Abstract
Two GRAS family transcription factors, SHORT-ROOT (SHR) and SCARECROW (SCR), are required for ground tissue and quiescent center formation in Arabidopsis roots. The action of SHR and SCR is regulated by two INDETERMINATE DOMAIN (IDD) family proteins, JACKDAW (JKD) and MAGPIE (MGP). Although the reciprocal interaction of these transcription factors is considered to be involved in the modulation of SHR and SCR action by JKD and MGP, the underlying mechanism remains unclear. In this study, we use a transient assay with Arabidopsis culture cells to show that the physical interaction of these transcription factors modulate their transcriptional activity. Transient expression of LUC reporter genes with the proximal sequences upstream from the ATG codon of SCR and MGP in protoplasts were activated by JKD. Moreover, promoter activities were enhanced further by the addition of SHR and SCR to JKD, but not by the combination of SHR and SCR in the absence of JKD. Yeast one-hybrid analysis showed that JKD binds to the SCR and MGP promoter sequences, indicating the existence of another binding sequences of JKD different from the previously determined IDD binding sequence. Our findings suggest that JKD directly regulates SCR and MGP expression in cooperation with SHR, SCR and MGP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Root cells display a radial pattern of concentric epidermal and ground tissue layers surrounding a central stele. The radial pattern of the ground tissue is the result of stereotypical divisions of cortex/endodermis initial (CEI) cells. The CEI cell first divides transversely, generating a new initial cell and a daughter cell. The CEI daughter (CEID) cell then divides longitudinally, generating the first cells of the endodermal and cortical cell lineages (Fig. 1A). In Arabidopsis, two GRAS family proteins, SCARECROW (SCR) and SHORTROOT (SHR) are involved in the radial pattern formation of the ground tissue (Di Laurenzio et al. 1996; Helariutta et al. 2000; Heidstra et al. 2004; Sena et al. 2004).
Localization of JKD, MGP, SCR and SHR transcripts and protein in Arabidopsis roots. A Schematic of the longitudinal section, and RNA localization of JKD, MGP, SCR and SHR in Arabidopsis roots. S stele, E endodermis, C cortex, CEI cortex/endodermis initial, QC quiescent center. B Diagram illustrating JKD, MGP, SCR and SHR protein localization and co-localization patterns in Arabidopsis roots
SHR mRNA is found exclusively in the stele (Fig. 1A), but SHR protein moves outward to cells adjacent to the stele (Helariutta et al. 2000; Nakajima et al. 2001). SCR is expressed in cells of the endodermis, CEI and the quiescent center (QC), where it has been shown to prevent stem cells differentiation in adjacent cells (Di Laurenzio et al. 1996). In scr mutant roots, ground tissue consists of a single layer with mixed endodermal and cortical identity, the QC is not properly specified, and SHR protein movement is not restricted to the endodermis (Di Laurenzio et al. 1996; Sabatini et al. 2003; Heidstra et al. 2004; Sena et al. 2004; Nakajima et al. 2001). These results suggest that SCR is required for asymmetric periclinal ground tissue division, maintenance of QC identity, and localization of SHR to endodermal cells. The SCR gene was shown to be a direct target of the SHR transcription factor, and the SCR protein binds to its own promoter in the presence of SHR (Levesque et al. 2006; Cui et al. 2007), demonstrating that SCR is controlled by a SHR/SCR-dependent feed-forward loop. Although scr mutants form a single layer of ground tissue with endodermal characteristics, in shr mutants the single layer of ground tissue has only cortical identity (Helariutta et al. 2000; Benfey et al. 1993). This demonstrates that endodermis specification is controlled through SHR targets independent of the SHR-SCR complex.
A recent meta-analysis approach identified two INDETERMINATE DOMAIN (IDD) family genes, MAGPIE (MGP) and NUTCRACKER (NUC) as direct targets of SHR (Levesque et al. 2006). Another IDD family protein, JACKDAW (JKD) was also identified as an important factor that acts in a SHR-dependent feed-forward loop to regulate the range of action of SHR and SCR. JKD and MGP were also shown to interact physically with both SCR and SHR (Welch et al. 2007).
Here we investigated the effect of interactions of JKD, MGP, SHR and SCR transcription factors on the transcriptional activity of these transcription factor complexes by transient assay using an Arabidopsis culture cell system. Our data demonstrate that, of these transcription factors, only JKD activates transcription from the promoters of SCR, MGP and JKD by itself. Moreover, we show activation of transcription from the SCR and MGP promoters by the combination of SCR and SHR proteins, only when JKD is present.
