Abstract
In higher plants, DNA transfer from the plastid (chloroplast) genome to the nucleus is a frequent, ongoing process. However, there has been uncertainty over whether this transfer occurs by a direct DNA mechanism or whether RNA intermediates are involved. Previous experiments utilising transplastomic Nicotiana tabacum (tp7 and tp17) enabled the detection of plastid-to-nucleus transfer in real time. To determine whether RNA intermediates are involved in this transfer, transplastomic lines (tpneoACG) were generated containing, in their plastid genomes, a nuclear promoter-driven kanamycin resistance gene (neo) with a start codon that required plastid RNA editing but otherwise identical to tp7 and tp17. Therefore it was expected that kanamycin resistance would only be acquired following RNA-mediated transfer of neo to the nucleus. Screening of tpneoACG progeny revealed several kanamycin-resistant plants, each of which contained the neo gene located in the nucleus. Surprisingly, neo was unedited in all these plants, indicating that neoACG was active in the absence of an edited start codon and suggesting that RNA intermediates were not involved in the transfers. However, analysis of tpneoACG revealed that only a low proportion of transcripts potentially able to mediate neo transfer were edited, thus precluding unequivocal conclusions regarding the role of RNA in plastid-to-nucleus transfer. The low proportion of edited transcripts was found to be due to predominant antisense neo transcripts, rather than to low editing efficiency of the sense transcripts. This study highlights a number of important considerations in the design of experiments utilising plastid RNA editing. The results also suggest that RNA editing sites reduce but do not eliminate functional plastid-to-nucleus gene transfer. This is relevant both in an evolutionary context and in placing RNA editing-dependent genes in the plastid genome as a means of transgene containment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In eukaryotes, plastids and mitochondria are the descendents of once free-living prokaryotic ancestors. Over time, these organelles have donated a large number of genes to the nucleus, such that many organellar proteins are now encoded in the nuclear genome (Adams and Palmer 2003; Keeling and Palmer 2008; Timmis et al. 2004). In addition, there are many nuclear genes derived from organellar genomes which are not involved in organellar functions (Deusch et al. 2008; Martin et al. 2002; Noutsos et al. 2007). Organellar genomes have therefore been an important source of genetic material for nuclear genome evolution. However, the functional relocation of a gene to the nuclear genome requires a number of steps. Firstly, there must be escape of the DNA sequence from the plastid and incorporation into the nuclear genome. The gene must then acquire nuclear regulatory sequences, different from those of the organelle, to enable appropriate gene expression. In addition, if genes are to retain their role in organellar biogenesis they must establish a means of targeting the encoded protein to the organelle; usually through acquisition of a targeting peptide-encoding sequence.Very rarely, the acquisition of these additional sequences may occur immediately on insertion as a consequence of the size of the integrant, the break points involved and the site of integration. However, it is more likely that activation will involve several complex post-insertional rearrangements. While functional organelle-to-nucleus gene transfer is relatively rare, non-functional DNA sequence transfer is much more common, with many nuclear genomes containing large amounts of mitochondrial and, in the case of plants, plastid DNA (Hazkani-Covo et al. 2010; Matsuo et al. 2005; Noutsos et al. 2005; Richly and Leister 2004a, b; Woischnik and Moraes 2002). These nuclear organelle sequences have been designated numts (nuclear integrants of mitochondrial DNA) and nupts (nuclear integrants of plastid DNA).
Organelle-to-nucleus sequence transfer could occur either by direct transfer of DNA or by transfer involving reverse-transcribed RNA. Some studies support a direct DNA mechanism because many numts and nupts are very long and include non-coding regions. For example, a 620 kb numt and a 131 kb nupt have been found in Arabidopsis and rice respectively, which are strongly suggestive of direct DNA transfer (Stupar et al. 2001; Yu et al. 2003). Whole genome studies have also indicated that any part of an organelle genome can be transferred to the nucleus and there is no evidence for overrepresentation of highly transcribed regions in numts and nupts (Matsuo et al. 2005; Richly and Leister 2004a, b; Woischnik and Moraes 2002). However, there are a number of examples of functional mitochondrial gene relocation to the nucleus where splicing and RNA editing appear to have occurred prior to transfer, suggesting the involvement of RNA intermediates (Adams et al. 2000; Grohmann et al. 1992; Nugent and Palmer 1991). Experimental evidence in support of a direct DNA mechanism has been found in yeast, where it has been shown that in a mutant strain with a high rate of mitochondrion-to-nucleus transfer, transfer occurs independently of an RNA intermediate (Shafer et al. 1999). However, it is possible that the mechanism of transfer is variable, so RNA-mediated transfer may also have an important role to play. Determining the mechanism(s) of transfer in other systems will help to shed light on this.
