Abstract
The developmental regulation of grasses lemma and palea and their relationship to the floral organs in dicots had been variously explicated and extensively debated. Here, we characterized a triangular hull mutant th1-1 from EMS-mutagenized Oryza sativa ssp. indica cv. 93-11. The th1-1 mutant exhibited obviously triangular hull with tortuous and slender lemma/palea. Using a map-based cloning strategy, the TH1 gene was narrowed down to a 60-kb region on the long arm of chromosome 2. Sequence verification revealed that the th1-1 mutant harbored 1-bp deletion in exon 2 of LOC_Os02g56610 which resulted in a frame-shift mutation. The RNA-interference transgenic plants of LOC_Os02g56610 displayed a similar phenotype to the th1 mutant. Consequently, LOC_Os02g56610 was identified as the TH1 gene which encoded 248 amino acids and contained a DUF640 domain. RT-PCR analysis and GUS staining showed that the transcripts of TH1 mainly accumulated in young inflorescence, lemma and palea of spikelet. These results suggested that TH1 was an important gene controlling the lemma and palea development in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grass (Poaceae) is one of the largest flowering plant families of angiosperms with ~10,000 species, including many important crops. Rice, a kind of grass, is one of most important food crop in the world. Spikelet is a fundamental unit of rice yield, the spikelet development and morphogenesis has profound influence on seed production. A typical spikelet of rice (Oryza sativa) consists of a pair of rudimental glumes, two empty glumes, a lemma and a palea, two lodicules, six stamens and one pistil. Among these floral organs, the development of lemma and palea dramatically influenced on the grain shape, grain size, even the grain yield in rice (Fan et al. 2006; Song et al. 2007; Shomura et al. 2008; Weng et al. 2008).
The rice fon1 and fon4 mutants (Suzaki et al. 2004; Chu et al. 2006) exhibited enlargement of floral meristems, which ultimately led to extra lodicules or lemma/palea-like organs, or homeotic conversion of organ identity (Suzaki et al. 2004; Chu et al. 2006), indicating that the floral meristem maintenance influenced the development of lemma and palea in rice. Similarly, mutations in the FONs homologs, the CLV1, CLV2 and CLV3 genes in Arabidopsis, caused enlargement of floral meristems, as well as inflorescence and shoot apex meristems, which led to increase of floral organs and flower number (Clark et al. 1993; Clark et al. 1995; Kayes and Clark 1998).
Recently, a few of genes, including LHS1, PAL1, DH1, REP1 GS3, GW2, qSW5 and GW5, had been identified to control the hull development and grain size in rice (Prasad et al. 2001; Luo et al. 2005; Fan et al. 2006; Song et al. 2007; Li et al. 2008; Shomura et al. 2008; Weng et al. 2008; Yuan et al. 2009). Nevertheless, the genetic regulation of the developmental draft of basic lemma and palea was not established in rice.
In this study, we identified the TH1 gene that controlled grain shape of rice. First, we isolated two mutants, the triangular hull1-1 (th1-1) and th1-2, from the ethyl methane sulphonate (EMS)-mutagenized population of the indica cv. 93-11 and the indica line CP78, respectively. Both mutants exhibited triangular hull and narrower spikelet hull width traits compared with its wild type. Further observation of the lemma and palea revealed that th1-1 showed thicker lemma and palea, and more bulges in the epidermis. Using the map-based cloning approach, we identified the TH1 gene that encoded an unknown functional protein with a DUF640 domain and expressed in young panicle, lemma, palea, gynoecia, ovary and young embryos. This work provided novel insight into lemma and palea development and grain traits in rice.
Materials and methods
Plant materials
All the requisite cultivars, including 93-11, Guichao 2, CP78, C418, IR24, Minghui 63, Zhenshan 97B, Milyang 46, Dianchao 3, Zhonghua 17, Asominori and genetic populations, were grown in the field of the Experimental Station of China Agricultural University.