Materials and methods
Construction of vectors for transient assay
Effector vectors with the DNA-binding domain of the yeast GAL4 transcription factor (GAL4BD) were constructed by introducing sequences for fusion proteins (JKD, MGP, SCR and SHR with GAL4BD) to downstream of cauliflower mosaic virus (CaMV) 35S promoter. First, ORFs of these transcription factor genes were amplified by PCR using primer sets listed in Table 1: for amplification of full length ORF coding sequences of JKD, MGP, SCR and SHR, primer sets of JKD full F/R, MGP F/R, SCR F/R and SHR F/R, respectively, were used. Amplified fragments were cloned into pCR-Blunt (Invitrogen) and cut by EcoRI and BamHI or EcoRI and SalI. The fragments were cloned into EcoRI and BamHI or EcoRI and SalI site of pGBT9 (Clontech) in frame with GAL4BD. The sequence for fusion proteins were amplified by PCR using the primer pair 5′-GCATACAATCAACTCCAAGC-3′ and 5′-GAAATTCGCCCGGAATTAGC-3′, and cloned into smaI site downstream of CaMV35S promoter in pUC19 vector (35S:pUC19, Maeo et al. 2009). To construct effector vectors without GAL4BD, PCR-amplified ORFs of these transcription factors were directly cloned into the SmaI site downstream of CaMV35S promoter of 35S:pUC19.
To construct various LUC reporter plasmids, the 1.5, 3.5, 2.5 and 3.5 kb sequence regions upstream of the ATG codon of SCR, MGP, SHR and JKD, respectively, were amplified by PCR using primer sets listed in Table 1, and amplified DNA sequences were introduced upstream of the luciferase reporter gene (LUC) in pBI221(pBI221-luc, Maeo et al. 2009). For detection of the activity of GAL4BD fusion proteins, pGLL, whose expression was driven by a chimeric promoter containing four copies of the GAL4 binding site fused upstream of a minimal 35S promoter (−98) fragment, was used. As an internal control, 35S:hRLUC was used (Maeo et al. 2009).
Transient assay
Protoplasts prepared from the Arabidopsis T87 suspension-cultured cells were transfected with plasmid DNA according to a methods by (Maeo et al. 2009). A suspension of protoplasts (150 μl; 105 protoplasts per ml) was co-transfected with 10 μg each of the LUC reporter and the effector plasmid DNAs, and 0.5 μg of the 35S:hRLUC internal control plasmid. To adjust the amount of total DNA, empty vector (35S:pUC19) was used (total amount of effector vectors were adjusted to 40 μg). The protoplasts were incubated at 22°C for 20 h before collection and measurement of reporter activities. The LUC and hRLUC activities were measured using the Dual-Luciferase Reporter Assay system (Promega). The LUC activity was normalized according to the hRLUC activity in each assay, and the relative ratio was determined by comparing this ratio with that obtained with the empty vector. The mean relative ratios were calculated from three to four independent experiments.
Yeast one-hybrid assay
The yeast one-hybrid assay was performed using the MATCHMAKER One-Hybrid system (Clontech). For amplification of JKD and MGP, the same primer pairs used for construction of effector vectors were used. For amplification of the N terminal half of JKD and C terminal half of JKD, primer sets of JKD full F/JKD ND R and JKD CD F/JKD full R, respectively, were used (Table 1).
The amplified ORF regions were cloned in frame after the transcriptional activation domain of the yeast GAL4 transcription factor (without DNA binding domain) in pGAD424. For construction of reporter plasmids, we amplified the promoter sequence of SCR (SCRp-up: from −1,500 to −500 bp, SCRp-down: from −1,000 to −1 bp from the start codon) and MGP (MGPp-3.5–3.0: from −3.5 to −3.0 kb, MGPp-3.0–2.5: from −3.0 to −2.5 kb and MGPp-2.5–2.0: from −2.5 to −2.0 kb) by PCR from Arabidopsis (Columbia) genomic DNA. Primer pairs used for amplification were SCRp1500F and SCRp500R for SCRp-up, SCRp1000F and SCRpR for SCRp-down, MGPp3500F and MGPp3000R for MGPp-3.5–3.0, MGPp3000F and MGPp2500R for MGPp-3.0–2.5 and MGPp2500F and MGPp2000R for MGPp-2.5.0–2.0 (Table 1).
Amplified PCR products were cloned upstream of the LacZ reporter gene in pLacZi vector (Clontech), and then, the promoter sequences and reporter genes were transferred to the genomic DNA of yeas strain BY5444 according to the manufacturer’s manual.
The vectors in pGAD424 were introduced into yeast which carry SCR or MGP promoter sequences, and X-gal activities were detected by filter assay according to the manufacturer’s manual (Clontech).