The mode of transfer may also have implications in the use of transplastomic plants in biotechnological applications. Because plastids are predominantly maternally inherited, transplastomic crop plants offer greatly enhanced transgene containment compared with nuclear transgenics (Maliga 2002). However, there are two mechanisms by which plastid transgenes can escape through pollen at low frequency: occasional paternal transmission of plastids (Ruf et al. 2007; Svab and Maliga 2007) and transfer of transgenes to the nuclear genome (Huang et al. 2003; Sheppard et al. 2008). The latter type of escape would not normally result in transgene expression due to the absence of a nuclear promoter, but fortuitous integration or subsequent rearrangement could bring a transgene into context with an existing nuclear promoter (Lloyd and Timmis 2011; Stegemann and Bock 2006). Furthermore, it has been shown that a plastid promoter can have weak nuclear activity (Cornelissen and Vandewiele 1989; Lloyd and Timmis 2011). Therefore, if strict containment of a transgene product is vital, further measures will be required to prevent expression following transfer to the nuclear genome. One possibility is the introduction of plastid RNA editing sites such that the transgene transcripts are dependent upon RNA editing in the plastid to encode a functional protein (Lutz et al. 2006). However, this is only a useful strategy if transfer to the nucleus occurs independently of an edited RNA intermediate.
Plastid-to-nucleus DNA sequence transfer has been detected experimentally using transplastomic tobacco lines that contain, in their plastid genomes, a selectable marker gene tailored for nuclear expression. In one set of experiments the neo gene, under the control of a nuclear promoter (35S) and containing a nuclear intron (STLS2), was inserted into the plastid genome to generate two independent transplastomic lines, tp7 and tp17, which are assumed to be genetically identical (Huang et al. 2003). This gene encodes neomycin phosphotransferase (NPTII), which confers resistance to kanamycin. Backcross progeny derived from tp7 or tp17 pollen parents were screened for kanamycin resistance, to reveal any kanamycin resistant progeny that had arisen by transfer of neo to the nucleus and its resulting expression. Such experiments determined a transfer frequency in the male germline of approximately 1 event for every 11,000–16,000 pollen grains (Huang et al. 2003; Sheppard et al. 2008). In another set of experiments, a similar transplastomic line (containing neo under the control of the 35S promoter in the plastid genome) was used to screen for plastid-to-nucleus sequence transfer in somatic cells and it was estimated that one transfer event occurred for every 5 million leaf cells (Stegemann et al. 2003).
For the kanamycin-resistant plants derived from tp7 and tp17, DNA blot analysis was used to partially characterise the integrants at the molecular level (Huang et al. 2004; Sheppard et al. 2008). It was found that the majority had very large regions of plastid DNA integrated into the nuclear genome along with neo, with the size of most integrants exceeding 20 kb. Due to the large size of integrants, it seems likely that transfer occurred by a direct DNA mechanism. However, northern analysis of tp7 and tp17 with double stranded DNA probes revealed highly abundant neo-hybridising transcripts of 3.3 and 2.2 kb (both larger than the mature neo gene transcript of 1.2 kb) which were assumed to be the result of transcriptional read-through from adjacent promoter(s) in the transplastome (Huang et al. 2003, 2004), so it was impossible to determine whether some integrants resulted from RNA-mediated transfer. Longer, less abundant transcripts that were undetectable by northern blotting may also exist and could potentially have given rise to the larger integrants.
The target of insertion of the transgenes in tp7 and tp17 is between trnV and ORF70B/ORF131—an arrangement that was intended to minimise read-through transcription from adjacent native plastid promoters. Thus, the abundant transcripts containing neo-hybridising regions in tp7 and tp17 were unexpected, particularly since Zoubenko et al. (1994) were unable to detect any read-through transcription of transgenes inserted into the same plastid genome location using the same transformation vector (pPRV111A). However, ORF70B/ORF131 genes are on complementary strands with the promoter of ORF70B initiating transcription towards neo in tp7 and tp17. The promoter of ORF131 and other adjacent promoters 3′ of neo initiate transcription on the opposite strand, away from the transgenes. Similarly, the promoter of trnV and adjacent promoters 5′ of neo promote transcription away from the transgenes. In view of the lack of detection of transcripts by Zoubenko et al. (1994), it is most probable that the abundant neo-hybridising transcripts arose from activity of the 35S promoter in the plastid.
Whatever is the cause of the abundant neo transcripts in tp7 and tp17, it is necessary to test whether they are responsible for a proportion of the gene transfer events reported by Huang et al. (2003) and Sheppard et al. (2008). Therefore, experiments were designed to test the mode of transfer directly by utilising plastid-specific C to U RNA editing, which occurs at well-defined sites of some plastid RNAs in higher plants. The specificity is conferred by the flanking sequence [reviewed by Bock (2000)]. By introducing several RNA editing sites into the neo cassette, experiments were designed to detect plastid-to-nucleus transfer only if it occurred via an RNA intermediate.
Results
Generation of tpneoACG
The experiments to detect RNA-mediated plastid-to-nucleus transfer were designed to be as similar as possible to previous experiments in which tp7 and tp17 were used to detect plastid-to-nucleus nucleic acid transfer (Huang et al. 2003; Sheppard et al. 2008). Minimally modified transplastomic lines were generated (tpneoACG) in which the neo gene contained several plastid RNA editing sites, one of which was required for the functionality of neo, but in all other respects tpneoACG was identical to tp7 and tp17. The experiments were designed in this way so that the results could be compared directly with previous experiments where the frequency of total plastid-to-nucleus transfer (both DNA and RNA-mediated) was measured. In this way the current experiments would not only determine whether RNA-mediated transfer occurs at an appreciable frequency but also, if it does, what proportion of transfer events involve an RNA intermediate. Given the abundant neo-hybridising transcripts in tp7 and tp17 that were revealed by northern blotting (Huang et al. 2003, 2004), it was expected that if plastid-to-nucleus transfer involving an RNA-intermediate occurs at a reasonable frequency, then it should be detectable with the tpneoACG system.