Identification of the th1-1 and th1-2 mutants
About 20,000 seeds of the indica cv. 93-11 and CP78, respectively, were EMS (ethyl methane sulphonate)-mutagenized and subsequently grown in the field to generate the EMS-mutagenesis populations according to Wu et al. (2005). The triangular hull 1-1 (th1-1) mutant and th1-2 were identified from the EMS-mutagenized population of the indica cv. 93-11 and the indica line CP78, respectively.
Histological analysis
Spikelets from th1-1 and 93-11 plants just before heading stage were fixed in FAA (70% ethanol, 5% glacial acetic acid, 3.7% formaldehyde), embedded in paraffin. Thin sections were prepared and stained with 1% fast green and the 1% safranine. For scanning electron microscopy (Liu et al. 2004), the lemma and palea of th1-1 and 93-11 spikelet just before heading stage was viewed.
Observation and quantitation of bulges
Bulges (a rounded projection and a bent or protruding part originated from the cuticular thickening on the epidermal cells) of the lemma and palea were observed by SEM (Prasad et al. 2005). Their numbers were quantified manually. Scan areas studied were 40,000 μm2 from proximal, middle and distal regions of the lemma and palea.
Map-based cloning of TH1
110 F2 progenies derived from the cross between the th1-1 mutant and an indica cv. Guichao 2, was used to preliminarily map the TH1 gene. To further map the TH1 gene, we generated a large F2 population with more than 15,000 progenies from the cross between th1-1 and a japonica cv. C418. 2,400 progenies from this F2 population showed triangular hulls at the heading stage, which were chosen for fine mapping of TH1.
RM3774 and RM3248 (McCouch et al. 2002), two SSR markers flanking the TH1 gene, were used to screen the recombinants. Three new polymorphic markers, including a CAPS (cleaved amplified polymorphic sequence) marker C1 and two SNP (single nucleotide polymorphism) markers C2 and C3 (Table S1), were developed based on the reference sequence (Nipponbare) to fine-map TH1. Fine mapping with these markers placed th1 within a 60-kb region between C1 and C2 markers. To find the discrepancy of genomic sequence between 93-11 and the th1 mutants, we sequenced the 60-kb region by PCR amplification using more than 40 pairs of primers (Data not shown).
Among these primers, HX1 (5′-gtactggcaaagcaagatgg-3; 5′-atcgggaagcagattcatcc-3′) were used to amplify the ORF sequence of LOC_Os02g56610 from th1-1, 93-11 and other cultivars. Moreover, to analyze the sequence of th1-2, another pair of primers HX2 (5′-catgtgtgtggcactgttgc-3; 5′-cactctagctacatagtgtgc-3′) were used to amplify the genomic sequence of LOC_Os02g56610 from th1-2 and CP78.
Vector construction and plant transformation
To generate the hairpin pTH1-RNAi construct, we cloned two copies of a 355-bp fragment (from +1 to +355 bases) of TH1 cDNA at inverted repeats, which was driven by the ubiquitin promoter, into the pJL1460 vector (Wang et al. 2004).
The pTH1::GUS construct was generated by cloning a 2,225-bp fragment harboring the endogenous TH1 promoter into the pCAMBIA1381 vector, which was amplified with the primers (5′-agcGAATTCctgccacaagatatgttcgg-3′ and 5′-acaGTCGACgcaacctgaatcacaagaagc-3′).
Both pTH1-RNAi and pTH1::GUS constructs were transformed into a japonica cv. Zhonghua 17 through the method of Microprojectile Bombardment (PDS-1000/He, BIORAD).
To generate the pAct1::TH1-GFP construct, we fused the TH1 coding region, the 744-bp fragment amplified by the primers (5′-acaCCCGGGaatggatcgtcaccatcacc-3′ and 5′-accTCTAGAcgggatgatgaactgcggc-3′), in-frame with GFP, which was driven by the Act1 promoter. The pAct1::TH1-GFP construct was introduced into onion epidermal cells through Microprojectile Bombardment (PDS-1000/He, BIORAD). The fluorescence of GFP was inspected under a confocal fluorescence microscope after incubation for 16–24 h at 26°C.