Results
Transcriptional activity of JKD and MGP
In wild-type Arabidopsis roots, expression of JKD and MGP overlap in the endodermis and CEI, whereas in the QC and cortex, only JKD is expressed (Fig. 1A, B; Welch et al. 2007; Levesque et al. 2006). JKD and MGP are reported to interact with each other, and they each interact with SCR and SHR as well (Welch et al. 2007). To assess whether the interaction of any or all of these transcription factors influences the transcriptional activity of JKD and MGP, we performed transient assays using Arabidopsis culture cells. JKD and MGP were fused to the GAL4 DNA binding domain (GAL4BD) gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter to construct effector vectors GBD JKD and GBD MGP (Fig. 2A). We also made constructs where JKD, MGP, SCR and SHR are under the control of the 35S promoter (JKD, MGP, SCR and SHR, Fig. 2A).
Transcriptional activity of JKD, MGP, SCR and SHR, in combination with other factors in an Arabidopsis protoplast system. A Schematic diagram of the reporter and effector plasmids used in transient assays. The GAL4-responsive reporter construct, pGLL, contains four copies of the GAL4 binding site and a minimal CaMV 35S promoter (35Smin), the gene for luciferase (LUC), and a nopaline synthase (NOS) terminator. Each effector construct contains a GAL4 DNA binding domain (GAL4BD) and either the JKD, MGP, SCR or SHR genes, driven by the CaMV35S promoter. Effector constructs without GAL4 DNA binding domain contained JKD, MGP, SCR or SHR genes driven by the CaMV 35S promoter. Transcriptional activity of B JKD fused with GAL4BD (GBD JKD), C GBD MGP, D GBD SCR and E GBD SHR in combination with other factors. As a control, an empty vector was used and all LUC activities are expressed relative to the control (value set at 1). Values shown are averages of results from three or four independent experiments. The error bars represent SD. Bars with asterisks are significantly different from control (*0.05 > P > 0.01; **P < 0.01)
When GBD JKD was transferred into protoplasts prepared from Arabidopsis T87 cells together with a luciferase reporter gene that contained four copies of the GAL4-responsive element (pGLL), a low level of activity was detected (Fig. 2B). By contrast, GBD MGP showed no transcriptional activity (Fig. 2C). Effectors without GAL4BD showed no activity by themselves (data not shown).
To determine whether addition of other transcription factors influence the level of JKD transcription activity, effector vectors without GAL4BD were transformed into the cells together with GBD JKD. Expression of reporter genes was about three times higher when GBD JKD was co-transformed with SHR than with GBD JKD by itself. On the other hand, addition of SCR showed only a slight increase in the activity of GBD JKD and transcriptional activity was slightly reduced with MGP. Maximum JKD activity was observed in the presence of both SHR and MGP. Interestingly, the increased GBD JKD activity observed by addition of SHR was severely attenuated when SCR was added as well (Fig. 2B).
Next, we examined the influence of additional effectors on the activity of MGP. Similar to what was found with JKD, the activity of GBD MGP was more than three times higher when SHR was added. In contrast with the results observed when MGP was added to GBD JKD, the addition of JKD to GBD MGP increased the activity to similar level of SHR. Addition of both SCR and SHR greatly increased the activity of MGP (about 30 times higher activity than MGP by itself). However, the addition of JKD along with SCR and SHR reduced the highest activity to basal levels (Fig. 2C). These results suggest that various combinations of transcription factors can influence transcriptional activity in both positive and negative directions.
Transcriptional activity of SCR and SHR
Because we observed the influence of protein interactions on the transcriptional activity of JKD and MGP, we also examined the activity of SCR and SHR. We constructed GBD SCR and GBD SHR effector plasmids in the same way as for GBD JKD and GBD MGP (Fig. 2D, E).
Significant activity for SCR by itself was not detected, and activity was not increased by the addition of any single factor. Elevated activity (about threefold) was observed when both JKD and MGP were added together. However, this increased activity was abrogated by the addition of SHR with JKD and MGP (Fig. 2D).
On the other hand, among the four transcription factors tested, SHR showed highest activity by itself, with about a 15-fold higher level than background (Fig. 2E). Because a large portion of SHR protein is excluded from the nucleus of plant cells in the absence of SCR (Sena et al. 2004; Cui et al. 2007), SHR might not show considerable transcriptional activity by itself in cells. In these experiments, the nuclear localization signal from the yeast vector most likely shuttles SHR protein to the nucleus. Interestingly, addition of any of the other factors, except for MGP, reduces the activity of SHR (Fig. 2E). Although SHR and SCR are considered to activate their target genes as a complex, the activity of both GBD SCR and GBD SHR were reduced in the presence of SHR and SCR, respectively.
