Abstract
“zebra” mutants have alternating green and chlorotic crossbands on leaf blades and are widely distributed in monocotyledonous crops. Most recently, we cloned the first responsible gene from rice, ZEBRA2, which also leads to the phenotype of rice preharvest sprouting. ZEBRA2, a single-copy gene in the rice genome, encodes a carotenoid isomerase (CRTISO), the key enzyme catalyzing the conversion of cis-lycopene to all-trans lycopene. ZEBRA2 shares high identity with known CRTISOs from other species. Expression analysis via both RT-PCR and ZEBRA2-promoter-β-glucuronidase (GUS) transgenic rice indicates that ZEBRA2 is predominantly expressed in mesophyll cells of mature leaves where active photosynthesis occurs. Consistent with the alteration in agronomic traits, the zebra2 mutant exhibits decreased photosynthetic rate and chlorophyll content. Mutation of the ZEBRA2 gene results in the accumulation of all-trans-lycopene precursor, prolycopene (7Z,9Z,7′Z,9′Z tetra cis-lycopene), in dark-grown zebra2 tissues. Light-grown zebra2 mutant exhibits the characteristic “zebra” phenotype and decreased level of lutein, the xanthophyll that is essential for efficient chl triplet quenching. More severe phenotype of the zebra2 mutant under high light intensity indicates that “zebra” phenotype might be caused by photooxidative damages. We conclude that ZEBRA2 is involved in photoprotection in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carotenoids fulfill an extraordinary variety of functions in plants (Cunningham and Gantt 1998). Besides the roles as membrane stabilizers in chloroplasts and light-harvesting pigments in photosystems, one important role of carotenoids is the protection of the photosynthetic apparatus from damage of reactive oxygen species (ROS) (Bartley and Scolnik 1995; Frank and Cogdell 1996; Havaux 1998; Niyogi 1999). Some carotenoids are precursors for the synthesis of abscisic acid (ABA), an important plant hormone regulating seed maturation, germination, and also adaptation to environmental stresses (Giraudat et al. 1994). As a result, blocking of the carotenoid biosynthesis pathway may result in ABA deficiency, leading to seed-dormancy failure and vivipary or preharvest sprouting (PHS) phenotypes (Fong et al. 1983; Groot and Karssen 1992; Leon-Kloosterziel et al. 1996).
In the carotenoid biosynthesis pathway, carotenoid isomerase (CRTISO) is the key enzyme that converts cis-lycopene to all-trans lycopene via cis-configuration intermediates (Breitenbach et al. 2001; Masamoto et al. 2001; Isaacson et al. 2002; Park et al. 2002). Interestingly, the CRTISO activity can partially be substituted by light in green tissues, i.e., photoisomerization (Isaacson et al. 2002; Park et al. 2002). Loss of CRTISO causes partial inhibition of lutein biosynthesis in light-grown tissues and appearance of yellow color in newly developed leaves in tomatoes. Also, delayed chlorophyll (chl) accumulation during photomorphogenesis (in an Arabidopsis CRTISO mutant ccr2) due to the lack of prolamellar body (PLB) has been observed, indicating its role in plastid development and photomorphogenesis (Isaacson et al. 2002; Park et al. 2002; Masamoto et al. 2004). Both crtiso mutants exhibit an orange color either in fruit (tomato) or in dark-grown tissue (Arabidopsis) due to the accumulation of the all-trans-lycopene precursor, prolycopene (7Z, 9Z, 7′Z, 9′Z tetra cis-lycopene)(Isaacson et al. 2002; Park et al. 2002). In addition to CRTISO, another isomerase in the carotenoid biosynthesis pathway Z-ISO has been identified in maize and Arabidopsis, which catalyzes the step upstream of CRTISO (Janick-Buckner et al. 2001; Li, et al. 2007, Chen et al. 2010). Mutation in Z-ISO resulted in conditional accumulation of 9, 15, 9′-tri-cis-ζ-carotene (the product of phytoene desaturase, PDS) in dark-grown tissues (Li, et al. 2007; Chen et al. 2010).
“zebra” mutants named by the striking feature of alternating green and yellow (or white) crossbands on leaf blades have been described in a number of monocotyledonous crops such as rice, maize, sorghum, and pearl millet (Coe et al. 1987; Werner and Burton 1991; Oki et al. 1997; He et al. 2000; Kusumi et al. 2000). The appearance of “zebra” phenotype depends to a great extent on environmental factors such as temperature and light, as well as developmental stages (He et al. 2000; Kusumi et al. 2000). The rice “zebra” mutant 1103s was shown to be thermo-sensitive and displayed the “zebra” phenotype only after induction by a cold shock in a ‘high (28°C)–low (20–23.1°C)–high(26°C or higher)’ temperature cycle (He et al. 2000), while another rice “zebra” mutant was dependent on both alternative light/dark growth condition and high light intensity (Kusumi et al. 2000), suggesting that this phenotype is regulated by both photoperiod and light intensity. Moreover, some “zebra” mutants from maize, rice, and pearl millet developed “zebra” phenotype leaves only within early developmental stages followed by a gradually disappearing of this phenotype (Werner and Burton 1991; He et al. 2000; Kusumi et al. 2000). In rice, fourteen non-allelic “zebra” mutants (zebra1-14) were identified, and the corresponding genes had been primarily mapped on different chromosomes (http://www.gramene.org/db/genes/search_gene?query=*zebra*&search_field=&species=&gene_type_id=&has_phenotype=&query_submit=Search&order_by=&page_no=1). Although some of these “zebra” mutants have long been used to determine genetic linkages in conventional breeding, the molecular mechanism underlying the phenotype is still enigmatic.