RNA isolation and RT-PCR
Total RNA was prepared from the inflorescences, the lemma, pelea, stamen, pistil, culm, flag leaf, rachis and uppermost node and young embryo with an RNAprep pure Plant Kit [TIANGEN BIOTECH (BEIJING) CO.LTD]. The first strand of cDNA was synthesized with Superscript II RT kit (INVITROGEN, USA) from total RNA. RT-PCR was carried out to amplify the TH1 (5′-ccatcatccactgttcatcc-3′; 5′-gatctcctcctccactgctc-3′) transcripts with 30 PCR cycles, using the first-strand cDNA as a template. Actin (5′-tccatcttggcatctctcag-3′; 5′-gtaccctcatcaggcatctg-3′) was also amplified as the control.
Histochemical GUS assays
The histochemical GUS assay was processed as previously described (Sieburth and Meyerowitz 1997). Samples for GUS staining were gently fixed by incubation in icy 90% acetone on ice for 20–30 min, then rinsed in 50 mM Na3PO4, pH7.2, 0.5 mM K3Fe[CN]6, and 0.5 mM K4Fe[CN]6. The samples were then placed in staining solution (50 mM Na3PO4, pH 7.2, 1 mM X-gluc, 0.5 mM K3Fe[CN]6, and 0.5 mM K4Fe[CN]6), vacuum infiltrated for 10 min, and incubated at 37°C overnight. Then the staining buffer was removed, and the samples were incubated in H2O for 5 min, and discolored in solution of 90% ethanol and 10% acetic acid.
Results
Identification of the th1 mutants
To understand the genetic regulation of lemma and palea development in rice, we identified a mutant that displayed triangular hull with a tortuous lemma and palea (Fig. 1A, B) from the ethyl methane sulphonate (EMS)-mutagenized population of an indica cv. 93-11. F1 plants, which were derived from the cross between this mutant and wild-type 93-11, showed the wild-type-like hull. 58 plants from the 240 F2 populations showed triangular hull. The segregation rate of wild-type (WT) and mutant plants was fit for the ratio of 3:1 and X 2C < X 20.05,1 = 3.84. These results indicated that the triangular hull was controlled by a single recessive gene, referred to as triangular hull 1(th1).
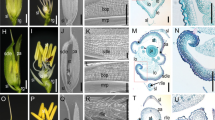
Phenotypes of the th1-1 mutant. A, B Spikelets in wild-type 93-11 (left) and th1-1 (right) just before heading. The th1-1 mutant exhibited triangular hull trait without other floral organs variation. Scale bar is 2 mm in A and 5 mm in B. C–F Scanning electron microscope (SEM) images of lemma and palea in 93-11 (C, E) and the th1-1 mutant (D, F). C, D SEM images of the spikelet morphology in the stage of the elongation of empty glumes, lemma and palea. Scale bar is 0.214 mm. E, F SEM images of the lemma/palea epidermis. The th1-1 mutant showed narrower spikelet hull width in D and more bulges in F. Scale bar is 75 μm. G, H Lemma and palea transverse sections of 93-11 (G) and th1-1 plants (H). Arrow points to fibrous Sclerenchyma. Scale bar is 80 μm. I, J Bulge density of lemma (I) and palea (J) in 93-11 and th1-1 was calculated by mean the number of bulges in a scan area of 40,000 μm2 (n = 5 area). K Spikelet hull width in 93-11 and th1-1 just before heading (n = 30 spikelets). L Comparison of spikelet hull thickness of transverse sections in 93-11 and th1-1 just before heading (n = 5 spikelets). M Seed set rate in 93-11 and th1-1 (n = 30 panicles). All data are given as mean ± SD. A Student’s t test is used to generate the P values in I–M
Observation of the lemma and palea of th1-1 using scanning electron microscopy (SEM) revealed that the th1-1 had narrower lemma and palea (Fig. 1C, D). Just before heading, we viewed the epidermis of lemma and palea using SEM, and found that the bulges in the th1-1 mutant were more than that of wild type (Fig. 1E, F). Further examination on transverse sections of lemma and palea showed that the number of fibrous sclerenchyma cells was increased in the th1-1 mutant (Fig. 1G, H). Quantitative analysis showed that the bulge densities on lemma and palea epidermis in the th1-1 mutant were increased by 43.57 and 49.52%, respectively (Fig. 1I, J). The width of the spikelet hull in th1-1 was markedly narrower (24.4%) than that of wild type (Fig. 1K), whereas the spikelet hull thickness of transverse sections became thicker (76.16%) than that of wild type (Fig. 1L). In addition, we also found that the seed set rate of th1-1 were obviously less (22.5%) than that of wild type (Fig. 1M).
