sequence was subcloned into the expression vector pCAMBIA1300-35S-GFP4; the pCAMBIA1300-35S-GFP4 (p1300-GFP) vector was used as a control. The leaves of 6-week-old tobacco plants growing under normal conditions were infiltrated with Agrobacterium cultures harboring one of the two transformation vectors. After 45–48 h, the infected leaves were placed on slides and visualized using a laser confocal microscope at an excitation wavelength of 488 nm. The primers used for qRT-PCR are shown in Table 1.
Measurement of the contents of ions, proline, soluble sugars, chlorophyll, and malondialdehyde
Seeds from homozygous Arabidopsis transgenic lines and controls were plated on 1/2MS plates and cultured in an illuminated incubator at 22 °C. After 10 days, seedlings were transferred to pots filled with vermiculite and grown at 22 °C and under normal light conditions. After another 14 days, plants were subjected to salt treatment with 200 mM NaCl solution. The treatments were terminated after 10 days, at which time the above-ground tissues were harvested to analyze the levels of ions (Zhao et al. 2009), proline (Troll and Lindsley 1955), soluble sugars (Dubois et al. 1956), chlorophyll (Arnon 1949), and malondialdehyde (MDA) (Heath and Packer 1968).
Expression of stress-related genes in transgenic Arabidopsis plants
Total RNA was extracted from transgenic homozygous Arabidopsis and control plants growing on 1/2MS medium for 15 days. The cDNA transcribed from the RNA was used in the qPCR analysis of stress-inducible genes in transgenic Arabidopsis plants (Zhao et al. 2009). The primers for these qPCR reactions are shown in ESM Table S1.
β-Glucurondiase staining and fluorescent quantitative assay
The TaSIP cDNA sequence was used to search the Chinese Spring Wheat Genome Sequence Variety library (http://www.cerealsdb.uk.net/). The full-length promoter region was extracted from the assembled sequence using a promoter prediction program. This predicted full-length promoter was divided into four segments, each containing different putative functional domains. Primers were designed to amplify each segment using PCR (Table 2).
The DNA extracted from RH8706-49 plants at the two-leaf stage was used in PCR reactions to amplify four TaSIP promoter segments, each extending different distances upstream from the start codon. The amplified regions ranged in size from 1,410 bp (segment A) for the full-length promoter to 1,176 bp (segment B), 579 bp (segment C), and 331 bp (segment D) for the three smaller segments, respectively (ESM Fig. S2). The PCR products for the four segments were cloned into the expression vector p1300-35 s-β-glucurondiase (GUS), and the resulting transformation vectors were used to transform Arabidopsis. Histochemical GUS staining of the transgenic plants and quantitative assays of GUS fluorescence were performed.
Results
Cloning and analysis of TaSIP cDNA sequence
Microarray analysis of the salt-tolerant RH8706-49 wheat line following salt treatment (our laboratory, unpublished data; Fig. 1a) and subsequent self-organizing mapping identified a salt-inducible probe (gb: BQ162513). A BLAST search against the National Center for Biotechnology Information (NCBI) expressed sequence tag (EST) libraries found several highly homologous wheat EST clones related to salt stress. The full-length cDNA sequence was built by assembling these clone sequences. The gene contained an 888-bp open reading frame. The predicted protein was 296 amino acids in length and contained no known putative conserved domain. It is 86 and 76 % homologous to two Brachypodium distachyon genes (LOC100836538 and LOC100840303) at the DNA level, and 79 and 62 % identical at the predicted protein level. These respective values are 82 and 78 % when compared with a Sorghum bicolor gene; 78 and 65 % and 71 and 57 % when compared with two Zea mays genes (LOC100279368 and LOC100273348, respectively); 75 and 61 % when compared with a rice gene (Os05g0199100). We named this gene TaSIP (Triticum aestivum salt-induced protein) and deposited the sequence in the GenBank database as HM205107. The TaSIP structure model was predicted using the I-TASSER server (Roy et al. 2010; Zhang 2008) (ESM Fig. S3).