Transcriptional activity of JKD, MGP, SCR and SHR on the SCR promoter
Expression of SCR is regulated directly by SHR and SCR (Levesque et al. 2006; Heidstra et al. 2004). Recent data showed that JKD is also required for maintenance of SCR expression particularly in the QC (Welch et al. 2007). To analyze the transcriptional regulation of SCR by these transcription factors, the 1.5 kb promoter region of SCR driving the luciferase gene was introduced and used as a reporter construct in transient assays in plant cells. This 1.5 kb sequence upstream of ATG was shown to be sufficient for accurate SCR expression when fused to a yellow fluorescent protein (YFP) reporter gene and transferred to Arabidopsis (Aida et al. 2004). We used effectors without GAL4BD hereafter.
Individually, SHR, SCR and MGP did not activate transcription from the SCR promoter, while JKD alone showed a slight increase in transcription from the SCR promoter (about five times higher than empty vector). Surprisingly, while the combination of SCR and SHR did not activate transcription from SCR promoter, JKD with SHR showed approximately three times the transcriptional activity of JKD alone (Fig. 3A). The combination of JKD, SCR and SHR did not significantly increase LUC activity compared to just JKD and SHR, but transcriptional activity was increased significantly when all four transcription factors are introduced (Fig. 3A). The localization of these factors is observed in CEI cells and endodermis (Fig. 1B), and these results suggest that the high expression of SCR in these tissues is activated by the combined action of these factors. In this experiment, LUC activity was detected only when JKD was present, indicating that feed-forward activation of SCR transcription by SHR and SCR requires JKD.
Activation of transcription from promoters of the JKD, MGP, SCR or SHR genes by JKD, MGP, SCR and SHR transcription factors alone or in various combinations with other factors. Transactivation of A SCR promoter, B MBP promoter, C JKD promoter and D SHR promoter, by JKD, MGP, SCR and SHR in combination with other factors. Schematic diagram of the reporter plasmid used in each transient assay is shown under each graph. As a control, an empty vector was used and all LUC activities are expressed relative to the control (value set at 1). Values shown are averages of results from three or four independent experiments. The error bars represent SD. Bars with asterisks are significantly different from control (*0.05 > P > 0.01; **P < 0.01)
Transcriptional activity of JKD, MGP, SCR and SHR on the MGP promoter
Meta-analysis identified MGP as one of the putative direct targets of SHR (Levesque et al. 2006), and SHR and SCR were found to bind to the promoter region of MGP. A Chromatin immunoprecipitation-polymerase chain reaction (ChIP-PCR) assays and promoter scanning indicated that both SHR and SCR bind to a region approximately 3.2 kb upstream of the translation start site of MGP (Cui et al. 2007). In addition, a 3.5 kb region upstream of MGP was determined to be sufficient for correct expression of MGP, when the sequence was fused to GFP (Miyashima et al. 2009). Therefore a 3.5 kb sequence upstream of the MGP start codon was used as a reporter sequence in transient assay.
When individual transcription factors were added, only JKD showed increased transcription activity from the MGP promoter, with a sevenfold increase over basal levels (Fig. 3B). The combination of SCR and SHR showed a slight but insignificant increase in activity (Fig. 3B). Although it has not been reported previously, this result indicates that JKD is a direct regulator of MGP. Interestingly, the addition of SCR, SHR or MGP suppressed JKD activity to basal level. Moreover, the highest level of activity from the MGP promoter occurs when the trio of JKD, SCR and SHR were used in the assay with transcription levels increased to nearly tenfold higher than basal, and slightly higher than JKD by itself (Fig. 3B). In marked contrast to the result with the SCR promoter, the addition of MGP completely eliminated the high transcriptional activity mediated by JKD, SHR and SCR together (Fig. 3B). These results indicate MGP itself may mediate a feed-back inhibition of its own promoter.
We obtained similar results to that with the 3.5 kb promoter assay above when the transient assay was carried out with the 1.5 kb sequence (from −3.5 to −2.0 kb from the MGP ATG) containing a putative binding site of SHR and SCR (data not shown). This result shows that the region from −3.5 to −2.0 kb from ATG is sufficient for the regulation of MGP expression by these factors.
