Abstract
Many eukaryotic genomes have experienced ancient whole-genome duplication (WGD) followed by massive gene loss. These eliminations were not random since some gene families were preferentially retained as duplicates. The gene balance hypothesis suggests that those genes with dosage reduction can imbalance their interacting partners or complex, resulting in decreased fitness. In Arabidopsis, the cytoplasmic ribosomal proteins (RP) are encoded by gene families with at least two members. We have focused our study on the two RPS6 genes in an attempt to understand why they have been retained as duplicates. We demonstrate that RPS6 function is vital for the plant. We also show that reducing the level of RPS6 accumulation (in the knock-out rps6a or rps6b single mutants, or in the double heterozygous RPS6A/rps6a,RPS6B/rps6b), confers a slow growth phenotype (haplodeficiency). Importantly, we demonstrate that the functions of two RPS6 genes are redundant and interchangeable. Finally, like in most other described Arabidopsis rp mutants, we observed that a reduced RPS6 level slightly alters the dorsoventral leaf patterning. Our results support the idea that the Arabidopsis RPS6 gene duplicates were evolutionarily retained in order to maintain an expression level necessary to sustain the translational demand of the cell, in agreement with the gene balance hypothesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gene duplication is one major source of functional innovation of species. During the evolution of Angiosperms, there have been several whole-genome duplication (WGD) events and many small-scale gene duplications (Bowers et al. 2003). These duplication events are thought to have fueled the explosive evolutionary diversification of angiosperms during the early Cretaceous (De Bodt et al. 2005). The Arabidopsis thaliana genome is thought to result from three rounds of whole-genome polyploidisations, followed by diploidization and massive gene loss (Simillion et al. 2002; Bowers et al. 2003). The fully sequenced genomes from different taxa suggest that such gene losses are apparently biased. Computational analyses reveal that not all functional categories of genes were equally affected. For example, Arabidopsis genes involved in development, in transcriptional regulation, in the proteasome and in signalling cascades have been preferentially retained after duplication (de Bodt et al. 2005). The 80 Arabidopsis cytoplasmic ribosomal proteins (RP) are encoded by small gene families, of two to seven paralogs, representing a total of 249 genes (Barakat et al. 2001). These genes belong to functional classes that are over-represented among those retained as duplicates (Blanc and Wolfe 2004; Seoighe and Gehring 2004; Maere et al. 2005). Interestingly, ribosomal genes are also duplicated in distant species such as the yeast Saccharomyces cerevisiae, in which the WGD has been followed by massive gene loss, and Paramecium tetraurelia. (Aury et al. 2006), pointing out a fundamental aspect of this genomic evolutionarily trend (Conant and Wolfe 2008).
There are three main hypotheses to explain survival of gene duplicates during their evolution: neofunctionalization, subfunctionalization, and the gene balance hypothesis (Freeling 2009). Neofunctionalization is the process whereby duplicated genes acquire divergent functions; subfunctionalization is the process whereby duplicated genes acquire divergent patterns of expression or keep only a subset of the ancestral functions; and the gene balance hypothesis postulates that duplicated genes are retained only because a reduction of their dosage would decrease fitness. This gene balance hypothesis can explain the observed post-WGD over-retention of duplicated genes which encode proteins that interact with partners, either in multi-subunit complexes or in regulatory cascades (Birchler and Veitia 2007, 2009; Freeling 2009; Papp et al. 2003; Veitia 2002).
In higher eukaryotes there are very few reported cases of RP with extra-ribosomal functions (Warner and McIntosh 2009). The best-known case in Arabidopsis is RPL10A, which participates in resistance to geminivirus (Carvalho et al. 2008; Rocha et al. 2008). Also, the comparatively mild sequence conservation in the Arabidopsis RPP2, RPL7 and RPS15A families could be a possible signature of extra-ribosomal function (Barakat et al. 2001). With the exception of these examples, neofunctionalization is not a major driving force for maintaining several copies of Arabidopsis RP genes.