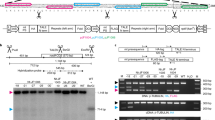
The tpneoACG transformation vector (pPRV111A::neoSTLS2ACG) was produced by modification of the vector pPRV111A::neoSTLS2 that was previously used to generate the tp7 and tp17 transplastomic lines. In both constructs, neo is under the regulatory control of the 35S promoter and the nuclear STLS2 intron is present within the neo open reading frame, such that kanamycin selection can be used to detect transfer of neo to the nucleus (Fig. 1). The two vectors contain the same regions of the plastid genome, such that neo is targeted for integration at the same plastome site in both experiments. Adjacent to neo in both transformation cassettes is aadA, which confers resistance to spectinomycin that was used for selection of transplastomic lines.
Experimental design. a The generalised structure of pPRV111A::neoSTLS2 (Huang et al. 2003) and pPRV111A::neoSTLS2ACG. The neo gene differs between these two vectors (shown in more detail in (b) and (c) respectively), but otherwise they are identical. Regions of identity to the plastid genome are shown in green, aadA (the plastid selectable marker) in black and neo (the nucleus-specific gene) in blue. b The structure of pPRV111A::neoSTLS2. In this construct neo is under the control of 35S promoter and terminator sequences and contains the STLS2 intron (Huang et al. 2003). The sequence of the first 12 bases of neo, beginning from the start codon, is shown along with its conceptual translation in the expanded version. c The structure of neo in pPRV111A::neoSTLS2ACG. In this construct neo is also under the control of 35S promoter and terminator sequences and contains the STLS2 intron. Orange regions are derived from plastid genes (see text for details) and contain three C to U RNA editing sites within their sequence contexts (shown in red). Neo sequence is shown in blue. The conceptual translation, after C to U editing, is also shown
The new vector (pPRV111A::neoSTLS2ACG) differed from pPRV111A::neoSTLS2 by two modifications to the neo gene (Fig. 1b, c). Firstly, the start codon of neo was removed and replaced with a 101 bp region derived from tobacco psbF and psbL genes which contained a single plastid C to U RNA editing site. This editing site was part of an ACG codon in frame with the neo open reading frame, so that editing would create an AUG start codon. In the absence of plastid RNA editing, neo transcripts lack a start codon and were not expected to be translated. The 101 bp region containing the editing site had previously been inserted into the tobacco plastid genome as a fusion with a kanamycin resistance gene under the control of a plastid promoter and the editing site was shown to be efficiently edited (~70%) in this sequence context (Chaudhuri et al. 1995). If transfer of neo to the nucleus occurs via an edited RNA intermediate, then the nuclear integrant will have a functional version of neo and the plant should be kanamycin-resistant. On the other hand, if transfer occurs by a direct DNA mechanism, then the nuclear integrant will contain neo without a start codon and the plant should be kanamycin-sensitive. In this way, kanamycin selection can be used to detect only those events where transfer of neo to the nucleus occurs via an RNA intermediate.
It is possible that a kanamycin-resistant plant could be obtained by a C to T mutation of the editing site prior to, during or after transfer to the nucleus, rather than by plastid RNA editing. To allow for this possibility, a 94 bp region of ndhB containing two plastid RNA editing sites was introduced into the 3′ UTR of neo (Fig. 1c). This 94 bp region had previously been inserted into the tobacco plastid genome in the 3′ UTR of an aadA gene under the control of a plastid promoter and the editing sites were efficiently edited (~95%) in this sequence context (Bock et al. 1996). If a kanamycin-resistant plant obtained from screening was indeed the result of RNA-mediated transfer, then these two additional sites would be expected to be edited also. The presence of three edited sites at a novel nuclear locus would rule out point mutation as an origin and unequivocally implicate an RNA intermediate.
Two independent transplastomic lines were generated using pPRV111A::neoSTLS2ACG, named tpneoACG5-3 and tpneoACG8-2. These lines were shown to be homoplasmic by DNA blot and PCR analysis (data not shown).
Screening for RNA-mediated transfer
To detect RNA-mediated plastid-to-nucleus transfer of neo, tpneoACG was used as a pollen parent in crosses with wildtype plants and 328,000 progeny screened for kanamycin resistance (Table 1). In the initial stages of screening there were several plants that appeared to be resistant but were still somewhat sickly. Therefore any plants that appeared resistant were transferred to non-selective medium in an attempt to recover all possible resistant plants. Amongst the 328,000 progeny screened, 10 putative resistant plants were obtained which were named kr5.1-kr5.10. After removal from selective medium, only 6 plants survived (kr5.1, kr5.2, kr5.3, kr5.4, kr5.6 and kr5.7). Presumably the other 4 plants died as a result of kanamycin exposure, so they may have been false positives or they may have been only partially kanamycin-resistant (see “Discussion”). This frequency of a plastid to nucleus transfer event of 1 in 55,000 seedlings is over threefolds lower than the 1 in 16,000 frequency observed for the gene without an editing requirement (Huang et al. 2003). Moreover, no plants with high levels of resistance to kanamycin were isolated indicating limited expression of the neo gene in the nucleus. The results suggest that the inclusion of editing sites in plastid transgenes may reduce, but not eliminate, functional transfer of genes from the plastid to the nucleus. DNA was prepared from the surviving plants (kr5.1-kr5.4, kr5.6 and kr5.7) and the neo gene was successfully amplified from all samples, indicating that plastid sequence transfer had occurred in each line. However, for each plant, all three editing sites remained unedited, indicating that the sequence transfers had not involved edited RNA intermediates. This result also indicated that the neo gene was somehow active in the absence of the intended translational start codon.