Transcriptional activity of JKD, MGP, SCR and SHR on the JKD and SHR promoter
JKD expression is not affected by SCR and SHR in early stages of embryo development, but in post-embryonic stages, JKD expression in the QC and ground tissue stem cells is enhanced by SCR and SHR (Welch et al. 2007). A 2.8 kb region upstream of JKD is sufficient to drive a reporter gene expression in a pattern that matches the in situ hybridization pattern (Welch et al. 2007). We used a slightly longer sequence, 3.5 kb upstream of the JKD start codon, to construct a reporter for a transient assay. A low level of activity, about twofold above basal, was detected when JKD alone was added to the assay, and the addition of other transcription factors by themselves had no effect on transcription from the reporter (Fig. 3C). In addition, the combination of JKD with any other factor did not reduce or increase activity levels relative to JKD alone. These results indicated that there is a feed-forward mechanism for JKD expression. Moreover, we did not observe regulation of JKD by SCR and SHR together (Fig. 3C). This result suggests that the expression of JKD might be regulated by factors other than the transcriptional regulators examined here.
We also tested whether SHR expression was controlled by these transcription factors by using a 2.5 kb region upstream of SHR fused to luciferase gene as a reporter. A previous study demonstrated that the SHR promoter is contained within this 2.5 kb region (Nakajima et al. 2001). None of these transcription factors, including SHR itself, had a large effect on transcription from the SHR promoter (Fig. 3D). These results agree with the fact that SHR is expressed exclusively in the stele, where the SCR, JKD and MGP transcription factors are not co-expressed (Fig. 1A). This finding also supports a previous report that SHR expression is not altered in scr or shr mutants (Helariutta et al. 2000).
JKD binds to the SCR and MGP promoter sequences
Since JKD by itself is able to activate the promoters of the SCR and MGP genes and transcription from its own promoter, we examined the direct interaction between JKD and the promoter region of SCR and MGP using a yeast one-hybrid assay.
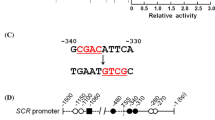
Two regions upstream of SCR, a sequence −1.5 to −0.5 kb from the ATG (SCRp-up) and the 1.0 kb sequence upstream of the ATG (SCRp-down) were placed upstream of LacZ gene to create reporter constructs, and integrated into the yeast genome (Fig. 4A). For prey vectors, full length JKD, the N-terminal half of JKD and C-terminal half of JKD were fused with the yeast GAL4 activation domain (GAL4 AD). The prey vectors were introduced into the yeast carrying the reporter genes, and DNA binding activity was estimated by X-gal staining assay.
JKD and MGP bind to the SCR and MGP promoter. A SCR promoter region from −1,000 to −1 bp (SCRp-down) and −1,500 to −500 bp (SCRp-up) were placed upstream of LacZ gene. IDD binding sequence contains four copies of the 11 bp IDD binding sequence (TTTGTCGTATT) in tandem placed upstream of LacZ gene. B, D X-gal staining assay. C MGP promoter region from −3.5 to −3.0 kbp MGPp-3.5–3.0), from −3.0 to −2.5 kbp (MGPp-3.0–2.5) and from −2.5 to −2.0kbp (MGPp-2.5–2.0) were placed upstream of LacZ gene. empty, empty vector pGAD424; JKD F, full length JKD; JKD N, N-terminal half of JKD; JKD C, C-terminal half of JKD; MGP, full length MGP
Full-length JKD protein was found to interact with the 1 kb region of SCR immediately upstream of the ATG, but no interaction was evident with the sequence from −1.5 to −0.5 kb (Fig. 4B). These results suggested that cis-acting elements recognized by JKD are present within a region 0.5 kb from the translation start site. Chip-qRT-PCR experiments indicated that SHR binding site is present between 209 and 528 bp upstream from ATG of SCR (Levesque et al. 2006). Therefore the JKD binding site may be close to that of SHR.
When JKD was divided into two parts, the N-terminal peptide was sufficient to bind the SCR promoter (Fig. 4B). Because JKD binds to the SCR promoter though N-terminal half, most of which consists of ID domain (IDD), we tested whether MGP also bind to SCR promoter. Although MGP did not activated transcription from SCR promoter, it does appear to bind to the SCR promoter in this assay (Fig. 4B).
To analyze the binding of JKD to the MGP promoter, we used a 1.5 kb sequence (−3.5 to −2.0 kb from the ATG), because we found that the entire 3.5 kb is too long to be effective in a yeast one-hybrid assay, and our preliminary data showed that JKD activated the reporter gene expression when the 1.5 kb region was used in transient assays (data not shown). The 1.5 kb region was further divided into three parts, sequences −3.5 to −3.0 kb, −3.0 to −2.5 kb and −2.5 to −2.0 kb from ATG, and these sequences were placed upstream of LacZ gene to create reporter construct (MGPp-3.5–3.0, MGPp-3.0–2.5 and MGPp-2.5–2.0).