In plants, many paralogous RP genes are distinguished by their specific developmental or conditional expression (McIntosh and Bonham-Smith 2006; Whittle and Krochko 2009). In Brassica napus, transcript profiling of the 996 putative RP genes unveiled a clear tissue-specific expression for many of them (Whittle and Krochko 2009). Eighty percent of the 249 Arabidopsis RP genes are transcribed (Barakat et al. 2001). Differences of expression between individual members of Arabidopsis RP gene families have been observed for the RPL11 (Williams and Sussex 1995) and the RPS5 genes families (Weijers et al. 2001). Therefore, there is good experimental evidence for a substantial subfunctionalization (partitioning of expression) among the RP genes of Arabidopsis and the closely related Brassica napus (Whittle and Krochko 2009).
The role of specific RP paralogs can be assayed by mutant analysis. So far, mutants in only fourteen RP genes have been examined. Most of these mutants display reduced growth and/or abnormal development (Byrne 2009). Some of these (rpl5a, rpl5b, rpl9, rpl10b, rpl24b and rpl28a) were also isolated in enhancer screens of the asymmetric leaves1 and/or asymmetric leaves2 mutations (Degenhardt and Bonham-Smith 2008; Pinon et al. 2008; Yao et al. 2008); some of these mutations phenotypically interact with the angustifolia3 mutation (Fujikura et al. 2009). The loss-of-function mutation in RPS5A is lethal and haplodeficient (Weijers et al. 2001), and the knock-out of RPL4A, RPL4D, RPS18A or RPS13A results in delayed growth and pointed leaves (Ito et al. 2000; Rosado et al. 2010; Van Lijsebettens et al. 1994). The disruption of RPL24B has pleiotropic effects on development, such as the alteration of the gynoecium apical-basal polarity, the leaf shape, and the cotyledon’s vascular pattern (Nishimura et al. 2004, 2005). In some cases, the mutation of a RP gene can confer a relatively specific phenotype. For example, the loss or reduced expression of RPS27A confers hypersensitivity to DNA damages (Revenkova et al. 1999). Also, the knock-out of RPL10A significantly increases susceptibility to geminivirus infection (Carvalho et al. 2008; Rocha et al. 2008), whereas a missense mutation allele in this gene semi-dominantly suppresses the dwarf phenotype of acaulis5 mutant (Imai et al. 2008).
Functional comparison within duplicated pairs of Arabidopsis RP genes have been examined between RPL23aA and RPL23aB (Degenhardt and Bonham-Smith 2008), between RPL5A and RPL5B (Fujikura et al. 2009; Yao et al. 2008) and between RPL4A and RPL4D (Rosado et al. 2010). The RNAi-triggered extinction of RPL23aA expression induces abnormal shoot development whereas the knock-out of RPL23aB does not confer any visible phenotype (Degenhardt and Bonham-Smith 2008). By contrast, mutants in RPL5A and RPL5B, and mutants in RPL4A and RPL4D, share the same phenotypes and appear functionally equivalent although this has not been demonstrated (Fujikura et al. 2009; Yao et al. 2008; Rosado et al. 2010). Based on the literature mentioned above, it appears that paralog-specific RP functions are not exceptions in plants.
Still, whole-genome statistical analyses support and favor the gene balance hypothesis—as in yeast (Papp et al. 2003)—over the subfunctionalization model, to explain the predominance of Arabidopsis RP genes as duplicates (Freeling 2009). Clearly, the analysis of more mutants is necessary to have a better understanding of the biological role of each plant RP duplicates.
In this work we have focused our attention on the two RPS6 genes in Arabidopsis. In mammals, Drosophila and yeast the RPS6 protein is part of an evolutionarily conserved TOR (Target Of Rapamycin)-pathway that controls cell growth and division (Hay and Sonenberg 2004; Meyuhas 2008). Several orthologs of this pathway are present in Arabidopsis (Deprost et al. 2005; Mahfouz et al. 2006; Menand et al. 2002).
Genetic analysis in mice, Drosophila melanogaster and Caenorhabditis elegans have unveiled the complex and sometimes unexpected consequences from reducing the RPS6 level. The development of Drosophila larvae hemizygous for strong loss-of-function rps6 alleles is delayed, accompanied by growth inhibition in most larval organs. However, the imaginal discs and lymph glands proceed to overgrow and in addition display variable melanotic tumor phenotypes; these Minute larvae eventually die (Stewart and Denell 1993; Watson et al. 1992). In mouse liver cells or T-lymphocytes the conditional RPS6 haplodeficiency prevents their high rate of proliferation (Sulic et al. 2005; Volarevic et al. 2000). This RPS6 haplodeficiency also blocks mouse embryonic development (Panić et al. 2006). Reducing ribosomal content is thought to trigger a p53-dependent checkpoint that reduces growth, activates apoptosis and also stimulates the proliferation of melanocytes (Panić et al. 2006; Sulic et al. 2005).