Neomycin phosphotransferase activity was measured in the kr5.X lines (X = the individually numbered plants from the screen) and found to be indistinguishable from wildtype (Table 2). In contrast, there was at least 80-folds higher activity (P < 0.001, ANOVA, Bonferroni corrected) in kr2.2 than in the kr5.X lines. Kr2.2 was derived from a screen to detect plastid-to-nucleus transfer in tp7 and contains a single nuclear copy of neo with an ATG start codon (Lloyd and Timmis 2011; Sheppard et al. 2008). NPTII protein level in kr5.X, as shown by NPTII ELISA, was also indistinguishable from wildtype (Table 2). These results indicate that the kr5.X lines are kanamycin-resistant, despite undetectable levels of both NPTII protein and NPTII activity. Therefore, very low levels of NPTII activity are sufficient to confer kanamycin resistance. This suggests that translation of the NPTII protein is greatly inhibited by the absence of the AUG start codon, although the presence of high levels of a less functional (presumably shorter) NPTII protein that is not recognised by the ELISA antibody cannot be ruled out. This reduction in NPTII activity may have reduced the ability of the screen to identify positive plants.
The presence of unedited neo sequences in the kr5.X plants indicates that transfer of neo to the nucleus occurred either by a direct DNA mechanism or via unedited RNA intermediates. To attempt to distinguish between these possibilities, the efficiency of RNA editing was analysed in tpneoACG. If the proportion of edited transcripts was close to 100%, this would effectively rule out transfer via unedited RNA as a possibility, thus showing that transfer occurred by a direct DNA mechanism.
To determine what proportion of neo RNA molecules were edited in tpneoACG plastids, total cDNA was synthesised using random hexamer primers, followed by PCR amplification of the neo cDNA including all three editing sites. Sequencing of this PCR product showed no clear evidence for editing at any of the three editing sites (Fig. 2). This indicated that it was not possible to distinguish between DNA and RNA-mediated transfer using the tpneoACG system, since the vast majority of transcripts were unedited.
Antisense neo transcripts in tpneoACG
The lack of detectable edited transcripts in tpneoACG could be due to a low editing efficiency but this is unlikely since the editing sites each have flanking sequence which has previously been shown to be sufficient for efficient editing to occur (Bock et al. 1996; Chaudhuri et al. 1995). The other possibility is that a large proportion of antisense transcripts are present, which would not be edited. This latter explanation is consistent with the activity of the ORF70B promoter which would transcribe the complementary strand of neo. Northern analysis of tp7 and tp17 (Huang et al. 2003, 2004) used a double-stranded DNA probe, so it is not known whether the neo-hybridising bands represented sense or antisense transcripts. To attempt to distinguish between the two possibilities for the lack of edited transcripts (low editing efficiency or high proportion of antisense transcripts), cDNA was synthesised from tpneoACG RNA using primers complementary to the neo sense strand, so that theoretically only neo sense transcripts would be represented in the cDNA population. If this revealed a high proportion of edited transcripts (as opposed to the results obtained with cDNA synthesised using random hexamer primers which do not distinguish between the two strands) then it could be concluded that highly abundant antisense transcripts were the main reason for the low representation of edited RNA species.
For technical reasons, the two editing regions were amplified and sequenced separately from cDNA samples synthesised using different neo primers. In contrast to the results obtained using random hexamer cDNA, editing was detected at each of the three editing sites, but the majority of transcripts remained unedited (Fig. 3). To confirm that the presence of detectable edited transcripts was the result of using neo-specific primers for reverse transcription, rather than different PCR primers, amplification and sequencing from random hexamer cDNA was repeated using the same PCR primers as those used for neo-specific cDNA. Again this revealed no detectable editing (data not shown).
When a neo primer was used for cDNA synthesis, neo sense transcripts should have been reverse-transcribed, but neo antisense transcripts should not. However, since reverse transcription was performed at a relatively low temperature (42°C), it is possible that the primer was able to bind fortuitously to a few bases on the antisense transcripts. If this was the case, then cDNA synthesis using a neo primer would have enriched for sense transcripts (compared to using random hexamer primers), but not eliminated antisense transcripts.
To test this possibility, the experiment using neo primers for reverse transcription was repeated for tpneoACG5-3 using a thermostable reverse transcriptase which allowed reverse transcription to be performed at 65°C. This resulted in a higher proportion of edited transcripts (Fig. 4), confirming that some antisense transcripts were reverse transcribed when a non-thermostable reverse transcriptase was used.
The results shown in Fig. 4 give a minimum estimate for the editing efficiency (i.e. the proportion of sense transcripts that are edited) at each of the three editing sites, since it is possible that some antisense transcripts may have been reverse transcribed, despite the stringent conditions used. The first editing site, with the same amount of flanking sequence as that used here, was previously reported to be edited with ~70% efficiency (Chaudhuri et al. 1995), consistent with the results shown in Fig. 4. The second and third editing sites, with the same amount of flanking sequence as that used here, were previously reported to be edited with ~95% efficiency (Bock et al. 1996). However, in Fig. 4, only around half of the transcripts are edited at each site. It is possible that the efficiency of editing of these sites in tpneoACG is only around 50%, which could be due to reduced accessibility of the editing sites in neo sense transcripts as a result of interactions with the abundant antisense transcripts or altered secondary structure.