Recently, we reported map-based cloning of the rice PHS3 gene from our extensive screening of preharvest sprouting mutants (Fang et al. 2008). Interestingly, the ZEBRA2 was also mapped to the same region and was further confirmed to be PHS3 by genetic crossing (data not shown). The zebra2-1 mutant, previously described as phs3-1, showed both the PHS and “zebra” phenotypes. In this paper, we provide the detailed data on characterization of ZEBRA2, as well as its biologic role in photoprotection.
Materials and methods
Plant materials
All the rice (Oryza sativa L. var. Nipponbare) plants used in this study were grown in the field unless specified otherwise. For carotenoid analysis, the zebra2-1 mutant, complemented and RNAi transgenic seedlings, and the corresponding wild-type controls were grown in a growth chamber (either under a 16-h light/8-h dark or under a 24-h dark regime), and shoots of ten-day-old seedlings were harvested. Carotenoids were also extracted from inner parts of mature stems at mature stage and leaves from shaded (approximately 40% of the light intensity of field condition) and non-shaded plants at tillering stage. For chlorophyll analysis, leaves were harvested at tillering stage. For detection of ZEBRA2 expression in different tissues, total RNA was extracted from shoots and roots of two-week-old seedlings, as well as flag leaves, stems and young panicles of plants at heading stage under natural long-day conditions. For histochemical analysis, flag leaves, roots, internodes, nodes, and flowers (at flowering stage) and immature seeds (at filling stage) were harvested from six ZEBRA2::GUS transgenic rice lines.
Pigments analysis and measurement of photosynthetic rate
Carotenoid analysis was carried out according to the reference (Fraser et al. 2000). Measurements were carried out on a HPLC 10AVP system (Shimadzu, http://www.shimadzu.com). A reverse-phase C30 column (250 × 4.6 mm, 5 μm) coupled to a 20 × 4.6 mm C30 guard column (YMC Inc., Wilmington, NC, USA) with the corresponding mobile phases and time programs were used in subsequent HPLC analysis (Fraser et al. 2000). Carotenoids were identified by their characteristic absorption spectra and typical retention time, by comparison with authentic standards and by referring to literatures (Isaacson et al. 2002; Park et al. 2002). Chl was extracted with 80% acetone and determined as previously described (Arnon 1949). Two independent HPLC runs were carried out. Also, relative abundance of each carotenoid component in the zebra2-1 mutant (or complemented lines) compared to that in the wild type was obtained by showing the ratio of each peak area (between the zebra2-1 mutant (or complemented lines) and the wild type) derived from HPLC chromatograms recorded at 430 or 460 nm.
Photosynthetic gas exchange measurements were performed, when the plants grown in the field reached tillering stage, by using a LI-6400 portable photosynthesis system (Li-cor, Lincoln, NE, USA). The parameters were controlled as following: CO2 concentration 400 μmol s−1, air flow 500 μmol s−1, 28°C and field humidity. A range of light intensities between 0 and 2,500 μmol photons m−2 s−1 were supplied from a LED source attached to the leaf chamber. Five measurements were performed.
Agronomic trait analysis
Agronomic traits (including plant height, seed setting rate and thousand-grain weight) of both zebra2-1 mutant and wild type were analyzed at mature stage with 15–20 replicates.
RNA extraction and RT-PCR
Extraction of total RNA (Chomczynski and Sacchi 1987) and synthesis of the first-strand cDNA (Luo et al. 2006) were performed according to previously described methods, respectively. To analyze the gene expression profile for chl, chloroplast biosynthetic and carotenogenic genes, total RNA was extracted from 2-week-old seedlings and tillering stage leaves of wild-type and zebra2-1 mutant plants, respectively. For RT-PCR analysis, 4 μg of total RNA was digested with DNase I and reverse-transcribed by using the M-MLV Reverse Transcriptase (Promega, USA). Product of the first-strand cDNA synthesis reaction was then used as a template for amplification in a PCR. For semiquantitative PCR, the PCR was run for 28 to 30 cycles at 94°C for 30 s, 55 to 58°C for 30 s and 72°C for 1 min, with an additional extension at 72°C for 8 min. ACTIN1 gene was used as an internal control for all RT-PCR analysis. PCR products were separated by electrophoresis on 1.2–3.0% (W/V) agarose gel. The quantitative RT-PCR was performed with Bio-Rad CFX96 Real-time System following the protocol as described previously (Tong et al. 2009). Primers used in the PCR were listed in Table S1. Experiments were repeated three times to obtain representative results.