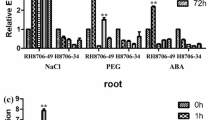
Expression of the newly identified Triticum aestivum salt-induced protein (TaSIP) gene in a salt-tolerant and salt-susceptible wheat line. a Microarray analysis of the salt-tolerant wheat cultivar RH8706-49 subjected to the salt treatment identified the salt-inducible probe BQ162513. b Expression of TaSIP in leaves of salt-tolerant RH8706-49 and salt-susceptible wheat cultivar RH8706-34 at 0, 1, 6, 12, 24, or 72 h following treatment with 170 mM NaCl, 15 % polyethylene glycol (PEG), and 50 μmol/L abscisic acid (ABA). c Expression of TaSIP in roots of salt-tolerant RH8706-49 and salt-susceptible RH8706-34 following treatment with NaCl, PEG, or ABA at 0, 1, 6, 12, 24 or 72 h. *P < 0.05 and **P < 0.01. Values are mean ± standard deviation (SD) (n = 3 independent experiments)
Analysis of TaSIP gene expression
Analysis using qPCR (Fig. 1b) revealed that TaSIP was induced in the leaves of salt-tolerant wheat cultivar RH8706-49 at most of the time points following NaCl treatment, with the highest TaSIP transcript level (4.63-fold higher than that of the untreated control) detected after 12 h of treatment. Following PEG treatment, gene expression fluctuated over time, reaching the highest level (9.23-fold higher than that of the untreated control) after 72 h of treatment. ABA treatment also induced gene expression at most of the time points following treatment, with the highest level (4.13-fold higher than that of the untreated control) being observed 1 h after treatment initiation. In the leaves of the non-salt-tolerant RH8706-34 plants, neither salt, PEG, nor ABA treatment induced a significant change in TaSIP transcript levels (Fig. 1b).
The expression of TaSIP in the roots of RH8706-49 did not follow the same trend as that in the leaves, although it was still induced in the salt-tolerant variety. Salt treatment induced expression of TaSIP in roots at most of the time points of treatment, with the highest expression level (3.38-fold higher than that of the control) observed 12 h after salt treatment. Under PEG stress, gene expression also fluctuated, with the expression levels at 12 and 72 h increased by 11.88- and 7.71-fold, respectively. For ABA treatment, gene expression also increased at most of the time points, reaching the highest level (4.42-fold higher than that of the control) after 72 h of treatment. As in the leaves, the expression of TaSIP in the roots of RH8706-34 under salt treatment did not change significantly (Fig. 1c).
Salt and drought tolerance of transgenic Arabidopsis
Arabidopsis plants transformed with TaSIP were selected for hygromycin resistance and selfed to generate the homozygous TaSIP transgenic lines L1, L2, and L6. WT plants were used as controls. Root growth was similar among the lines before salt treatment, but a significant difference in root length was found between the transgenic and WT plants after 6 days of treatment with 170 or 200 mM NaCl (Fig. 2a, b). For example, after treatment with 170 mm NaCl, the average root length of WT and transgenic plants was 0.84 and 1.42 cm, respectively. The TaSIP transgenic plants produced roots that were 1.7- or 1.4-fold longer than those of the WT plants watered with 170 or 200 mM NaCl, respectively.
Salt and drought tolerance of TaSIP transgenic Arabidopsis plants. a, b Root growth before and after salt treatment in transgenic and control plants. WT Wild type (control), L1, L2, L6 three transgenic TaSIP lines. *P < 0.05 and **P < 0.01, according to the t test. c Phenotypes of WT and L1, L2, and L6TaSIP transgenic plants either in the absence of salt stress (0 mM) or following salt stress imposed by watering the plants with either 180 or 200 mM NaCl for 0, 8, and 12 days. d Phenotypes of WT and L1, L2, and L6TaSIP transgenic plants (L1, L2, and L6) either without simulated drought stress (0 %) or following drought stress imposed by watering the plants with either 15 or 20 % PEG for 0, 8, and 12 days. Values are the mean ± SD (n = 3 independent experiments)
When grown in soil under control conditions, transgenic and WT Arabidopsis plants grew similarly. After exposure to treatment with 180 or 200 mM NaCl for 8 or 12 days, the transgenic plants maintained relatively strong growth; however, the non-transgenic WT plants appeared wilted, and the bolts were stunted (Fig. 2c). After treatment with 15 or 20 % PEG solution for 8 or 12 days, the transgenic plants continued to grow, but the WT plants wilted and turned yellow (Fig. 2d). These results showed that overexpression of the wheat TaSIP gene enhanced the salt and drought tolerance of transgenic Arabidopsis plants.