Isolation of the TH1 gene
In order to map the TH1 gene, we generated a population of 110 F2 plants from a cross between th1-1 and an indica cv. Guichao 2. Analysis of genetic linkage between 72 simple sequence repeat (SSR) markers and triangular hull phenotype showed that the TH1 gene was linked with the SSR marker RM8024 (Fig. 2A) on the long arm of chromosome 2.
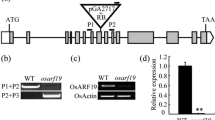
Map-based cloning of TH1. A Genetic mapping located th1 between marker RM3774 and RM3248; cM, centimorgan. B Fine mapping placed th1 in the region of 60-kb, between the a CAPS (cleavable amplified polymorphic sequence) marker C1 and a SNP (single nucleotide polymorphism) marker in the BAC AP004081; Within this 60-kb region, there were six predicted genes based on the genomic sequence of Nipponbare, the fifth gene (LOC_Os02g56610) corresponded to th1 by genetic complement tests; r, recombinant; scale bar is 5 kb. C Schematic structure of candidate gene and mutation site in th1-1. Three exons and two introns were shown. A 1-bp deletion (∆C583) was observed within exon 2 of LOC_Os02g56610 in the th1-1 mutant, which resulted in reading frame shift. Empty box exon; thin line intron; filled box ORF; atg start code; tga stop code. D The 93-11 TH1 gene predicted a 1,586 bp transcript including a 747-bp CDS sequence encoding a 248-amino-acid expression protein, while th1-1 predicted a 1,673-bp transcript including a 834 bp coding DNA sequence because of a 1-bp deletion, from gagacgca to gagacga (the letter c with double underlines), which led to reading fame shift and amino acid changing (additional coding nucleotides and Italic amino acids in the frame)
To further map the TH1 gene, we generated a large F2 population with more than 15,000 progenies from the cross between th1-1 and a japonica cv. C418. RM3774 and RM3248, two SSR markers flanking the TH1 gene, were used to screen the recombinants from this F2 population, with 65 recombinants for RM3774 and 142 for RM3248 (Fig. 2A).
To fine-map TH1, we developed three new polymorphic markers, including a CAPS (cleaved amplified polymorphic sequence) marker C1 and two SNP (single nucleotide polymorphism) markers C2 and C3 (Table S1), based on the reference sequence (Nipponbare). TH1 was finally narrowed down within a 60-kb region between C1 and C2 markers in the BAC clone AP004081 (Fig. 2B; Table S1).
Within this 60-kb region, there were six predicted genes based on the genomic sequence of Nipponbare (the TIGR Rice Genome Annotation Database) (Fig. 2B). Comparing the genomic sequence of the mapping region (~60-kb) between the 93-11 and th1-1, we found 1-bp deletion in exon 2 of LOC_Os02g56610, which resulted in a frame-shift mutation (Fig. 2C, D).