The inactivation of the RPS6 gene in adult Caenorhabditis elegans induces sterility (Maeda et al. 2001), but it also increases lifespan and thermal-stress resistance, as do other genes involved in mRNA translation (Hansen et al. 2007). In yeast, the deletion of RPS6A or RPS6B reduces the cell size and growth rate (Chiocchetti et al. 2007). Additionally, the deletion of RPS6B, but not RPS6A, significantly extends its replicative life span, possibly by altering the aging process through the modulation of translation (Chiocchetti et al. 2007). This is intriguing because RPS6A is more highly expressed than RPS6B, and because RPS6A and RPS6B have identical protein sequences. Thus, in all organisms tested so far the complete lack of RPS6 is lethal. However, the compromised cell growth, proliferation and survival when RPS6 expression is lowered have often more complex origins than just the general slowing down of ribosomal activity. In animal cells, cellular checkpoints and/or shifts in either metabolism or developmental routes explain some of the mutant phenotypes. Additionally, in yeast, there are paralog-specific RPS6 functions (Komili et al. 2007).
In this work, we have taken a genetic approach to assess the roles of the two RPS6 genes on Arabidopsis growth and development. We have also tested whether the two RPS6 genes are functionally equivalent.
Results
Isolation and molecular characterization of the rps6a and rps6b mutants
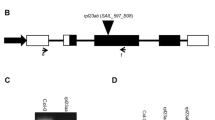
For RPS6A (At4g31700) and RPS6B (At5g10360), we selected homozygous T-DNA insertion mutant lines (Fig. 1a, b). These two lines are hereafter referred to as rps6a and rps6b, respectively. The RPS6B gene was previously annotated emb3010; however, we believe this embryonic lethality is due to a separate gene mutation, since the homozygous rps6b mutant we have isolated is viable (see below).
Molecular characterization of the rps6a and rps6b mutants. a RPS6 genes structure and location of T-DNA insertions. The T-DNA insertions are indicated by triangles above the genes. The star symbolizes the left border of the T-DNA insertion; the grey bar above the genes correspond to the probes used for the Southern blots analysis (b). b Southern blots analysis of the rps6 mutants. The black arrow indicates the wild-type band. c Expression analysis of the RPS6A and RPS6B in WT and rps6 mutants. RNA were extracted from 14-day-old plantlets and analyzed by quantitative real-time RT–PCR. The levels of expression have been normalized to that of the WT. Results are the mean ± standard deviation of technical triplicates of three independent biological replicates. d Northern blots analysis of rps6 mutants. The northerns blots were hybridized with a probe specific for the RPSA (left) or the RPS6B (right) transcripts. The 25S hybridization signal served as a loading control. The two stars indicate two new bands revealed by the RPS6B probe
We quantitatively analyzed RPS6 mRNA accumulation by real-time RT–PCR. Figure 1c shows that in the rps6a mutant no RPS6A mRNA is detected. In the rps6b mutant a signal is detected that likely corresponds to either one or both of the two new transcripts observed in northern blot (Fig. 1d) (note that the WT band has disappeared). These new transcripts may have started inside the T-DNA and it is highly probable that they do not code for a functional RPS6B. We believe therefore that both the rps6a and rps6b mutations are non-functional alleles. Note that for both rps6 mutants the level of mRNA accumulation of the non-mutated paralogue resembles the WT level (Fig. 1c).
RPS6A and RPS6B are fully redundant during formation of gametes
RPS6 is an essential protein for animal growth, development and survival. In plants, only one study has shown that the partial suppression of Arabidopsis RPS6 expression (with a transgenic antisense construct) alters shoot development (Morimoto et al. 2002). However the phenotypes were variable, and it is not clear how strong the suppressive effect was on the two RPS6 genes. To assess the role of RPS6 genes in plant development, we screened for a rps6a,rps6b double mutant.