When MGPp-3.5–3.0 was used as a reporter, we could detect a faint signal with full length of JKD and MGP, and the N-terminal region of JKD in X-gal staining assay. Chip-qRT-PCR experiments indicated that SHR bind to around −3.2 kb upstream of translational start site of MGP gene (Cui et al. 2007), indicating JKD binding site may be close to that of SHR as same as in the case of SCR promoter.
Previously, we showed that two zinc fingers of the maize ID1 protein, as well as another maize ID1 homolog, bind to an 11 bp consensus sequence (Kozaki et al. 2004). Here we observed that JKD and MGP also bind to the 11 bp ID1-binding consensus sequence in yeast (Fig. 4B). However, we could not find this 11 bp sequence in the 0.5 kb promoter region of SCR or MGP, indicating that JKD and MGP recognize sequences other than the 11 bp.
Discussion
Current research suggests that transcriptional regulators often form protein complexes and act in combination with each other to mediate tissue specific expression of numerous genes and to modulate the levels of gene transcripts (Stobe et al. 2009). However, the molecular mechanisms that specify how complexes of transcription factors control target gene expression are poorly understood (Pan et al. 2010). Several ID family proteins, JKD (AtIDD10), MGP (AtIDD3) and NUC (AtIDD8) recently were shown to play important roles in radial pattern formation in the Arabidopsis root meristem by interacting with the GRAS family transcriptional regulatory proteins, SHR and SCR (Welch et al. 2007). In this report, we investigated the transcription activities of these putative regulatory factors and the effects of their reciprocal interaction and activities by transient assay in Arabidopsis culture cells.
A previous study showed that SHR and SCR cooperatively regulate several downstream genes, including SCR and MGP (Levesque et al. 2006; Cui et al. 2007). Transient assays with SCR and MGP promoters showed that even in the presence of both SCR and SHR proteins, there was little or no apparent transcription from either the SCR promoter or the MGP promoter. Here we find that enhanced activity is observed only when JKD is present, presumably due to interaction between these factors (Figs. 3, 5). These results agree with another finding that showed SCR expression is not detected in the QC in jkd mutants (Welch et al. 2007). However, low levels of SCR mRNA were still detected in the ground tissue of jkd mutants (Welch et al. 2007), thus indicating that other factors are involved in controlling SCR expression in this region of the root.
Model illustrating action of JKD, MGP, SCR and SHR on promoters of SCR, MGP and a putative target gene. JKD directly binds to promoters of SCR and MGP, and activates these genes. SCR and SHR enhance the transcription of SCR and MGP only when JKD is present. MGP enhances the SCR promoter along with other factors. On the other hand, MGP suppresses the MGP transcription activated by other factors. The combination of MGP, SCR and SHR will activate the putative target gene (X) which promotes cell division, while JKD will suppress the activated transcription of the putative target gene
Transcription activity from the SCR promoter was enhanced when SHR and JKD were added together and feed-forward regulation by SCR on the SCR promoter was observed only when MGP was present along with SHR and JKD, presumably acting in a ternary complex (Fig. 3A).
On the other hand, whereas JKD alone activated transcription from the MGP promoter, both SHR and SCR each individually suppressed activity from the MGP promoter. Only the combination of both SHR and SCR together along with JKD caused activity to be enhanced slightly compared to transcription mediated by JKD alone (Fig. 3B). These results agree with the report that MGP expression is eliminated in both scr and shr mutants (Levesque et al. 2006). The combination of SHR, SCR and JKD, which exist in the QC where MGP is not expressed (Welch et al. 2007), activated the MGP promoter to the highest level in this system. The expression of MGP is probably strongly repressed in the QC by other unknown factors. The highest activity by the combination of SHR, SCR and JKD was suppressed to basal levels by the addition of MGP, indicating that feedback regulation is in effect (Fig. 5). The combination of these four factors is observed in the CEI and endodermal cells, where MGP is highly expressed (Welch et al. 2007). It is possible that MGP expression needs to be maintained at an adequate level by feedback regulation.
In contrast to what was observed with the SCR and MGP promoters in this system, neither the JKD nor SHR promoters were regulated by SCR and SHR. JKD transcription is reported to be initiated early in ground tissue development by a SHR- and SCR-independent mechanism; later, maintenance of JKD expression becomes dependent on SHR and SCR (Welch et al. 2007). Our results show that JKD itself has a slight positive effect on its own promoter (Fig. 3C). Although the 3.5 kb sequence upstream of JKD was shown to be sufficient for JKD expression in the QC and ground tissues (Welch et al. 2007), 5′ sequences further upstream might be required for regulation by SCR and SHR, or unknown factors might be required for SHR/SCR-dependent regulation of the JKD promoter as in the case of the SCR and MGP promoters. Recently, it was reported that SHR and SCR together activate the transcription of micro RNAs (miRNA) 165 and 166 (miR 165/166) in the endodermis. miR165 and miR166 suppress the expression of the Class III HD-ZIP transcription factor PHABULOSA (PHB), which suppress the expression of JKD (Miyashima et al. 2011; Carlsbecker et al. 2010). These results indicate that SHR and SCR indirectly regulate the expression of JKD. Further analysis will clarify the regulation of JKD expression. The SHR promoter was not activated by any of the transcription factors described here (Fig. 3D). This finding was not unexpected because SHR is expressed in the stele where SCR, MGP and JKD are not present (Fig. 1A). Moreover, SHR does not appear to regulate expression from its own promoter.