Vector constructions and plant transformation
To create the ZEBRA2::GUS construct, the 1,571-bp promoter region of ZEBRA2 was fused to the GUS coding sequence in CAMBIA1301Z (CAMBIA). For the ZEBRA2 RNAi construct, a 417-bp fragment from 465 to 881 bp of the ZEBRA2 ORF was inserted as a SalI/XbaI fragment in sense orientation downstream of the potato (Solanum tuberosum L.) GA20 oxidase intron into pUC-RNAi (Luo et al. 2006). The same fragment was inserted in antisense orientation into the SpeI/XhoI sites of pUC-RNAi already carrying the sense fragment. Subsequently, the fragment comprising sense and antisense fragments of ZEBRA2 interspersed by potato GA20 oxidase intron was excised from pUC-RNAi using the flanking PstI and inserted into a pXQAct plasmid between rice ACTIN1 promoter and Ocs terminator, yielding the binary construct. Construction of complementation and plant transformations for all the constructs were essentially performed as previously described (Liu et al. 2007). The primers used for generating the ZEBRA2::GUS and RNAi constructs are listed in Table S1.
Histochemical analysis
Histochemical assay for GUS activity in transgenic plants was performed as described (Jefferson 1989). Various organs of ZEBRA2::GUS transgenic rice were incubated in a solution (50 mM sodium phosphate buffer, pH 7.0, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 0.1% Triton X-100 and 1 mM X-Gluc), directly or after hand-cutting, at 37°C for 12 h followed by incubation in gradient concentrations of ethanol to remove chl.
Phylogenetic analysis
A neighbor-joining tree was built using MEGA version 3.1 (Kumar et al. 2004), adopting Poisson correction distance, and was presented using traditional rectangular TreeView. Support for the tree obtained was assessed using the bootstrap method with 1,000 replicates.
Results
Phenotypes of the zebra2-1 mutant
Four zebra2 alleles were originated from different mutations, and zebra2-1, previously described as phs3-1, which showed a PHS phenotype, was selected for further studies (Fang et al. 2008). Under field conditions, the zebra2-1 mutant began to exhibit alternating transverse green and yellow sectors on leaves at early tillering stage (Fig. 1A). At late tillering stage, the yellow sectors expanded and turned white, and the newly grown leaves were completely yellow (Fig. 1B, C). In addition, the stem of zebra2-1 mutant, especially the part around basal nodes at maturation (Fig. 1D), callus derived from zebra2-1 embryos (Fig. 1E) and the dark-grown zebra2-1 seedlings showed an orange color (Fig. 1F). Interestingly, the inner part of the stem from the zebra2-1 mutant also showed an orange color; however, the surface of stem from zebra2-1 appeared as normally pale green as seen with the wild type (Fig. 1G). The zebra2-1mutant also showed altered agronomic traits, including plant height, seed setting rate, and thousand-grain weight (Fig. 2).
Morphological characteristics of the zebra2-1 mutant. (A) Phenotypes at early tillering stage (the wild type is on the left and zebra2-1 mutant right, the same below); (B) Phenotypes at late tillering stage; (C) Magnified leaves from (B); (D) Stems at maturation; (E) Callus derived from embryos; (F) Dark-grown seedlings; (G) Inner parts of stems exposed to sunlight
The zebra2-1 mutant shows decreased photosynthetic rate and reduced chl content
To evaluate whether and to what extent the ZEBRA2 mutation affects photosynthesis, light curves of photosynthetic rates were measured (Fig. 3A). In all light intensities, the photosynthetic rate of yellow leaf sectors of zebra2-1 was approximately 27–42% of the wild-type leaves, and it reached a maximum at a light intensity of 500 μmol photons m−2 s−1, while in the green sectors of zebra2-1 leaves, the photosynthetic rate was higher than that of the yellow sectors, but still significantly lower than that of the wild type.
For better understanding of the decreased photosynthetic rate of zebra2-1 mutant, chl content was measured (Fig. 3B). The total chl content in green and yellow leaf sectors was approximately 75 and 36% of that in wild-type leaves, respectively. However, the chl a/chl b ratio was not altered (Fig. 3C).
ZEBRA2 encodes an CRTISO in rice
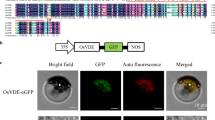
The identity of ZEBRA2 was confirmed by genetic complementation and RNAi experiments. The preharvest sprouting and “zebra” phenotypes were rescued when the genomic ZEBRA2 was introduced, while the RNAi transgenic lines exhibited the “zebra” phenotype (Fang et al. 2008 and Fig. S1). To comprehensively prove that CRTISO gene is indeed attributed to the zebra2-1 phenotype, the expression profiles regarding both size and abundance of CRTISO transcript in the zebra2-1 mutant, complemented and RNAi lines were analyzed by semiquantitative PCR and quantitative real-time PCR, respectively. The CRTISO transcript in zebra2-1 mutants is 24 bp shorter than that of the wild type due to an alteration in the splicing site, while both of the transcripts were observed in the complemented line, indicating the wild-type CRTISO gene has been successfully transformed into the zebra2-1 mutant and well expressed (Fig. S2). The quantitative real-time PCR was conducted to verify the abundance of CRTISO in those lines accordingly (Fig. S3). The abundance of CRTISO transcript in the complemented line is comparable to (or even higher than) that in the wild type, while the transcript in zebra2-1 mutants and RNAi lines is significantly reduced compared to the wild type (Fig. S3). Taken together, we conclude that the “zebra” phenotype can indeed be attributed to the mutation in CRTISO/ZEBRA2 gene.