Salt tolerance of RNAi knockdown transgenic rice plants
Transgenic rice plants overexpressing the empty vector, the wheat TaSIP gene or an RNAi construct of the rice OsSIP gene were generated, and T1-generation plants were analyzed for salt tolerance. Both the overexpressing and the RNAi knockdown transgenic lines grew well and had green leaves under normal conditions (Fig. 3a), with no obvious physiological differences from transgenic plants under normal growth conditions. After a 7-day treatment with 170 mM NaCl, plants of transgenic rice lines L3, L4, and L9 grew well, whereas control plants wilted. More severe wilting occurred in the RNAi lines (K2, K3, K5) than in the control plants, resulting in the death of several of the RNAi-expressing plants (Fig. 3b). These results confirmed that there was an association between OsSIP gene expression and the level of salt tolerance. Analysis by qPCR revealed that the OsSIP transcript level in the three individual RNAi lines was reduced to 48, 41, and 55 % of that of non-transgenic plants (Fig. 3c). These results confirm that RNAi knockdown reduced the expression level of the rice gene OsSIP, which is homologous to TaSIP, and that those RNAi transgenic rice plants became highly susceptible to salt stress as a consequence.
Salt tolerance of transgenic rice that express OsSIP (a homologue of wheat TaSIP) and RNA interference (RNAi) rice plants. Phenotype of OsSIP-overexpressing (L3, L4, L9) and RNAi (K2, K3, K5) plants: a under normal growth conditions, b after 7 days of 170 mM NaCl stress. Control plants (CK) were transformed with empty vector pTCK303. c Analysis of RNAi rice plants using qPCR. **P < 0.01. Values are the mean ± SD (n = 3 independent experiments)
Subcellular localization of TaSIP in transgenic tobacco plants
In tobacco leaves infiltrated with Agrobacterium harboring a TaSIP-GFP fusion construct, microscopic analysis revealed that fluorescence was mainly localized to the cell membrane (Fig. 4a–c). In cells inoculated with the empty p1300-GFP vector plasmid, fluorescence was localized to the nucleus and the cell membrane (Fig. 4d). These results indicate that TaSIP was mainly localized on the cell membrane.
Subcellular localization of TaSIP in tobacco plants by immunofluorescence assay. a Fluorescence image of TaSIP-GFP protein, b bright-field microscopic image of TaSIP-GFP-expressing cell, c merged image of the fluorescence and bright-field microscopic images of TaSIP-GFP, d Fluorescence image of GFP in control plants. GFP Green fluorescent protein
Contents of ions, proline, soluble sugars, chlorophyll, and MDA
Under normal culture conditions, no differences in the contents of Na+, K+, or Ca2+ was detected between plants overexpressing TaSIP and the WT control. Although both lines showed some increase in Na+ content after treatment with 200 mM NaCl, the content of Na+ in the TaSIP transgenic plants was significantly lower than that in the WT plants. Following NaCl treatment, the Na+ levels increased by 13.19-fold in the WT plants and by 5.13-fold in the TaSIP transgenic plants compared with the levels before salt treatment. There were no differences in K+ content between the transgenic and WT plants with or without salt treatment (Fig. 5b). Consequently, the TaSIP-overexpressing lines had a 1.61-fold higher K+/Na+ ratio than the WT plants after salt stress (P < 0.05).