The binding of JKD to the SCR and MGP promoters was demonstrated by transient assay and yeast one-hybrid analysis (Figs. 3A,B, 4B,D). MGP also interacted with the SCR and MGP promoter in yeast (Fig. 4B), but it did not activate transcription from the SCR or MGP promoters (Fig. 3A). Binding to the SCR and MGP promoter was mediated by the N-terminal half of the JKD protein, which contains nearly the entire ID domain including all four zinc fingers (Fig. 4B). This suggests that the ID domain is involved in binding of sequences upstream of SCR and MGP. We previously demonstrated that IDD family proteins bind to an 11 bp of consensus sequence via two internal zinc fingers of the ID domain (Kozaki et al. 2004). This 11 bp sequence is not present in the 1.5 kb SCR or MGP promoter sequence used here, suggesting that IDD proteins have the potential to bind other sequences.
JKD was also shown to affect intracellular SHR localization in the QC (Welch et al. 2007). GFP-SHR was found to localize to the cytoplasm in root cells of jkd mutants, in contrast to the nuclear-localization of GFP-SHR in the QC of wild-type root cells. This was explained as mislocalization of SHR protein resulting from reduced SCR expression due to the absence of JKD (Welch et al. 2007). In transient assays, transcription activity of JKD was markedly enhanced by the presence of SHR (Figs. 2B, 3A), further supporting the idea that JKD directly controls the nuclear localization of SHR, even in the absence of SCR.
In contrast to JKD, MGP alone showed no appreciable transcriptional activity (Figs. 2C, 3A), which may explain why the mpg single mutant does not show a visible phenotype (Welch et al. 2007). Although MGP did not show transcription activity by itself, it regulates the transcription of downstream genes through interaction with other transcription factors, in different way from JKD.
Major phenotypes observed in ground tissue of jkd mutants include an increase in cell numbers within layers of the cortex and endodermis, and ectopic periclinal divisions in cortical cells. The ectopic periclinal divisions found in cortical cells of the jkd mutant were not observed in jkd scr or jkd shr double mutants. Further, increased numbers of cortical and endodermal cells in the jkd mutant was complemented by suppression of MGP expression, indicating that JKD counteracts a MGP-dependent cell division-promoting activity that is also dependent on SHR and SCR (Welch et al. 2007). The opposing effects of JDK and MGP were clearly observed in the GBD MGP experiment described here; i.e., the activity of GBD MGP increases dramatically in the presence of both SHR and SCR, and activity is reduced to basal levels by the addition of JKD (Fig. 2C). Moreover, the high transcription activity level shown by SHR and SCR together is markedly decreased in the absence of either SHR or SCR. If cell division-promoting genes are under this type of regulation, their gene expression is usually repressed by JKD such that, in the absence of JKD, gene expression and cell division are enhanced by the presence of both SCR and SHR (Fig. 5).
It is a challenge to analyze how a complex of several transcription factors can control specific target genes because one transcription factor could activate or repress several signaling pathways and, in turn, could be regulated by a complicated network of inter-related signaling pathways. In this context, the transient assays used here is an effective tool in dissecting the mechanisms of gene regulation by multimeric transcription factor protein complexes of interest because other factors that might otherwise confound this analysis are excluded.
In Arabidopsis, trichome cell fate determination is known to utilize a lateral inhibition mechanism that relies on the interplay of several transcription factors. GLABRA1 (GL1), an R2R3 MYB transcription factor (Oppenheimer et al. 1991), GLABRA3 (GL3), a basic helix-loop-helix (bHLH) transcription factor (Payne et al. 2000), and TRANSPARENT TESTAGLABRA1 (TTG1), a WD40 protein (Walker et al. 1999), are believed to form an activator complex that controls the transcription of GLABRA2 (GL2), a homeodomain protein (Rerie et al. 1994), which in turn specifies trichome formation in shoots (Ishida et al. 2007). A recent study using the Arabidopsis mesophyll protoplast transfection assay revealed that in addition to the protein–protein interaction between GL1-GL3, concurrent binding of GL1 and GL3 to the promoter of GL2 via their own DNA-binding domains is probably required to activate GL2 (Wang and Chen 2008).