ZEBRA2 is a single-copy gene in the rice genome and predicted to be 1,761 bp long in cDNA sequence (from ATG to TAG) and composed of 13 exons. This gene structure is consistent with CRTISOs in tomato and Arabidopsis (Isaacson et al. 2002; Park et al. 2002). The ZEBRA2 ORF encodes a 586-amino acid protein with a 49-amino acid transit peptide at N-terminal, which was predicted to localize the protein to chloroplasts (data not shown).
To determine the evolutionary relationship between ZEBRA2 and CRTISO orthologs from other organisms, an unrooted tree was built using neighbor-joining method based on full-length protein sequences (Fig. 4). Phylogenetic analysis indicated that the CRTISO orthologs were clearly divided into two groups: group I comprised members from higher plants and group II from prokaryotic organisms. The rice ZEBRA2 belongs to group I and shows closer phylogenetic relationship to CRTISOs from grass family including maize and sorghum than to other monocotyledonous species, such as Oncidium cv. Gower Ramsey, or dicotyledonous species, indicating its evolutionary process. ZEBRA2 also shares considerable identity (56–59%) with bacterial carotenoid desaturase and FAD-dependent oxidoreductase, suggesting its involvement in similar redox reactions.
Phylogenetic analysis of ZEBRA2. Neighbor-joining tree was built on the full-length protein sequences of rice ZEBRA2/CRTISO and other representative CRTISO orthologs from diverse organisms. The scale bar is an indicator of genetic distance based on branch length. Group I includes the rice ZEBRA2/CRTISO and CRTISOs from maize (ACO71189 for ZmCRTISO 1 and NP_001148055 for ZmCRTISO 2, respectively), sorghum (XP_002449729), Arabidopsis (NP_172167), tomato (Q8S4R4), carrot (Daucus carota) (Q2VEX9), and Oncidium (Q52QW3). Group II includes CRTISOs from Prochlorococcus (Prochlorococcus marinus str. NATL2A) (YP_291830), Synechococcus (Synechococcus elongatus PCC 6301) (YP_171014) and Synechocystis (Synechocystis sp. PCC 6803) (NP_442727), Nostoc (Nostoc punctiforme PCC 73102) PDS (ZP_00108188), and Anabaena (Anabaena variabilis ATCC 29413) FAD-dependent oxidoreductase (YP_323615)
Carotenoid biosynthesis was impaired in the zebra2-1 mutant
Since ZEBRA2 is a single-copy gene in the rice genome, and if ZEBRA2 is a functional CRTISO in rice, mutations in ZEBRA2 gene should cause alterations in the carotenoid composition. To test this possibility, HPLC was performed using various tissues, and peaks were identified by their absorbance spectra (Fig. S4). Under light-growing conditions, both the zebra2-1 and wild-type plants showed very similar carotenoid profiles except for decreased lutein and α-carotene levels, and an increased zeaxanthin level in the zebra2-1 mutant (Fig. 5A and Table S2). No statistically significant changes for other carotenoid components were likely due to the compensatory photoisomerization of cis-carotenoid occurring in light (Isaacson et al. 2002; Park et al. 2002). To avoid the interference from photoisomerization, we further investigated the carotenoid composition of zebra2-1 mutant grown in the darkness. In etiolated zebra2-1 seedlings, the major carotenoids that accumulated were prolycopene (7Z, 9Z, 7′Z, 9′Z tetra cis-lycopene), the all-trans-lycopene precursor, and carotenoids in cis-configuration (Fig. 5B). However, we only detected the accumulation of neoxanthin, violaxanthin, and lutein at low levels in both the wild type and complemented etiolated tissues under 430 nm wavelength (Fig. 5B). Those cis-carotenoids are at high level in the zebra2-1 mutant but at very low level in the wild-type plant, resulting in more than 10,000 times of relative abundance in zebra2-1 mutant compared to the wild type (Table S3). These results were consistent with reports from two other CRTISO mutants, ccr2, an Arabidopsis mutant that accumulated prolycopene in etiolated seedlings, and tangerine, a tomato mutant that accumulated prolycopene as the major carotenoid in fruits (Isaacson et al. 2002; Park et al. 2002).
HPLC chromatograms recorded at 430 nm of light-grown (A) and etiolated (B) wild-type, zebra2-1 mutant, and complementation transgenic seedlings (Comp.), as well as (C) stems of the wild-type and zebra2-1 mutant. Peak 1, neoxanthin; peak 2, violaxanthin; peak 3, chl b; peak 4, lutein; peak 5, α-carotene; peak 6, chl a; peak 7, β-carotene isomer 1; peak 8, β-carotene isomer 2; peak 9, ζ-carotene isomer 1; peak 10, ζ-carotene isomer 2; peak 11, prolycopene; peak 12, cis-lycopene isomer 1; peak 13, neurosporene isomer 1; peak 14, cis-lycopene isomer 2; peak 15, neurosporene isomer 2; peak 16, zeaxanthin
The complemented transgenic plant and the wild type had comparable carotenoid compositions both in light-grown leaves and in etiolated seedlings (Fig. 5A, B and Table S2, S3), indicating the wild-type genomic ZEBRA2 can also rescue the zebra2-1 phenotype regarding the carotenoid composition.