Physiological analyses in TaSIP transgenic and WT Arabidopsis plants before and after salt treatment. a Na+ content, b K+ content, c Ca2+ content, d K+/Na+ ratio, e proline content, f soluble sugar content, g chlorophyll content, h malondialdehyde content. * P < 0.05, ** P < 0.01. Values are the mean ± SD (n = 3 independent experiments)
The other osmotic compounds tested were present at significantly different levels in WT and transgenic TaSIP plants. Following treatment with 200 mM NaCl, the levels of Ca2+, proline, and soluble sugars were all higher in the TaSIP transgenic plants than in WT plants. In salt-stressed WT plants, the contents of proline, soluble sugars, and chlorophyll increased by 1.24-, 1.04- and 0.67-fold, respectively, relative to those in the untreated WT plants. In the salt-treated TaSIP transgenic plants, the levels of these three compounds increased by 1.45-, 1.38-, and 0.91-fold, with all of these levels being significant increases compared with those in WT plants. The MDA content increased by 1.41- and 1.21-fold in treated WT and transgenic plants, respectively, but the change was only significant in the WT plants (Fig. 5).
Expression of salt-stress-related genes in transgenic Arabidopsis plants
Nine genes that are known to be closely associated with salt tolerance in Arabidopsis—SOS2, SOS3, FRY1, SAD1, COR15a, RD29B, KIN2, ADH, and P5CS—were selected for expression analysis. Following salt stress, SOS2, SOS3, COR15a, and P5CS transcripts were more abundant in salt-treated transgenic Arabidopsis plants overexpressing TaSIP than in WT plants (Fig. 6).
Relative expression of nine stress-related genes—SOS2, SOS3, FRY1, SAD1, COR15a, RD29B, KIN2, ADH, and P5CS—in TaSIP transgenic Arabidopsis plants under salt stress conditions. Comparable WT plants were used as a control, with the transcript level in WT plants arbitrarily assigned a value of 1. ** P < 0.01. Values are the mean ± SD (n = 3 independent experiments)
Analysis of different tissues carrying full-length or segments of the TaSIP promoter by GUS staining and the quantitation of fluorescence.
Staining of plants that expressed the full-length TaSIP promoter construct with GU- generated signals in roots, stems, leaves, flowers, and seed pods, with stronger staining in meristematic tissues. These results indicated a lack of tissue-specific expression, but with some preference for expression in meristematic tissues (Fig. 7a–e). Quantitation of the GUS fluorescence confirmed that the full-length promoter showed the strongest activity in leaves under control and untreated conditions and following treatment with 150 mM NaCl and the weakest activity in roots; it was significantly more active in stem, leaf, and flower tissues following salt treatment (Fig. 7f).
β-Glucuronidase (GUS) staining and quantitative fluorescence analysis of TaSIP transgenic plants harboring different promoter segments and in different tissues carrying the full-length promoter. GUS staining of: a mature seed pods, b roots, c stems, d leaves, e flowers. f Fluorescence quantitation of GUS activity in different tissues of TaSIP transgenic plants. g–j GUS staining of TaSIP transgenic plants with promoter regions of 1,410 bp (g; segment A), 1,176 bp (h; segment B), 579 bp (I; segment C), and 331 bp (j; segment D). A schematic of these segments is shown in ESM Fig. S2. k Quantitative analysis of GUS signal in TaSIP transgenic plants with the 1,410-, 1,176-, and 579-bp promoter segments. *P < 0.05 and **P < 0.01. Values are the mean ± SD (n = 3 independent experiments)
When transgenic plants with different promoter segments (ESM Fig S2) were stained for GUS activity, the full-length promoter (segment A) produced the deepest GUS staining (Fig. 7g), followed by the segment B promoter region (Fig. 7h). Faint GUS activity was detected for segment C, which indicated that it is also a key functional region (Fig. 7i). No GUS staining occurred in the transgenic plants with segment D (Fig. 7j).