Alignment of the genomic sequence of six indica cultivars (Teqing, Guichao 2, IR24, Minghui 63, Zhenshan 97B and Milyang 46) and four japonica cultivars (Dianchao 3, C418, Zhonghua 17 and Asominori) further confirmed that the 1-bp deletion of LOC_02g56610 occurred only in the th1-1 mutant (Data not shown).
Sequence analysis revealed that the LOC_Os02g56610 gene was consisted of 2,100 bp, and its full-length cDNA (AK111446) contained three exons and two introns in 93-11 (Fig. 2C). The CDS located in the second exon and encoded a 248 amino-acid protein with a DUF640 domain according to prediction by NCBI and TIGR Web site. Among DUF640 protein family, there were ten predicted LIGHT SENSITIVE HYPOCOTYLS (LSH) proteins in Arabidopsis thaliana and six homologs in rice, which had 50–65% and 50–60% amino acids sequence identity, respectively. Most of the homologs were hypothetical or predicted proteins in rice.
In addition to th1-1, we identified another triangular hull mutant from EMS-mutagenized an indica line CP78, referred as th1-2 (Fig. 3A, B). The th1-2 displayed similar triangular hull as th1-1, and the width of the spikelet hull of th1-2 was markedly narrower (22.4%) than that of wild type (Fig. 3C). The F1 plants derived from the cross of th1-1 with th1-2 displayed triangular hull phenotypes which were similar to the th1-1 and th1-2 mutants (Fig. 3D), suggesting that these two mutants were allelic.
Phenotypes and sequence of the th1-2 mutant. A, B Spikelet of the th1-2 mutant plant (right) and wild-type CP78 plant (left). The th1-2 mutant exhibited triangular hull trait without other floral organs variation. Scale bar is 2 mm in A and 5 mm in B. C Spikelet hull width in CP78 and th1-2 just before heading (n = 30 spikelets). D Allelic test of the th1-1 (right) and the th1-2 (left). The spikelet hull of the F1 plants (middle) derived from the cross of th1-1 with th1-2 displayed triangular hull phenotypes which were similar to the th1-1 and th1-2 mutants. Scale bar is 2 mm. E Sequence analysis of th1-2 and CP78. The underlined nucleotides is initiator codon, the nucleotides in the frame is stop codon. All data are given as mean ± SD. A Student’s t test is used to generate the P values in C
Further sequence analysis of the th1-2 mutant and the corresponding wild-type CP78 showed that the th1-2 mutant contained a section of 1,166-bp nucleotide deletion covering the whole ORF in the LOC_Os02g56610 gene (Fig. 3E).
RNAi-mediated knockdown of TH1
To verify whether triangular hull phenotype was caused by a loss-of-function mutation of the LOC_Os02g56610 gene, we generated a hairpin RNAi construct by cloning two copies of a 355-bp fragment (from +1 to +355 bases) of TH1 cDNA at inverted repeats, which was driven by the ubiquitin promoter, into the pJL1460 vector (referred to as pTH1-RNAi) (Wang et al. 2004). The construct was introduced into a japonica cv. Zhonghua 17 and the transgenic lines were thoroughly surveyed.
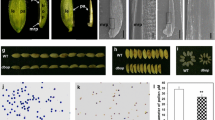
Seven out of 16 lines similarly displayed th1-like phenotypes. The transgenic lines L1 and L2 were representatively shown in Fig. 4. L1 and L2 exhibited triangular hull phenotype with a tortuous lemma and palea (Fig. 4A). Further examination at the histological level showed that the density of bulges in lemma and palea epidermis in the transgenic plants was increased (Fig. 4B–E). To confirm whether the phenotype found in the transgenic plants were in virtue of down-regulation of the LOC_Os02g56610 expression, we detected the endogenous expression level of LOC_Os02g56610 by RT-PCR method. As shown in Fig. 4, the transcripts in the lemma and palea of the transgenic plants were distinctly decreased in comparison with the control plants (Zhonghua 17 with an empty plasmid).