Interestingly, carotenoids extracted from inner parts of the zebra2-1 stem were very similar to those from etiolated zebra2-1 seedlings (Fig. 5C and Table S4), which accumulate high levels of prolycopene and its precursors. This suggests that the orange color of these two tissues was caused by high level of carotenoids in cis-configuration.
Expression pattern of ZEBRA2
To determine the expression pattern of ZEBRA2, mRNA level in various tissues from wild-type plants, including young shoots, young roots, stems, young panicles, and mature leaves was detected. ZEBRA2 is expressed in all tissues/organs tested with its highest expression in mature leaves and relatively low expression in the other tissues (Fig. 6).
To get more expression information of ZEBRA2, a binary vector containing the uidA gene (β-glucuronidase, GUS) driven by the ZEBRA2 promoter (ZEBRA2::GUS) was constructed, and 13 independent (ZEBRA2::GUS) transgenic lines were generated by Agrobacterium-mediated transformation. Six independent ZEBRA2::GUS transgenic lines were used to examine GUS expression. Histochemical localization revealed that the GUS expression was exclusively restricted to mesophyll cells in leaves and vascular bundles in roots, respectively (Fig. 7A, B). In immature seeds, GUS expression was found in the seed coat and embryos but not in the endosperms (Fig. 7C). However, GUS activities were observed in all tissues of internodes, nodes, and flowers (Fig. 7D–F).
Expression pattern of stress-responsive and carotenogenic genes in the zebra2-1 mutant
As it has been shown that PSY3 was up-regulated by abiotic stimuli including salt and drought stresses (Welsch et al. 2008), the normal light condition for wild-type rice growing might be a stress condition for the zebra2 mutants due to inefficient photoprotective systems (Fang et al. 2008), PSY3 (encoding phytoene synthase 3) might be up-regulated in the zebra2-1 mutant. Therefore, we analyzed by quantitative real-time PCR to explore this possibility. Also included in this assay were genes ZEBRA2, PSY1, and PSY2 (encoding phytoene synthase 1 and 2, respectively), PDS (encoding phytoene desaturase), ZDS (encoding zeta-carotene desaturase), LycB (encoding lycopene β-cyclase), LycE (encoding lycopene ε-cyclase), and VxDE (encoding violaxanthin de-epoxidase) to check whether the ZEBRA2 mutation affects the expression of other carotenogenic genes (Fig. 8A). The transcripts of upstream genes including PSY1, PSY2, PDS, ZEBRA2, and LycE were reduced in the zebra2-1 mutant compared to those in the wild type. However, none of the transcripts of downstream genes exhibited difference between wild-type and the zebra2-1 mutant. It is not surprising that there is no significant difference of PSY3 expression between the wild-type and the zebra2-1 mutant because PSY3 was up-regulated by increased ABA level upon salt or drought treatment (Welsch et al. 2008) while the ABA level was decreased in the zebra2-1 mutant (Fang et al. 2008).
Expression analyses of some carotenogenic and stress-responsive genes (A) and genes of chl and chloroplast development (B) in leaves of wild-type (WT) and zebra2-1 mutant. ACTIN1 cDNA was used as a control. Reproducible results were obtained from three independent experiments. PSY, phytoene synthase; LycB, lycopene β-cyclase; LycE, lycopene ε-cyclase; PDS, phytoene desaturase; VxDE, violaxanthin de-epoxidase; ZDS, ζ-carotene desaturase; HEMA, gene encoding glutamyl-tRNA reductase; POR, gene encoding NADPH:protochlorophyllide oxidoreductase; YGL1, gene encoding chl synthase; CAO, gene encoding chlorophyllide a oxygenase; NYC1, gene encoding chl b reductase; CAB; gene encoding the Lhcb1 light-harvesting chl a/b-protein of the PS II; RS, gene encoding the small subunit of Rubisco; FZ, gene encoding a plastid division protein FtsZ; SA, gene encoding a sigma factor of a plastid RNA polymerase
Expression of genes for chl metabolism and chloroplast development in the zebra2-1 mutant
The chl metabolism and chloroplast development might be disturbed in the zebra2-1 mutant since it displayed a chlorosis phenotype. We examined the expression of genes for the chl metabolism including HEMA (encoding glutamyl-tRNA reductase), POR (encoding NADPH:protochlorophyllide oxidoreductase), YGL1 (encoding chl synthase), CAO (encoding chlorophyllide a oxygenase), NYC1 (encoding chl b reductase), and those for chloroplast development including CAB (encoding the Lhcb1 light-harvesting chl a/b-protein of PS II), RS (encoding the small subunit of Rubisco), FZ (encoding a plastid division protein FtsZ), and SA (encoding a sigma factor of a plastid RNA polymerase) (Fig. 8B). Except for the expression of HEMA and POR, two upstream pathway genes in chl biosynthesis that were slightly increased, expressions of most genes for chl metabolism and chloroplast development were relatively low in the zebra2-1 mutant compared to the wild type. The disturbance of chl metabolism and chloroplast development in zebra2-1 mutant might account for the decreased chl content and abnormality in chloroplast development (Fig. 3B and Fang et al. 2008).