To investigate the influence of secondary structure, the Mfold program was used to analyse the structure of the inefficiently edited neo transcript. We used the efficiently edited endogenous ndhB transcript, and the efficiently edited aadA:ndhB transcript reported by Bock et al. (1996) for comparison. Computational structure prediction of long RNAs (>700 nt) is poor, with prediction sensitivity often well below 50% (Deigan et al. 2009; Tsang and Wiese 2010). Therefore, as well as for full length transcripts, secondary structure was determined for a smaller (~600 nt) region of the RNA molecules covering the ndhB editing sites and the flanking 250 nt on either side.
The secondary structure of the editing site region in the efficiently edited aadA:ndhB transcript and the inefficiently edited neo transcripts were essentially identical when predicted using the ~600 nt analysis (Fig. S1) but the analysis showed a different structure for the endogenous ndhB transcript (Fig. S1). Structures predicted in the full length analysis differed for each transcript (Fig. S2); however no relationship was evident between the degree of base pairing surrounding the edited bases and the efficiency of editing (Fig. S2). An alternative possibility is that editing occurs efficiently in neo sense transcripts but that the primer used for reverse transcription prior to sequencing the second and third editing sites is particularly prone to binding the antisense transcripts.
Discussion
In an attempt to distinguish between two possible modes of plastid-to-nucleus transfer (direct DNA transfer and RNA-mediated transfer), transplastomic lines were generated which contained, in their plastid genomes, a neo gene designed for nuclear expression but with several plastid RNA editing sites. One of these editing sites was designed so that editing would be required to create a translational start codon for neo. Surprisingly, neo was found to be active without this site being edited. A total of 328,000 progeny from backcrossed tpneoACG plants were screened and 6 kanamycin-resistant progeny were identified, each of which contained one or more copies of the experimental neo gene in its nucleus. Sequencing indicated that none of these arose from transfer involving an edited RNA intermediate. However, sequencing of tpneoACG cDNA indicated that the majority of neo transcripts were antisense and hence unable to be edited.As a result the screen was not able to distinguish definitively between direct DNA and RNA-mediated transfer. Nonetheless edited sense strand RNAs were present and none of these mediated gene transfer. The finding that all the kr5.X plants contained unedited versions of neo precludes other explanations of the acquisition of resistance. How then is neo expressed in these plants when it lacks a translational start codon? Clearly the NPTII levels are low and cannot be detected by ELISA so translation is inefficient. Translation initiation from a downstream AUG codon seems unlikely, as the first in-frame AUG occurs almost halfway through the coding sequence, making it unlikely that a functional protein would be produced. Alternatively, translation may be initiated from a non-AUG codon, as ACG, CUG, AUC and GUG codons have been shown to be capable of initiating translation in plants (Depeiges et al. 2006; Kobayashi et al. 2002; Wamboldt et al. 2009). It is also remotely conceivable that some transcripts are edited in the cytoplasm, perhaps by precursors of the nuclear-encoded chloroplast RNA editing enzymes on their way to the chloroplast. However, these were not detectable in mRNA from leaf tissue of selected kr5.X plants using a variety of RT–PCR strategies (results not shown). It is assumed that the intron in the kr5.X lines is efficiently spliced but this was not tested experimentally as the additional sequences in pPRV111A::neoSTLS2ACG compared with pPRV111A::neoSTLS2 are considered very unlikely to influence splicing.
Previous screens that were not designed to distinguish between direct DNA transfer and RNA-mediated transfer measured a transfer frequency of 1 event for every 11,000–16,000 male gametes (Huang et al. 2003; Sheppard et al. 2008). However, the current screen revealed only 6 resistant plants out of 328,000 tested, corresponding to a frequency of 1 in 55,000. A possible reason for the lower proportion of resistant plants in this screen is that even though the neo gene is able to confer some level of resistance in the absence of editing, the phenotype is more subtle and the level of resistance may not always be enough to overcome the selection regime used. The lack of detectable NPTII protein in the kr5.X lines supports this idea. In this case the 6 resistant plants obtained may have had multiple nuclear insertions of neo, or neo may have inserted into genomic regions that allowed higher expression than other sites. In this way the 4 putative kanamycin-resistant plants that died could have had a nuclear insertion of neo that made them less sensitive to kanamycin than wildtype, but not sufficiently resistant to survive the practical experimental screen. Presumably some other plants had nuclear insertions of neo that could not be identified by the screening procedure.