Functional complementation for the TH1 gene. A Spikelets morphology of a transgenic plant with pTH1-RNAi. Spikelet of a Zhonghua 17 plant harboring an empty plasmid (ZH17) was on the left. Spikelet of two transgenic plant lines harboring with the construct pTH1-RNAi (L1 and L2) were in the middle and on the right, respectively. Scale bar is 2.5 mm. B Bulge density of lemma (left) and palea (right) in 93-11 and th1-1 was calculated by mean the number of bulges in a scan area of 40,000 μm2 (n = 5 area). C–E SEM images of the lemma and palea epidermis from ZH17 C, L1 D and L2 E. Scale bar is 50 μm. F RT-PCR analysis in the lemma and palea of transgenic plants L1, L2, CP1, CP2 and ZH17. Reduction of the endogenous TH1 was detected in the L1 and L2 that exhibited th1-like phenotype, but not in CP1 and CP2 that exhibited wild-type phenotype. All data are given as mean ± SD. A Student’s t test is used to generate the P values in B
The remaining nine lines, including CP1 and CP2, exhibited wildtype-like phenotype. Further RT-PCR analysis showed that the expression of endogenesis LOC_Os02g56610 in these lines was similar to the control plant (Fig. 4F). The expression level of the LOC_Os02g56610 gene in the CP1 and CP2 lines were representatively shown in Fig. 4.
Taken together, these results demonstrated that down-regulation of the LOC_Os02g56610 gene caused the phenotypes which were similar to the th1 mutants, suggesting that the LOC_Os02g56610 gene corresponded to TH1 and that mutations in LOC_Os02g56610 were responsible for the triangular hull phenotypes in the th1 mutants.
Subcellular localization and expression patterns of TH1
To investigate the subcellular localization of TH1, we constructed a vector of the TH1-GFP fusion gene driven by the Act1 promoter. Transient expression of the construct in the epidermal cells of onion showed that the TH1-GFP fusion protein localized to the nucleus (Fig. 5A–C), indicating that the TH1 was a nuclear protein.
The expression pattern of the TH1 gene. A–C The TH1-GFP fusion protein was localized to the nucleus. The photographs were taken in dark field for green fluorescence B, in bright light for the morphology of the cell A, and in combination of the cells C. Scale bar is 1 μm. D–F The expression of TH1 was detected to express in the lemma and palea. Scale bar is 1.0 cm in D and E, 0.5 cm in F. G, H RT-PCR analysis in inflorescences at various developmental stages indicated by rachis length G and RT-PCR analysis in various organs. H le lemma; pa palea; st stamen; pi pistil; cu culm; fl flag leaf; no uppermost node; rad rachis without spikelets; yem young embryo
To study the tissue specificity of the TH1 expression, we introduced the construct consisting of the TH1 promoter regions fused to the GUS reporter gene into the japonica cv. Zhonghua 17. The GUS expression was detected in the lemma and palea (Fig. 5D–F), not in stamen, rachis branches and flag leaf (Data not shown), indicating that TH1 was mainly accumulated in lemma and palea.
Further analysis of the expression patterns of TH1 by RT-PCR showed that the TH1 transcript was strongly accumulated in reproductive organs including inflorescence primordium (rachis length <0.5 cm), young panicle (rachis length from 0.5 to 6 cm), lemma, palea, uppermost node, gynoecia combined ovary and young embryo (10–15 days after pollination) (Fig. 5G). However, no TH1 transcript was detected in stamens, rachis and flag leaf (Fig. 5H). These results were consistent with the expression patterns observed by GUS staining. Taken together, we concluded that the TH1 gene expressed temporally on the periods of inflorescence development and spatially on the lemma, palea, pistil and young embryo.