The “zebra” phenotype was enhanced by high-intensity light
To further confirm that the “zebra” phenotype was caused by photooxidation, the zebra2-1 mutant and wild-type control plants were grown under different light intensities. Under high light intensity (field condition with the maximum light intensity of 3,000 μmol photons m−2 s−1), zebra2-1 exhibited its characteristic phenotype, while the shaded zebra2-1 mutant plants (with 40% of the light intensity of field condition) showed a much less severe phenotype (Fig. 9A), indicating that the “zebra” phenotype was enhanced by high-intensity light. Consistent with the chlorosis pattern under different light intensities, the zebra2-1 plant under non-shaded light condition showed a lower chl and carotenoids level than the shaded one as well as the wild type (Fig. 9B and Table S5). However, the zebra2-1 mutants had distinguished lower lutein levels than the wild type, regardless of the light intensities. In contrast, the zebra2-1 mutant exhibited higher zeaxanthin level than the wild type (Fig. 9B and Table S5).
Phenotypes and carotenoid profiles of wild-type and zebra2-1 mutant plants under different light intensities. (A) phenotypes under field light and shaded conditions (the plants in front were shaded while those behind were not. All the zebra2-1 mutants were grown on the left and wild type on the right, no matter shaded or not shaded); (B) HPLC chromatograms recorded at 460 nm. Peak 1, neoxanthin; peak 2, violaxanthin; peak 3, chl b; peak 4, lutein; peak 5, chl a; peak 6, β-carotene isomer 1; peak 7, β-carotene isomer 2; peak 8, zeaxanthin
Discussion
“zebra” mutants from different plant species were first reported more than seven decades ago (Hayes 1932; Kadam et al. 1940; Coe et al. 1987; Kusumi et al. 2000), but the molecular mechanisms underlying remain unclear. In rice, fourteen non-allelic “zebra” mutants were identified, and the corresponding genes were mapped on different chromosomes. ZEBRA2 has been the first gene cloned responsible for the “zebra” phenotype among grasses. The identity of ZEBRA2 was fully confirmed by transgenic complementation and RNAi experiments, RT-PCR, as well as carotenoid analysis (Fig. 5, Fig. S2, S3 and Table S2, S4).
ZEBRA2 shares high identity with the known CRTISOs, containing the FAD-binding domain and oxidoreductase domain, a feature of other CRTISOs which have been biochemically characterized (Isaacson et al. 2002; Park et al. 2002; Isaacson et al. 2004), indicating the phylogenic conservation in the gene structure. Furthermore, ZEBRA2 exhibited the conserved gene structure as shown in CRTISOs from tomato and Arabidopsis, which conferred a similar phenotype (carotenoids profile) when the homologous gene was mutated. Therefore, we conclude ZEBRA2 is a functional OsCRTISO in rice. Unlike other mutants blocked at early steps of the carotenoid biosynthetic pathway, which show lethal albino phenotype (Hable et al. 1998; Conti et al. 2004; Dong et al. 2007; Qin et al. 2007), the zebra2-1 mutant is variegated and non-lethal. Since CRTISO activity can be partially substituted by light in green tissues (Isaacson et al. 2002; Park et al. 2002), the non-lethal phenotype of the zebra2-1 mutant may be attributed to the compensatory effects of photoisomerization.
However, CRTISO is not the sole protein responsible for isomerization reactions in the plant carotenoid biosynthesis pathway. A maize gene, Y9, encodes a product essential for the cis-to-trans conversion of the 15-cis-bond in 9,15,9′-tri-cis-ζ-carotene to 9,9′-di-cis-ζ-carotene (Janick-Buckner et al. 2001; Li et al. 2007), the reaction occurring upstream of CRTISO. Both CRTISO and Z-ISO activities can be partially compensated by photoisomerization, which was reflected from the facts of “zebra”/striping phenotype and reduced carotenoids in light-grown tissues from these loss-of-function mutants, respectively (Figs. 5A, B, 9B and Janick-Buckner et al. 2001). The CRTISO was shown to be specific for adjacent cis-double bonds at 7, 9 and 7′, 9′ positions, while Z-ISO catalyzes substrates with single cis-double bond, suggesting their distinct/complementary roles in carotenoid isomerizations (Janick-Buckner et al. 2001; Isaacson et al. 2002,2004; Park et al. 2002; Li, et al. 2007). Interestingly, CRTISO might have evolved from bacterial PDS (CRTI), while Z-ISO might originate from a bacterial gene NnrU that is required for denitrification (Isaacson et al. 2002, 2004; Park et al. 2002; Chen et al. 2010).