Quantitation of GUS fluorescence after ABA treatment revealed that segment E, the region 1,176–1,410 bp upstream of the start codon, contained an ABA-responsive element. The observation that segments B and C responded to salt stress suggested that a salt-stress response element lies in the region 579–1,176 bp upstream of the exon (Fig. 7k). This result is in agreement with earlier reports that the region 579–1,176 bp upstream of the start codon contains a salt-stress and drought-responsive element (CCGAC) (Bastola et al. 1998; Takumi et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2005), which is universal among drought- and high-salt-responsive promoters. Several key salt-stress-responsive elements have been identified in this region of the TaSIP promoter, such as multiple ABA response elements (A/GCGT) (Liu et al. 1998; Uno et al. 2000; Chung et al. 2005) in the region 1,176–1,410 bp upstream of the start codon. Based on these results, we concluded that the TaSIP promoter contains key elements that regulate the activity of the promoter in response to salt stress and ABA signaling.
Discussion
The use of the cDNA microarray hybridization technique has proven to be valuable for the identification of new genes, including those that are either upregulated or downregulated in plants subjected to stressful conditions. By applying this technique to our wheat lines, we were able to determine that the TaSIP gene was significantly induced after a 12-h salt treatment. Analysis of a full-length TaSIP cDNA failed to reveal any similarity in terms of amino acid sequence to any known proteins. The localization experiment provided evidence that the gene encodes a protein that is localized to the cell membrane, and the presence of a transmembrane domain from the 59th to the 81st amino acid in the TaSIP protein was predicted (using the online tool Prediction of Transmembrane Helices in Proteins; www.cbs.dtu.dk/services/TMHMM/). The amino acid sequences of TaSIP and six homologous proteins [Brachypodium distachyon proteins (LOC100836538 and LOC100840303), Sorghum bicolor protein, Zea mays proteins (LOC100279368 and LOC100273348) and rice protein (Os05g0199100)] were aligned and compared using the MegAlign function of DNAStar (ESM Fig. S4). The substantial extent to which the expression of the TaSIP gene was induced by salt in the salt-tolerant mutant RH8706-49 suggests that TaSIP may participate in pathways responding to salt stress and thus confer salt tolerance to the isogenic line.
Plants rapidly accumulate high levels of ABA following exposure to stresses, such as high salt, drought, and low temperature. The role of the phytohormone ABA in restricting plant growth under stressful conditions is well documented (Hirayama and Shinozaki 2007). The signaling pathways triggered by ABA activate stress-related gene expression, leading to physiological and biochemical responses (Beck et al. 2007) that include the regulation of the stomatal opening. When the salt-tolerant wheat mutant RH8706-49 was exposed to treatment with 50 μM ABA, the expression level of TaSIP was highly induced, suggesting that TaSIP may participate in transduction of the ABA signal.
Plant tolerance to salt and drought stresses results from the activation of several signal pathways and the additive affects of several discrete physiological changes. Several molecular and physiological experiments were undertaken to explore the mechanism of stress tolerance conferred by TaSIP. The experiment with the PEG-induced drought treatment showed that TaSIP also confers drought tolerance. When plants are exposed to salt stress, the osmotic pressure inside cells is reduced, causing water to be lost to the surroundings. In our study, overexpression of the TaSIP gene resulted in greater root growth, which could enhance water uptake from deeper soil strata. Compared with WT plants, transgenic plants that overexpressed TaSIP had a greater capacity to export Na+, which would reduce cellular Na+ toxicity and osmotic pressure. These plants also had a higher K+/Na+ ratio, which is an important property for salt tolerance (Shabala and Cuin 2008). Calcium-dependent protein kinases (CDPKs) are unique calmodulin-independent Ca2+ sensors with both kinase activity and Ca2+-binding activity. When subjected to treatment with 200 mM NaCl, TaSIP transgenic lines contained a higher level of Ca2+. This increase in Ca2+ may activate the CDPKs, which are known to regulate ion channels and stomatal closure in order to reduce the evaporation rate and maintain water balance within cells.