Discussion
TH1 regulates lemma and palea development
In recent years, interest in genetic regulation of inflorescence and flower development in grasses has been increased (Bommert et al. 2005). However, only a few mutants and genes related to the lemma and palea development were identified in rice (Yoshimura et al. 1997; Prasad et al. 2001; Luo et al. 2005; Li et al. 2008; Yuan et al. 2009). In this study, we characterized two triangular hull mutants, th1-1 and th1-2. The allelic test demonstrated that the th1-1 and th1-2 were allelic. Observation at a histological level showed that the number of fibrous sclerenchyma cells of lemma and palea was increased in the mutants. Using a map-based cloning strategy, we identified the TH1 gene that regulated the shape and development of the lemma and palea. Sequence verification revealed that the th1-1 mutant harbored 1-bp deletion in exon 2 which resulted in a frame-shift mutation, and th1-2 mutant harbored a section of 1,166-bp nucleotide deletion covering the whole ORF of the TH1 gene, which caused triangular hull mutation. These results demonstrated that TH1 was a novel gene controlling lemma and palea development in rice.
TH1 is a new gene encoded the protein with a DUF 640 domain
The lemma and palea of cereals, corresponded to sepals of dicotyledon, are important components of grass floret, which determine the grain shape and protect the kernel from the attacking of pathogens and insect (Bowman 1997; Ambrose et al. 2000; Jeon et al. 2000; Prasad et al. 2001; Abebe et al. 2004). Up to date, four mutants (lhs1, pal1, dh1 and rep1) of lemma and palea were identified in rice (Prasad et al. 2001; Luo et al. 2005; Li et al. 2008; Yuan et al. 2009). The lhs mutant exhibited an elongated leafy palea and lemma, an open hull, decreased stamen number, and increased the number of carpel (Jeon et al. 2000; Prasad et al. 2001). The pal1 mutant showed palealess spikelets, and palea was substituted by two leaf-like organs (Luo et al. 2005). The dh1 florets showed a degenerated hull with naked stamens and pistils which replaced by velum-like or filamentous organs (Li et al. 2008). The development of palea in the rep1 mutant was markedly retarded (Yuan et al. 2009). LHS1 belonged to MADS-box gene family (Prasad et al. 2001), PAL1 was a DNA-binding protein gene (Luo et al. 2005), the DH1 gene was a member of LOB genes family and expressed only at axillary bud and young ear (Li et al. 2008), the REP1 transcript was a TCP family transcription factor and mainly accumulated in 0.5–4 cm young inflorescence (Yuan et al. 2009).
The th1 mutants identified in this study displayed tortuous and slender lemma/palea, and did not show floral organs degeneration or homeotic transformation. The TH1 gene encoded a 248 amino-acid protein with a DUF640 domain. The TH1 gene transcript was mainly accumulated in 0.5–6 cm young inflorescence, lemma and palea of the spikelets, not in stamen, rachis branches and flag leaf. Taken together, we inferred that TH1 was a new gene which had the function on regulating the lemma and palea development in rice.
TH1 controls grain shape through regulating the cell division and extension of lemma and palea
The grain shape is a complicated trait which plays an important role in influencing yield and quality of rice. Recently, four key genes or QTLs for grain shape, GS3 (Fan et al. 2006), GW2 (Song et al. 2007), GW5 and qSW5 (Shomura et al. 2008; Weng et al. 2008), had been identified in rice. GW2 controlled rice grain width and weight, and encoded an unknown RING-type E3 ubiquitin ligase (Song et al. 2007). GS3 was a negative regulator and mainly regulated length and weight of grain (Fan et al. 2006). The deletion of qSW5 resulted in a significant increase in seed size owing to an increase in cell number in the outer glume of the rice flower, while GW5 that encoded a novel nuclear protein acted in the ubiquitin–proteasome pathway to regulate cell division during seed development (Shomura et al. 2008; Weng et al. 2008). The TH1 gene identified in this study controlled the grain shape and development of lemma and palea. Loss-function or down-regulation of the TH1 gene caused tortuous and slender lemma/palea and the decrease of grain weight. Examination at the histological level showed that the th1-1 mutant exhibited smaller cell (Fig. 1E, F, I, J), closer cell arrangement and more fibrous sclerenchyma cells on the epidermis of the lemma and palea comparison with the wild type 93-11(Fig. 1G, H, L). Therefore, we speculated that the mutation of TH1 might suppress cell division and extension of lemma and palea, which caused to decrease the width of lemma and palea.