The long-standing issue of leaf-variegated mutants, e.g., the zebra2-1 mutant in this study, is the formation of leaves with green and yellow/white sectors. Cloning and functional analysis of genes responsible for leaf variegation in Arabidopsis revealed that it might be caused by a defect in various metabolic pathways related to organelle (mitochondria or chloroplast) functions (Wetzel et al. 1994; Carol et al. 1999; Chen et al. 2000; Takechi et al. 2000; Naested et al. 2004; Rosso et al. 2006). It has been reported repeatedly that an altered carotenoid pool results in loss (or at least partial loss) of photoprotection (Pogson et al. 1998; Niyogi 1999). Lutein is the most abundant carotenoid in photosynthetic apparatus of higher plants and essential for efficient chl triplet quenching (Formaggio et al. 2001; Dall’Osto et al. 2006). However, in agreement with the down-regulation of several carotenogenic genes in the zebra2-1 mutant (Fig. 8A), the zebra2-1 mutant shows a dramatically decreased lutein level in light-grown leaves (Figs. 5A, 9B). Possibly due to the lack of this kind of photoprotective pigment, photooxidative damages to the photosynthetic apparatus occurred in zebra2-1 mutant, and this was also supported by the ROS accumulation, the decreased Fv/Fm and NPQ as well as reduction in the PS II core proteins (Fang et al. 2008). Furthermore, the “zebra” phenotype was enhanced under high light intensity (Fig. 9A) accompanied by decreased chl level (Fig. 9B), indicating that it was caused by photooxidative damages. It was documented that zeaxanthin was accumulated via so-called “xanthophyll cycle” under excess light, and zeaxanthin was proposed to mediate the harmless dissipation of excess energy as heat (Demmig-Adams and Adams 1996). We could also see zeaxanthin accumulation in the zebra2-1 mutant plant under light condition that is normal for the wild-type but might be a stress for the zebra2-1 mutant (Fig. 5A and Table S2), and this accumulation in the zebra2-1 mutant was enhanced when exposed to higher light intensities (Fig. 9B and Table S5). However, the enhanced photooxidative stress under higher light intensity may be not efficiently relieved due to limited protective carotenoid pool in the zebra2-1 mutant. Taken together, we propose that inactivation of ZEBRA2 mainly causes a dramatic change in carotenoids (especially lutein) content, and the decrease in these photoprotective pigments leads to low efficiency in scavenging ROS in the PS II followed by the accumulation of ROS that results in photoinhibition and photobleaching. However, the mechanism how the segregation of green and yellow tissues occurs precisely is still unknown. The production of variegation may be a common phenotype of interorganellar compensation and cytoplasmic sorting processes (Sakamoto 2003), ensuring that some cells (containing normal chloroplasts) can avoid photooxidative damage and form the green sectors while other cells are photooxidized and form the white sectors (containing defective chloroplasts) (Aluru and Rodermel 2004). In monocotyledonous plants, such as rice, the leaf emerges from the leaf sheath and has a linear gradient of cellular and chloroplast development between leaf base and leaf tip; therefore, the “zebra” phenotypes in rice are thought to be formed in response to the occurrence of periodic changes in some environmental factors (light and/or temperature) during a specific stage of leaf cell development (Kusumi et al. 2000). CRTISO may be involved in this series of events, and further study about environmental and developmental factors that influence the expression of CRTISO will help us clarify the “zebra” phenotype.
References
Aluru MR, Rodermel SR (2004) Control of chloroplast redox by the IMMUTANS terminal oxidase. Physiol Plant 120:4–11
Arnon D (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24:15
Bartley GE, Scolnik PA (1995) Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7:1027–1038
Breitenbach J, Vioque A, Sandmann G (2001) Gene sll0033 from Synechocystis 6803 encodes a carotene isomerase involved in the biosynthesis of all-E lycopene. Z Naturforsch 56:915–917
Carol P, Stevenson D, Bisanz C et al (1999) Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11:57–68
Chen M, Choi Y, Voytas DF, Rodermel S (2000) Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J 22:303–313
Chen Y, Li F, Wurtzel ET (2010) Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol 153:66–79
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
Coe EH, Hoisington DA, Neuffer MG (1987) Linkage map of corn (maize). MNL 61:116–123
Conti A, Pancaldi S, Fambrini M, Michelotti V, Bonora A, Salvini M, Pugliesi C (2004) A deficiency at the gene coding for zeta-carotene desaturase characterizes the sunflower non dormant-1 mutant. Plant Cell Physiol 45:445–455
Cunningham FX, Gantt E (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 49:557–583
Dall’Osto L, Lico C, Alric J, Giuliano G, Havaux M, Bassi R (2006) Lutein is needed for efficient chlorophyll triplet quenching in the major LHCII antenna complex of higher plants and effective photoprotection in vivo under strong light. BMC Plant Biol 6:32
Demmig-Adams B, Adams WW III (1996) The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci 1:21–26
Dong H, Deng Y, Mu J et al (2007) The Arabidopsis Spontaneous Cell Death1 gene, encoding a zeta-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signalling. Cell Res 17:458–470