This study highlights a number of important considerations for the design of future experiments. The neo gene used here was found to be active in the absence of the intended start codon, so it was necessary to sequence the neo gene in each resistant plant to determine the status of the editing sites. Most importantly, tpneoACG was found to have a high proportion of antisense neo transcripts relative to sense transcripts, such that the majority of neo transcripts were unedited. Therefore it was not possible to distinguish between DNA and RNA-mediated transfer. Consequently, the relative contribution of transcription from the two strands is an important aspect to test; though generating transplastomic lines still represents a large investment. Therefore, future experiments should be designed to avoid this problem. One approach could be to include a number of editing sites, in both sense and antisense orientations, such that RNA-mediated transfer events from both DNA strands will retain unique editing signatures. An alternative approach, which is particularly relevant in the context of plastid RNA editing use as a biological containment strategy, would be to place neo (with the 35S promoter) downstream of a strong plastid promoter to ensure high levels of sense neo transcripts and reduce the influence of antisense transcripts. Having high levels of neo transcripts would also presumably maximise the chances of RNA-mediated transfer occurring. One problem with this approach is that the promoter should be active in the tissue where plastid-to-nucleus transfer occurs, something that is still uncertain. The frequency of plastid-to-nucleus transfer appears highest in the male germline, but experiments have so far been unable to determine a specific stage of pollen development at which transfer occurs (Sheppard et al. 2008). Therefore, what constitutes an appropriate promoter is not known. An alternative would be to screen for transfer in somatic tissues, although it is likely that this would be less relevant than germline transfer in evolutionary terms.
In biotechnological applications, the incorporation of RNA editing sites in plastid transgenes, such that editing is required for functionality, is an approach that may help to minimise functional transgene escape through pollen. In our experiments, we have shown that functional transfer of neo to the nucleus occurs less frequently when such plastid RNA editing sites are included (this study), than when they are not (Huang et al. 2003; Sheppard et al. 2008). To our knowledge, this is the first direct evidence for the utility of such an approach. However, it is important to note that the modification, which was intended to make translation dependent on plastid RNA editing, did not abolish nuclear activity in this instance. Therefore, in an applied setting, the gene in question should first be tested to ensure that the modification, with incorporation of plastid RNA editing sites, provides a sufficient reduction in nuclear expression.
The partial activity of the modified neo gene in the nucleus, despite the intended reliance on plastid RNA editing, may also be relevant in an evolutionary context. It would generally be assumed that a requirement for RNA editing would create a barrier to functional organelle-to-nucleus gene transfer. As well as the usual requirements for obtaining nuclear regulatory sequences, functional transfer of such a gene would presumably also require mutation of the editing site(s) so that editing is no longer required. The results presented here indicate that a gene which is reliant on plastid RNA editing may be partially active in the nucleus without mutation occurring. Therefore, the presence of RNA editing sites may not be a significant barrier to functional gene transfer.
Materials and methods
Generation of pPRV111A::neoSTLS2ACG
Firstly, pPRV111A::neoSTLS2 was modified in two ways: the G in the neo start codon was changed to a C to introduce an AvrII restriction site and 4 bases (ATCT) were introduced downstream of the neo stop codon to introduce a BglII site. In order to achieve this, neo was amplified from pPRV111A::neoSTLS2 using the primers neoF + Avr2 and neoR + Bgl2 which contained the intended modifications. An A-tailing reaction was performed with this PCR product which was then cloned into pGemT Easy (Promega) according to the manufacturer’s instructions to generate pGemTEasy + neo. The 1 kb fragment resulting from fully digesting pGemTEasy + neo with XhoI followed by partial digestion with PpuMI was purified and cloned into pPRV111A::neoSTLS2 which had been digested with XhoI and PpuMI to generate pPRV111A::neoSTLS2 + RS.
A 94 bp region of ndhB was amplified from tobacco genomic DNA using the primers ndhBF + Bgl2 and ndhBR + Bgl2, digested with BglII and cloned into pPRV111A::neoSTLS2 + RS which had been digested with BglII to generate pPRV111A::neoSTLS2 + ndhB.
A 101 bp region of psbF and psbL was amplified from tobacco genomic DNA using the primers psbF + Xho1 and psbR + Nhe1, digested with XhoI and NheI and cloned into pPRV111A::neoSTLS2 + ndhB which had been digested with XhoI and AvrII to generate pPRV111A::neoSTLS2ACG.
Plastid transformation
Transplastomic plants were isolated by selecting for spectinomycin and streptomycin-resistant shoots following particle bombardment of wildtype N. tabacum cv W38 leaves as described (Iamtham and Day 2000).
Plant growth conditions and kanamycin selection
Nicotiana tabacum plants grown in soil were kept in a controlled environment chamber with a 14 h light/10 h dark and 25°C day/18°C night growth regime.
Screening of wt ♀ x tpneoACG ♂ progeny was performed by plating surface-sterilised seeds on 150 mm plates containing 80 mL of 0.5 × MS salt medium (Murashige and Skoog 1962) containing 150 μg mL−1 kanamycin at a density of approximately 2,000 seeds per plate. After around 2–4 weeks any plants that appeared resistant were transferred to 0.5 × MS salt medium without kanamycin. Surviving plants were later transferred to soil.
For protein assays, progeny from self-fertilised seed capsules of the original hemizygous kr5.X parent plants were grown on 0.5 × MS salt medium containing 150 μg mL−1 kanamycin. For kr2.2, plants hemizygous for the neo gene were grown on the same medium. In both cases, kanamycin resistant seedlings were transferred to soil for further growth.
DNA preparation
DNA extraction from tobacco leaf tissue was performed using a DNeasy Plant Mini kit (Qiagen) according to the manufacturer’s instructions.
Protein assays
An NPTII ELISA kit (Agdia, IN, USA) was used to determine NPTII protein levels according to the manufacturer’s instructions.