In the selfing of a double heterozygote RPS6A/rps6a,RPS6B/rps6b, we randomly selected 90 seedlings that we PCR-screened in order to detect rps6a/rps6a and rps6b/rps6b homozygous mutants. We isolated five rps6a/rps6a and twelve rps6b/rps6b mutants (however no double-mutants were isolated). Southern blot analysis of the 17 mutant lines with RPS6A and RPS6B-specific probes (figure S1) confirms that neither rps6a nor rps6b mutants carry a mutant allele of the paralogous gene. This result indicates that the double homozygous recessive mutant is lethal.
In order to unequivocally demonstrate that the rps6a and rps6b mutations are synthetically lethal during gamete formation, we conducted a tetrad analysis by means of the quartet1 (qrt1) genetic background (Preuss et al. 1994). In the anthers of the qrt1/qrt1 mutant the four pollen grains produced by each meiosis are not separated (tetrad). This feature was suggested to allow testing synthetic lethality (Copenhaver et al. 2000).
We introgressed different combinations of rps6a and rps6b alleles in the qrt1/qrt1 background and counted the number of dead and live pollen grains in mature tetrads. We analyzed the pollen of four RPS6A/rps6a,RPS6B/rps6b double heterozygotes, one RPS6A/rps6a and two RPS6B/rps6b simple heterozygous plants in addition to the qrt1/qrt1 parental line (Fig. 2a, b).
Analyse of pollen mortality in tetrads and ovule abortion in siliques. a Histogram of the different classes of pollen tetrads after the Alexander staining. The number of tetrads (n) observed for each plant is indicated above the bars, the genotype of which is indicated on the abscissa. b Scheme and pictures of the three segregating classes of pollen tetrads generated by a (RPS6A/rps6a, RPS6B/rps6b) double heterozygous plant. The RPS6A and RPS6B genes are located at the bottom of chromosome 4 (yellow) and at the top of chromosome 5 (blue), respectively. Neither of these two genes are genetically linked to its centromere. The + and − signs correspond to the WT and mutant alleles, respectively. The Alexander staining allows distinguishing alive (pink) from dead (no staining) pollen grains. PD, parental ditype; TT, tetratype; NPD, non-parental ditype. c Picture of siliques with aborted ovules. Aborted ovules are visible in the siliques of the double heterozygote (arrowheads) but not in the WT (Col0) or in the rps6a and rps6b single mutants. The double heterozygotes n°1 and n°2 are F1 plants from reciprocal crosses between the rps6a and rps6b parental lines
In the tetrads produced by the qrt1/qrt1 control, ~6% of the tetrads contain one dead pollen grain (with very few containing two or more dead grains). This fraction represents the background of pollen mortality. In the tetrads of the RPS6A/rps6a and RPS6B/rps6b simple heterozygous plants, the level of dead pollen grains is similar to the qrt1/qrt1 WT control. This indicates that pollen viability is not compromised when one of the two RPS6 proteins is missing. By contrast, the RPS6A/rps6a,RPS6B/rps6b double heterozygotes produce ~20% (n = 206) of the tetrads with no dead pollen grains (parental ditype, PD), ~60% (n = 586) with one dead grain (tetratype, T) and ~20% (n = 186) with two dead grains (non-parental ditype, NDP) (numbers are from Fig. 2a, b for the different classes of pollen tetrads). As expected for the segregation of two mutations that are located on different chromosomes and that are genetically unlinked to their centromeres (Fig. 2b), the ratio DP/NDP = 1 (χ2 = 1) and T is <66.7% of the tetrads (Fincham et al. 1979). These data demonstrate that meiocytes which inherit the rps6a,rps6b double mutation do not generate viable pollen grains.
Since we did not find any plants that inherited the double mutation from one parent (see above), and that siliques of the double heterozygotes segregate aborted ovules (Fig. 2c), the lethality also occurs in female gametes. As a corollary, these results demonstrate that the RPS6A and RPS6B genes display fully redundant functions during gametogenesis because the rps6a and rps6b single mutants are fully fertile.
RPS6A and RPS6B are limiting effectors of plant growth
Many mutations in ribosomal proteins alter the growth of yeast, Drosophila and Arabidopsis. We have monitored the effects of rps6 mutations on Arabidopsis plants grown in soil or in vitro.