Fang J, Chai C, Qian Q et al (2008) Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J 54:177–189
Fong F, Smith JD, Koehler DE (1983) Early events in maize seed development: 1-methyl-3-phenyl-5-(3-[trifluoromethyl]phenyl)-4-(1H)-pyridinone induction of vivipary. Plant Physiol 73:899–901
Formaggio E, Cinque G, Bassi R (2001) Functional architecture of the major light-harvesting complex from higher plants. J Mol Biol 314:1157–1166
Frank HA, Cogdell RJ (1996) Carotenoids in photosynthesis. Photochem Photobiol 63:257–264
Fraser PD, Pinto ME, Holloway DE, Bramley PM (2000) Technical advance: application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J 24:551–558
Giraudat J, Parcy F, Bertauche N et al (1994) Current advances in abscisic acid action and signalling. Plant Mol Biol 26:1557–1577
Groot S, Karssen C (1992) Dormancy and germination of abscisic acid-deficient tomato seeds: studies with the sitiens mutant. Plant Physiol 99:952–958
Hable WE, Oishi KK, Schumaker KS (1998) Viviparous-5 encodes phytoene desaturase, an enzyme essential for abscisic acid (ABA) accumulation and seed development in maize. Mol Gen Genet 257:167–176
Havaux M (1998) Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci 3:147–151
Hayes HK (1932) Zebra crossbands. J Hered 23:415–419
He R, Ding Y, Yu J, Gao W (2000) The changes of chlorophyll content and several enzyme activities in zebra-leaf rice. J Wuhan Univer (Nat Sci Ed) 46:761–765
Isaacson T, Ronen G, Zamir D, Hirschberg J (2002) Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell 14:333–342
Isaacson T, Ohad I, Beyer P, Hirschberg J (2004) Analysis in vitro of the enzyme CRTISO establishes a poly-cis-carotenoid biosynthesis pathway in plants. Plant Physiol 136:4246–4255
Janick-Buckner D, O’Neal J, Joyce E, Bucker B (2001) Genetic and biochemical analysis of the y9 gene of maize, a carotenoid biosynthetic gene. Maydica 46:41–46
Jefferson RA (1989) The GUS reporter gene system. Nature 342:837–838
Kadam BS, Patel SM, Kalkarni RK (1940) Consequences of in-breeding in bajra. J Hered 31:201–207
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Kusumi K, Komori H, Satoh H, Iba K (2000) Characterization of a zebra mutant of rice with increased susceptibility to light stress. Plant Cell Physiol 41:158–164
Leon-Kloosterziel KM, Gil MA, Ruijs GJ et al (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10:655–661
Li F, Murillo C, Wurtzel E (2007) Maize Y9 encodes a product essential for 15-cis-zeta-carotene isomerization. Plant Physiol 144:1181–1189
Liu XQ, Bai XQ, Wang XJ, Chu CC (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164:969–979
Luo A, Qian Q, Yin H et al (2006) EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol 47:181–191
Masamoto K, Wada H, Kaneko T, Takaichi S (2001) Identification of a gene required for cis-to-trans carotene isomerization in carotenogenesis of the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 42:1398–1402
Masamoto K, Hisatomi S, Sakurai I, Gombos Z, Wada H (2004) Requirement of carotene isomerization for the assembly of photosystem II in Synechocystis sp. PCC 6803. Plant Cell Physiol 45:1325–1329
Naested H, Holm A, Jenkins T et al (2004) Arabidopsis VARIEGATED 3 encodes a chloroplast-targeted, zinc-finger protein required for chloroplast and palisade cell development. J Cell Sci 117:4807–4818
Niyogi KK (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50:333–359
Oki S, Gu X, Kofoid KD, Liang GH (1997) A light-intensity sensitive chlorophyll mutant in sorghum. Hereditas 126:239–245
Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14:321–332
Pogson BJ, Niyogi KK, Bjorkman O, DellaPenna D (1998) Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc Natl Acad Sci USA 95:13324–13329
Qin G, Gu H, Ma L, Peng Y, Deng XW, Chen Z, Qu LJ (2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17:471–482
Rosso D, Ivanov AG, Fu A et al (2006) IMMUTANS does not act as a stress-induced safety valve in the protection of the photosynthetic apparatus of Arabidopsis during steady-state photosynthesis. Plant Physiol 142:574–585
Sakamoto W (2003) Leaf-variegated mutations and their responsible genes in Arabidopsis thaliana. Genes. Genet Syst 78:1–9
Takechi K, Sodmergen Murata M, Motoyoshi F, Sakamoto W (2000) The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol 41:1334–1346
Tong H, Jin Y, Liu W et al (2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58:803–816
Welsch R, Wüst F, Bär C, Al-Babili S, Beyer P (2008) A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol 147:367–380
Werner B, Burton G (1991) Dominant zebra mutant in pearl millet. J Hered 82:68–69
Wetzel CM, Jiang CZ, Meehan LJ, Voytas DF, Rodermel SR (1994) Nuclear-organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J 6:161–175
Acknowledgments
This work was kindly supported by grants from National Natural Science Foundation of China (30825029, 30621001, and 30871508) and Ministry of Science and Technology (2009CB118506) to C. Chu.
Author information
Authors and Affiliations
Corresponding author
Additional information
Chenglin Chai, Jun Fang contribute equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chai, C., Fang, J., Liu, Y. et al. ZEBRA2, encoding a carotenoid isomerase, is involved in photoprotection in rice. Plant Mol Biol 75, 211–221 (2011). https://doi.org/10.1007/s11103-010-9719-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-010-9719-z