An NPTII dot blot assay was used to detect NPTII activity. This assay was adapted from Curtis et al. (1995), with the following adjustments: blocking solution, 10 mM adenosine 5′-triphosphate (ATP) sodium salt (Sigma, St. Louis, MO), 50 mM tetrasodium pyrophosphate; extraction buffer, 100 mM Tris pH 6.8, 10% glycerol, 5% mercaptoethanol; reactions contained 20 μL protein extract (3 μg total protein) and 20 μL reaction mix (100 mM Tris pH 7.5, 50 mM MgCl2, 400 mM NH4Cl, 10 mM KF, 20 mM DTT, 0.5 mg mL−1 neomycin and 20 μCi [32P]-ATP). A primary wash (1% SDS, 50 μg mL−1 proteinase K) for 20 min at 60°C was included. The kr2.2 sample comprised 1.25% kr2.2 protein and 98.75% wt protein. For quantitative analysis kr2.2 protein was diluted with wt protein to generate a standard curve. Detection and quantification of phosphorylated neomycin was performed using a Typhoon Trio imaging system and ImageQuant TL software (GE Healthcare, Buckinghamshire, UK). Means are shown as a percentage of the neomycin phosphorylation seen in line kr2.2 and are an average of three technical replicates ±95% CI. Statistical analysis used Microsoft Excel and Prism 5 (GraphPad Software, Inc, CA, USA).
For both protein assays, each sample comprised protein extracted from a minimum of three plants.
RNA preparation and cDNA synthesis
RNA extraction from tobacco leaf tissue was performed using an RNeasy Plant Mini kit (Qiagen) and genomic DNA contamination removed either using a TURBO DNA-free kit (Ambion) or by treatment with bovine DNase I (Roche) followed by cleanup using an RNeasy MinElute Cleanup Kit (Qiagen).
For regular cDNA synthesis, reverse transcription was performed using an Advantage RT-for-PCR kit (Clontech) with 20 pMol primer (random hexamer or neo-specific) per reaction. For cDNA synthesis using a thermostable reverse transcriptase, reverse transcription was performed using a ThermoScript RT–PCR System (Invitrogen) with reactions performed at 65°C. All kits were used in accordance with the manufacturers’ instructions. The neo primer used for analysis of the psbL editing site was TneoR1 and the neo primer used for analysis of the ndhB editing sites was neoR2(AL).
PCR and sequencing
PCR of pPRV111A::neoSTLS2 using the primers neoF + Avr2 and neoR + Bgl2 was performed with PfuTurbo DNA polymerase (Stratagene) according to the manufacturer’s instructions. All other PCRs were performed using Taq DNA polymerase (New England Biolabs or Roche) according to the manufacturers’ instructions.
For sequencing of tpneoACG editing sites, the primers used to amplify the entire neo gene were psbF + Xho1 and neoR2(AL). The primers used to amplify the psbL editing site on its own were psbF + Xho1 and TneoR1 and the primers used to amplify the ndhB editing sites on their own were NPTF2 and neoR2(AL). In all cases psbF + Xho1 was used for sequencing the psbL editing site and neoF3 was used for sequencing the ndhB editing sites.
Prior to sequencing, PCR products were purified using either a PCR Purification Kit (Qiagen) or a Gel Extraction Kit (Qiagen) according to the manufacturer’s instructions.
Sequencing was performed using BigDye Terminator v3.1 (Applied Biosystems). Each 20 μL reaction contained 1 μL Ready Reaction Mix, 3 μL Sequencing Buffer, 5 pMol primer and template DNA. Thermal cycling was performed with an initial denaturation step at 96°C for 2 min followed by 26 cycles of 96°C for 30 s, 50°C for 15 s and 60°C for 4 min. Extension products were purified by isopropanol precipitation and analysed using a 3730 DNA Analyzer (Applied Biosystems).
Primer sequences are given in Table 3.
RNA structural analysis
Secondary structure was determined for full length transcripts and a 593 nt section of each transcript comprising a 93 nt region of ndhB containing the two editing sites and the flanking 250 nt either side of this region. Structural analysis was performed using the Mfold web server V 3.5 (Zuker 2003) with default settings.