Whether grown in vitro or in soil, both the rps6a and rps6b mutants display a slightly delayed growth (Fig. 3). The leaves of the rps6a and rps6b mutants are smaller than in WT; they are also slightly more elongated and pointed (Fig. 3a and S2). This phenotype has already been observed in mutants for others ribosomal proteins (Degenhardt and Bonham-Smith 2008; Ito et al. 2000; Nishimura et al. 2005; Pinon et al. 2008; van Lijsebettens et al. 1994; Yao et al. 2008). At bolting, the inflorescence of the two homozygous single mutant lines grows slower than the WT, but they eventually reach a WT size.
Leaf and root phenotype of the rps6 mutants. a, b Picture of 10-day-old WT, mutant and (rps6a X rps6b)F1 seedlings. Note that the growth of leaves (a) and roots (b) in the mutants and the double heterozygote is delayed. Arrowheads in a indicate the pointed leaves in mutant seedlings. c Primary root length of 11-day-old seedlings. Error bar represents the standard error of the mean. d Primary root growth speed. Root lengths were measured after 4, 6, 8 and 11 days of in vitro culture. Speed was determined as the ratio between ∆ length and ∆ time (mm/day) of ~30 individuals for each genotype
As for the shoot, the root growth of the rps6a and rps6b mutants is delayed (Fig. 3b, c). The rate of root growth is in accordance to the phenotype: the rps6a mutant grows slower than the rps6b (Fig. 3d). For all growth traits examined so far, the rps6a mutant has a slightly stronger phenotype than that of the rps6b mutant; this is more visible in vitro than in soil. Here we confirm that the mutant phenotype of the rps6a and rps6b lines is rescued by a T-DNA carrying the corresponding genomic wild-type RPS6 allele (Figure S2).
Both the rps6a and rps6b mutations are fully recessive (data not shown). Interestingly, the rps6a and rps6b mutants do not complement each other: the rps6a/RPS6A,rps6b/RPS6B double heterozygote has a similar Rps6 mutant phenotype (Fig. 3a, b, c). This non-allelic non-complementation involving two null alleles (derived from genes coding for almost identical proteins, Figure S3) indicates that the Rps6 mutant phenotype is due to a combined haploinsufficiency. We thus conclude that the RPS6 protein represents a limiting factor for plant growth.
The RPS6 proteins have redundant functions in vegetative tissues
In order to ascertain the functional equivalency of the two RPS6 genes in vegetative tissues, we performed reciprocal complementation tests between the two paralogous genes. In a first experiment, we crossed the rps6a single mutant with an rps6b single mutant complemented with a T-DNA construct carrying the WT RPS6B allele (rps6bC). This complemented line contains probably more than four copies of the transgenic RPS6B allele (see below Fig. 4a). In the F2 progeny of this cross we selected in vitro seedlings with a WT phenotype. In this sub-population we PCR-selected seventeen rps6a homozygous seedlings for southern-blot analyse. We found that all of them carry the RPS6B transgene, and five (n°25, 49, 59, 73 and 86) were also homozygous for the rps6b mutation (Fig. 4a). We believe these rps6a/rps6a,rps6b/rps6b double mutants were complemented most probably because they contain several RPS6B transgenes (Fig. 4a, bottom). This result shows that the RPS6B transgene fully complemented the rps6a as well as the rps6a/rps6a,rps6b/rps6b double mutations.
Reciprocal complementation between the RPS6 genes. a Southern blots analysis of the F2 progeny from the (rps6bC X rps6a) cross. The rps6bC line is an rps6b line complemented with a T-DNA carrying the WT RPS6B gene. The seventeen F2 seedlings have a WT phenotype and were PCR-selected for the rps6a/rps6a mutation. The top southern blot has been hybridized with an RPS6A probe and the bottom southern with the RPS6B probe. The underlined numbers correspond to the rps6a/rps6a, rps6b/rps6b double mutants F2. b Southern blots analysis of the F2 progeny from the (rps6aC X rps6b) cross. The rps6aC line is an rps6a line complemented with a T-DNA carrying the WT RPS6A gene. The sixteen F2 seedlings have a WT phenotype. The top southern blot has been hybridized with an RPS6A probe and the bottom southern with the RPS6B probe. The underlined number corresponds to an F2 rps6a/rps6a, rps6b/rps6b double mutant
In a parallel experiment, we performed the reciprocal test; i.e. we crossed the rps6b single mutant with the rps6a single mutant complemented with a T-DNA construct carrying the WT RPS6A allele, and selected F2 seedlings with a WT phenotype. Southern blot analysis of these F2 plants identified nine rps6a homozygous mutants, and one rps6a/rps6a,rps6/rps6b double mutant (n°74, Fig. 4b). As in the previous experiment, the rps6a/rps6a,rps6/rps6b double mutant was most likely complemented because it contains several RPS6A transgenes (Fig. 4b, top). Since these mutants had a WT phenotype (data not shown), we conclude that they were fully complemented by the RPS6A transgene.