Studies of Hox transcription factors, which play fundamental roles in controlling morphogenetic diversity along the anterior-posterior body axis in animals by regulating distinct sets of target genes (Mann and Morata 2000), revealed how Hox proteins regulate target gene expression in cooperation with other transcription factors. Although Hox proteins are active in a large number of cells, individual Hox proteins control cell differentiation and pattern formation and, consequently, target gene expression in a highly localized manner, sometimes even only in a single cell(Hersh et al. 2007). In Drosophila, a recent study of the regulation of the cell death gene reaper (rpr) by the Hox protein Deformed (Dfd) revealed that local activation of rpr expression in the anterior part of the maxillary segment is achieved through combinatorial interaction of Dfd with at least eight functionally diverse transcriptional regulators on a minimal enhancer for the Hox response elements (HREs). The authors suggest that a large number of transcription factors could dictate transcriptional output in combination with the respective Hox protein by binding selectively and independently to cis-regulatory sequences within HREs of target genes. Additionally, their model explains how Hox proteins can act as repressors in one context and as activators in another, because the combined transcriptional output is dependent on the regulatory activity of all transcription factors assembled on the enhancer (Stobe et al. 2009).
Our results support the model described above. We show that reciprocal interaction of transcription factors JKD, MGP, SCR and SHR differentially control target genes, depending on specific promoters. We found also that these transcription factors can act as both activator and repressor depending upon its interaction partner (or partners) and the promoter targets.
Taken together, it is important to elucidate how these transcription factors interact with promoter sequences and other transcription factors to clarify the mechanism used by these transcription factors to regulate the expression of their respective target genes. Understanding how the SCR-SHR pathway controls the ground tissue patterning will help define some of the principles involved in multiple transcription factor gene regulation.
References
Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119(1):109–120
Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119(1):57–70
Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindfren O, Moreno-Risueno MA, Vaten A, Thitamadee S, Campilho A, Sebastian J, Bowman JL, Helariutta Y, Benfey PN (2010) Cell signaling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465:316–321
Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316(5823):421–425
Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86(3):423–433
Heidstra R, Welch D, Scheres B (2004) Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev 18(16):1964–1969
Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101(5):555–567
Hersh BM, Nelson CE, Stoll SJ, Norton JE, Albert TJ, Carroll SB (2007) The UBX-regulated network in the haltere imaginal disc of D. melanogaster. Dev Biol 302 (2):717–727
Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, Okada K, Wada T (2007) Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19(8):2531–2543
Kozaki A, Hake S, Colasanti J (2004) The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res 32(5):1710–1720
Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, Scheres B, Benfey PN (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4(5):e143
Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60(3):476–487
Mann RS, Morata G (2000) The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol 16:243–271
Miyashima S, Hashimoto T, Nakajima K (2009) ARGONAUTE1 acts in Arabidopsis root radial pattern formation independently of the SHR/SCR pathway. Plant Cell Physiol 50(3):626–634
Miyashima S, Koi S, Hashimoto T, Nakajima K (2011) Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138:2303–2313
Nakajima K, Sena G, Nawy T, Benfey PN (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413(6853):307–311
Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67(3):483–493
Pan Y, Tsai CJ, Ma B, Nussinov R (2010) Mechanisms of transcription factor selectivity. Trends Genet 26(2):75–83
Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156(3):1349–1362
Rerie WG, Feldmann KA, Marks MD (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8(12):1388–1399
Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17(3):354–358
Sena G, Jung JW, Benfey PN (2004) A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131(12):2817–2826
Stobe P, Stein MA, Habring-Muller A, Bezdan D, Fuchs AL, Hueber SD, Wu H, Lohmann I (2009) Multifactorial regulation of a hox target gene. PLoS Genet 5(3):e1000412
Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11(7):1337–1350
Wang S, Chen JG (2008) Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol 49(12):1792–1804
Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B (2007) Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev 21(17):2196–2204
Acknowledgments
We thank K. Maeo and M. Takagi for providing expert technical advice and plasmids, and the RIKEN BioResource Center for Arabidopsis T87 suspension-cultured cells.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ogasawara, H., Kaimi, R., Colasanti, J. et al. Activity of transcription factor JACKDAW is essential for SHR/SCR-dependent activation of SCARECROW and MAGPIE and is modulated by reciprocal interactions with MAGPIE, SCARECROW and SHORT ROOT. Plant Mol Biol 77, 489 (2011). https://doi.org/10.1007/s11103-011-9826-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-011-9826-5