References
Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29:380–395
Adams KL, Daley DO, Qiu YL, Whelan J, Palmer JD (2000) Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408:354–357
Bock R (2000) Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie 82:549–557
Bock R, Hermann M, Kossel H (1996) In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J 15:5052–5059
Chaudhuri S, Carrer H, Maliga P (1995) Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J 14:2951–2957
Cornelissen M, Vandewiele M (1989) Nuclear transcriptional activity of the tobacco plastid psbA promoter. Nucleic Acids Res 17:19–29
Curtis IS, Power JB, Davey MR (1995) NPTII assays for measuring gene expression and enzyme activity in transgenic plants. In: Jones H (ed) Methods in molecular biology; plant gene transfer and expression protocols. Humana Press Inc, Clifton, pp 149–159
Deigan KE, Li TW, Mathews DH, Weeks KM (2009) Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA 106:97–102
Depeiges A, Degroote F, Espagnol MC, Picard G (2006) Translation initiation by non-AUG codons in Arabidopsis thaliana transgenic plants. Plant Cell Rep 25:55–61
Deusch O, Landan G, Roettger M, Gruenheit N, Kowallik KV, Allen JF, Martin W, Dagan T (2008) Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol Biol Evol 25:748–761
Grohmann L, Brennicke A, Schuster W (1992) The mitochondrial gene encoding ribosomal protein S12 has been translocated to the nuclear genome in Oenothera. Nucleic Acids Res 20:5641–5646
Hazkani-Covo E, Zeller RM, Martin W (2010) Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet 6. doi:10.1371/journal.pgen.1000834
Huang CY, Ayliffe MA, Timmis JN (2003) Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 422:72–76
Huang CY, Ayliffe MA, Timmis JN (2004) Simple and complex nuclear loci created by newly transferred chloroplast DNA in tobacco. Proc Natl Acad Sci USA 101:9710–9715
Iamtham S, Day A (2000) Removal of antibiotic resistance genes from transgenic tobacco plastids. Nat Biotechnol 18:1172–1176
Keeling PJ, Palmer JD (2008) Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9:605–618
Kobayashi Y, Dokiya Y, Kumazawa Y, Sugita M (2002) Non-AUG translation initiation of mRNA encoding plastid-targeted phage-type RNA polymerase in Nicotiana sylvestris. Biochem Biophys Res Commun 299:57–61
Lloyd AH, Timmis JN (2011) The origin and characterization of new nuclear genes originating from a cytoplasmic organellar genome. Mol Biol Evol. doi:10.1093/molbev/msr021
Lutz KA, Bosacchi MH, Maliga P (2006) Plastid marker-gene excision by transiently expressed CRE recombinase. Plant J 45:447–456
Maliga P (2002) Engineering the plastid genome of higher plants. Curr Opin Plant Biol 5:164–172
Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99:12246–12251
Matsuo M, Ito Y, Yamauchi R, Obokata J (2005) The rice nuclear genome continuously integrates, shuffles, and eliminates the chloroplast genome to cause chloroplast-nuclear DNA flux. Plant Cell 17:665–675
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Noutsos C, Richly E, Leister D (2005) Generation and evolutionary fate of insertions of organelle DNA in the nuclear genomes of flowering plants. Genome Res 15:616–628
Noutsos C, Kleine T, Armbruster U, DalCorso G, Leister D (2007) Nuclear insertions of organellar DNA can create novel patches of functional exon sequences. Trends Genet 23:597–601
Nugent JM, Palmer JD (1991) RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell 66:473–481
Richly E, Leister D (2004a) NUMTs in sequenced eukaryotic genomes. Mol Biol Evol 21:1081–1084
Richly E, Leister D (2004b) NUPTs in sequenced eukaryotes and their genomic organization in relation to NUMTs. Mol Biol Evol 21:1972–1980
Ruf S, Karcher D, Bock R (2007) Determining the transgene containment level provided by chloroplast transformation. Proc Natl Acad Sci USA 104:6998–7002
Shafer KS, Hanekamp T, White KH, Thorsness PE (1999) Mechanisms of mitochondrial DNA escape to the nucleus in the yeast Saccharomyces cerevisiae. Curr Genet 36:183–194
Sheppard AE, Ayliffe MA, Blatch L, Day A, Delaney SK, Khairul-Fahmy N, Li Y, Madesis P, Pryor AJ, Timmis JN (2008) Transfer of plastid DNA to the nucleus is elevated during male gametogenesis in tobacco. Plant Physiol 148:328–336
Stegemann S, Bock R (2006) Experimental reconstruction of functional gene transfer from the tobacco plastid genome to the nucleus. Plant Cell 18:2869–2878
Stegemann S, Hartmann S, Ruf S, Bock R (2003) High-frequency gene transfer from the chloroplast genome to the nucleus. Proc Natl Acad Sci USA 100:8828–8833
Stupar RM, Lilly JW, Town CD, Cheng Z, Kaul S, Buell CR, Jiang J (2001) Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: implication of potential sequencing errors caused by large-unit repeats. Proc Natl Acad Sci USA 98:5099–5103
Svab Z, Maliga P (2007) Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc Natl Acad Sci USA 104:7003–7008
Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5:123–135
Tsang HH, Wiese KC (2010) SARNA-predict: accuracy improvement of RNA secondary Structure prediction using permutation-based simulated annealing. IEEE/ACM Trans Comput Biol Bioinf 7:727–740
Wamboldt Y, Mohammed S, Elowsky C, Wittgren C, de Paula WBM, Mackenzie SA (2009) Participation of leaky ribosome scanning in protein dual targeting by alternative translation initiation in higher plants. Plant Cell 21:157–167
Woischnik M, Moraes CT (2002) Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res 12:885–893
Yu YS, Rambo T, Currie J, Saski C, Kim HR, Collura K, Thompson S, Simmons J, Yang TJ, Nah G et al (2003) In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300:1566–1569
Zoubenko OV, Allison LA, Svab Z, Maliga P (1994) Efficient targeting of foreign genes into the tobacco plastid genome. Nucleic Acids Res 22:3819–3824
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgments
We thank Yuan Li for technical assistance. This research was supported under the Australian Research Council’s Discovery Projects funding scheme (project numbers DP0557496 and DP0667006). AD was supported by the BBSRC (UK).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sheppard, A.E., Madesis, P., Lloyd, A.H. et al. Introducing an RNA editing requirement into a plastid-localised transgene reduces but does not eliminate functional gene transfer to the nucleus. Plant Mol Biol 76, 299–309 (2011). https://doi.org/10.1007/s11103-011-9764-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-011-9764-2