TaSIP-overexpressing transgenic plants also exhibited higher levels of proline and soluble sugars than the WT plants. Plants accumulate proline and soluble sugars in order to decrease the intracellular osmotic pressure, which is a physiological change that alleviates damage from salt stress (Khatkar and Kuhad 2000; Muthukumarasamy et al. 2000; Singh et al. 2000; Jain et al. 2001). In addition, the level of MDA in salt-stressed TaSIP transgenic plants was lower than those in salt-stressed WT plants, indicating that the salt-treated transgenic plants experienced less severe membrane damage from lipid peroxidation than the salt-treated WT plants. Banu et al. (2009) reported that plants could ameliorate apoptosis triggered by salt stress by suppressing the accumulation of reactive oxygen species and the peroxidation of lipids, as seen in the case of TaSIP transgenics. Overall, improvement in the physiological properties of the TaSIP transgenic plants confirmed the function of TaSIP in salt tolerance and can explain the physiological changes seen in these experiments.
The possible signal pathways linked to TaSIP can be deduced using the qPCR data, which showed the induction of SOS2, SOS3, COR15a, and P5CS in transgenic plants that overexpressed TaSIP. Under salt stress, cellular ion balance is regulated through the salt overly sensitive (SOS) signal transduction pathway, which includes the enzymes SOS2 and SOS3 that directly control the K+/Na+ ion balance. In this study, salt stress was shown to increase the levels of both SOS2 and SOS3 transcripts in TaSIP-overexpressing plants. Expression of the COR15a gene, which encodes a component of a CDPK signaling pathway, can be increased upon exposure to cold, ABA, or salt stress. Given that COR15a is upregulated in TaSIP-overexpressing plants under salt stress, we can speculate that salt tolerance regulated by the TaSIP gene may occur through this reported COR15a gene. Our study also showed that both the expression of P5CS and the content of proline were increased in TaSIP transgenic plants. The P5CS enzyme not only participates in the regulation of ABA signal transduction, but is also a key enzyme in proline biosynthesis (Igarashi et al. 1997; Strizhov et al. 1997). The levels of proline measured and P5CS expression data indicated that the expression of the TaSIP and P5CS genes were somehow coordinated. On the basis of these results, we postulate that the TaSIP gene may enhance salt tolerance via the SOS and ABA signal transduction pathways.
In our study, we used GUS staining to analyze the tissue-specific expression of TaSIP and to identify regulatory regions of the TaSIP promoter. Transgenic plants carrying either the full-length promoter or the region 579–1,176 bp upstream of the start codon showed the deepest GUS staining. When the transgenic plants carrying the full-length promoter were treated with 150 mM NaCl, expression of the GUS gene driven by the TaSIP promoter was enhanced in all types of tissue examined, indicating that salt stress activated the TaSIP promoter. However, among the different plant tissues, leaves exhibited the highest level of GUS protein, suggesting that different tissues may contain separate cis- or trans-acting elements that regulate TaSIP expression. No GUS staining was observed in plants carrying the 331-bp upstream promoter segment, which contains a TATA box and a transcription initiation site but no CAAT box. It is generally accepted that the CAAT box can increase the frequency of transcription initiation, and thus this segment may have been insufficient to act as a reliable promoter.
To summarize, we have identified a gene for salt tolerance in a salt-tolerant isogenic wheat line. Although we have not completely resolved the molecular basis of the improved stress tolerance in TaSIP-overexpressing Arabidopsis and rice, our data clearly suggest that the wheat TaSIP gene is involved in plant responses to salt and drought stress. Further study of the role of the wheat TaSIP gene in response to different stresses may lead to a deeper understanding of the integrated reactions of salt-tolerant plants to multiple abiotic stresses.