The seed set rate of the th1-1 mutant was obvious lower than that of the wild-type 93-11. The transgenic plants of pTH1-RNAi exhibited th1-like phenotypes with triangular hull and no filled seed. These results demonstrated that disruption of TH1 may severely affect the hull development, causing defect in fertility of rice. Further analysis of the TH1 function is important for understanding molecular mechanism of the grain development in rice.
References
Abebe T, Skadsen RW, Kaeppler HF (2004) Cloning and identification of highly expressed genes in barley lemma and palea. Crop Sci 44:942–950
Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RF (2000) Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5:569–579
Bommert P, Nagasawa NS, Jackson D, Hirano HY (2005) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46:69–78
Bowman JL (1997) Evolutionary conservation of angiosperm flower development at the molecular and genetic levels. J Biosci 22:515–527
Chu HW, Qian Q, Liang WQ et al (2006) The FLORAL ORGAN NUMBER4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol 142:1039–1052
Clark SE, Running MP, Meyerowitz EM (1993) CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119:397–418
Clark SE, Running MP, Meyerowitz EM (1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121:2057–2067
Fan CC, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, Li XH, Zhang QF (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112:1164–1171
Jeon JS, Jang S, Lee S et al (2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12:871–884
Kayes JM, Clark SE (1998) CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125:3843–3851
Li A, Zhang Y, Wu X et al (2008) DH1, a LOB domain-like protein required for glume formation in rice. Plant Mol Biol 66:491–502
Liu F, Ni WM, Griffith ME et al (2004) The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell 16:5–20
Luo Q, Zhou KD, Zhao XF, Zeng QC, Xia HG, Zhai WX, Xu JC, Wu XJ, Yang HS, Zhu LH (2005) Identification and fine mapping of a mutant gene for palealess spikelet in rice. Planta 221:222–230
McCouch SR, Teytelman L, Xu YB et al (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
Prasad K, Sriram P, Kumar CS, Kushalapa K, Vijayraghavan U (2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211:281–290
Prasad K, Sriram P, Vijayraghavan U (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43:915–928
Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet 40:1023–1028
Sieburth LE, Meyerowitz EM (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9:355–365
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630
Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano H-Y (2004) The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131:5649–5657
Wang Z, Chen CB, Xu YY, Jiang RX, Han Y, Xu ZH, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22:409–417
Weng JF, Gu SH, Wan XY et al (2008) Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res 18:1199–1209
Wu JL, Wu CJ, Lei CL et al (2005) Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol Biol 59:85–97
Yoshimura A, Ideta O, Iwata N (1997) Linkage map of phenotype and RFLP markers in rice. Plant Mol Biol 35:49–60
Yuan Z, Gao S, Xue DW, Luo D, Li LT, Ding SY, Yao X, Wilson ZA, Qian Q, Zhang DB (2009) RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol 149:235–244
Acknowledgments
We thank Prof. Zhizhong Gong for his kindest help. This work was supported by Self-Regulated Projects of State Key Laboratory of Plant Physiology and Biochemistry, and by grants from the National Program of Transgenic Variety Development (2008ZX08009-004, 2008ZX08009-008).
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiaojiao Li and Lianjun Sun contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, X., Sun, L., Tan, L. et al. TH1, a DUF640 domain-like gene controls lemma and palea development in rice. Plant Mol Biol 78, 351–359 (2012). https://doi.org/10.1007/s11103-011-9868-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-011-9868-8