References
Arnon DI (1949) Copper enzymes in isolated chloroplast polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Banu NA, Hoque A, Watanabe-Sugimoto M, Matsuoka K, Nakamura Y, Shimoishi Y, Murata Y (2009) Proline and glycinebetaine induce antioxidant defense gene expression and suppress cell death in cultured tobacco cells under salt stress. J Plant Physiol 166(2):146–156
Bastola DR, Pethe VV, Winicov I (1998) Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol Biol 38:1123–1135
Beck EH, Fettig S, Knake C, Hartig K, Bhattarai T (2007) Specific and unspecific responses of plants to cold and drought stress. J Biosci 32:501–510
Chung HJ, Fu HY, Thomas TL (2005) Abscisic acid-inducible nuclear proteins bind to bipartite promoter elements required for ABA response and embryo-regulated expression of the carrot Dc3 gene. Planta 220:424–433
Clough SL, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356
Flowers TJ, Troke PF, Yeo AR (1977) The mechanism of salt tolerance in halophytes. Annu Rev Plant Physiol 28:89–121
Ge RC, Chen GP, Zhao BC (2007) Cloning and functional characterization of a wheat serine/threonine kinase gene (TaSTK) related to salt-resistance. Plant Sci 173:55–60
Greenway H, Munns R (1980) Mechanisms of salt tolerance in nonhalophytes. Annu Rev Plant Physiol 31:149–190
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198
Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12:343–351
Igarashi Y, Yoshiba Y, Sanada Y, Yamaguchi-Shinozaki K, Wada K, Shinozaki K (1997) Characterization of the gene for delta1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativa L. Plant Mol Biol 33:857–865
Ishitani M, Xiong L, Stevenson B, Zhu JK (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9:1935–1949
Jain M, Mathur G, Koul S, Sarin NB (2001) Ameliorative effects of proline on salt stress-induced lipid peroxidation in cell lines of groundnut (Arachis hypogaea L.). Plant Cell Rep 20:463–468
Khatkar D, Kuhad MS (2000) Short-term salinity induced changes in two wheat cultivars at different growth stages. Biol Plant 43:629–632
Liu S, Kriz A, Duncan D, Widholm J (1998) Abscisic acid-regulated Glb1 transient expression in cultured maize P3377 cells. Plant Cell Rep 17:650–655
Muthukumarasamy M, Gupta SD, Pannerselvam R (2000) Enhancement of peroxidase, polyphenol oxidase and superoxide dismutase activities by triadimefon in NaCl stressed Raphanus sativus L. Biol Plant 43:317–320
Parida AK, Das AB (2005) Salt tolerance and salinity effects on plants. Ecotoxicol Environ Saf 60:324–349
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protocols 5:725–738
Sairam RK, Tyagi A (2004) Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci 86:407–421
Shabala S, Cuin TA (2008) Potassium transport and plant salt tolerance. Physiol Plant 133:651–669
Singh SK, Sharma HC, Goswami AM, Datta SP, Singh SP (2000) In vitro growth and leaf composition of grapevine cultivars as affected by sodium chloride. Biol Plant 43:283–286
Strizhov N, Ábrahám E, Ökrész L, Blicking S, Zilberstein A, Schell J, Koncz C, Szabados L (1997) Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidposis. Plant J 12:557–569
Takumi S, Koike A, Nakata M, Kume S, Ohno R, Nakamura C (2003) Cold-specific and light-stimulated expression of a wheat (Triticum aestivum L.) Cor gene Wcor15 encoding a chloroplast-targeted protein. J Exp Bot 54:2265–2274
Troll W, Lindsley J (1955) A photometric method for the determination of proline. J Biol Chem 215:655–660
Uno Y, Furihata T, Ade H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97:11632–11637
Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10:88–94
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40
Zhao Q, Zhao YJ, Zhao BC (2009) Cloning and functional analysis of wheat V-H+-ATPase subunit genes. Plant Mol Biol 69:33–46
Zhou W, Li Y, Zhao BC (2009) Overexpression of TaSTRG gene improves salt and drought tolerance in rice. J Plant Physiol 166:1660–1671
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Du, HY., Shen, YZ. & Huang, ZJ. Function of the wheat TaSIP gene in enhancing drought and salt tolerance in transgenic Arabidopsis and rice. Plant Mol Biol 81, 417–429 (2013). https://doi.org/10.1007/s11103-013-0011-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0011-x