These two reciprocal experiments clearly demonstrate that the biological function of the Arabidopsis RPS6A and RPS6B genes are qualitatively equivalent and interchangeable. Therefore, the Rps6 mutant phenotype of the single mutants and of the double heterozygotes is probably due to reduced dosage of RPS6 genes (i.e. haploinsufficiency).
Reduced root meristem activity in the rps6A and rps6B mutants
In order to understand the origin of the root growth defect we measured cell size in the WT and mutant lines. Compared to the WT, epidermal cells in the differentiated zone of the rps6 mutant root are not significantly shorter (Fig. 5a). We therefore assumed that the rps6 mutations reduce the activity of the primary root meristem.
Origin of the root growth defects. a Root epidermal cells length in the differentiated zone of 8-day-old seedlings. Values are the average of 30 primary roots. b Pictures of wild-type and mutant meristems. The proximal zone (PM) is defined has the zone between the stem cell niche and the transition zone (TZ). The arrowheads indicate the TZ between the PM and the elongation-differentiation zone. In the insets are shown a closer view of the TZ in the cortex. Scale bar: 100 μm. c Root-meristem cell number. The number of cortex cells in the proximal zone were counted on ~30 primary roots for each genotype. Two independent experiments gave similar results
The meristem activity and thus the root growth rate are correlated with the size of the root meristem (Dello Ioio et al. 2007). We therefore measured the size of the meristematic zone of the primary root, defined by the number of cortex cells per file, between the quiescent centre and the elongation zone (Dello Ioio et al. 2007). By using this cellular index we found that both the rps6a and rps6b mutants have half as many cortex cells in their primary root meristem as the WT; the rps6a appears to be more deficient than the rps6b (Fig. 5b, c). These observations clearly indicate that the root meristem of the rps6 mutants is less active than in the WT. Altogether, these results show that the slow root growth in rps6 mutants is due to a reduced meristem activity.
The plant growth delay is correlated to the total amount of RPS6
Our genetic analysis strongly suggests that the RPS6 proteins are limiting factors of growth. We thus quantified the level of RPS6 accumulation in seedlings. Proteins were extracted from seedlings grown 13 days in vitro and analyzed in western blots with a polyclonal antibody raised against a maize RPS6 (Williams et al. 2003). As shown in Fig. 6a, this anti-RPS6maize antibody recognizes both the Arabidopsis RPS6A and RPS6B at the expected size (30kD). In the rps6a mutant the RPS6 signal intensity is only ~15% of the WT level (Fig. 6b). Unexpectedly, in spite of the observation that the rps6 mutation does not allow the production of RPS6B (see above), the RPS6 signal in rps6b seedlings is close to the WT level (Fig. 6b). We have repeatedly observed this western blot pattern in three independent experiments. The simplest hypothesis to explain why the rps6b seedlings have a mutant phenotype without displaying a reduced western blot signal with the anti-RPS6maize antibody is that this antibody has a stronger affinity for RPS6A than for RPS6B. Assuming (from Fig. 6a) that ~85% of the WT signal revealed by this antibody is due to RPS6A and therefore ~15% is due to RPS6B, we expect that in the double heterozygote the total RPS6 signal is around 50% of the WT; this is what we observe (Fig. 6b). This explanation is only valid if there are no mutual compensatory increases in the expression of the remaining WT RPS6 paralogous gene carried by two rps6 simple mutants. Figure 1c confirms that this is the case, at least at the transcriptional level: by comparison to the WT, the level of RPS6B mRNA accumulation is not increased in the rps6a mutant, and vice versa. This indicates that reducing dosages of RPS6 genes results in reduced accumulation of RPS6.
Level of RPS6 accumulation and polysome analysis of rps6 mutants. a RPS6 immunodetection on proteins extracted from 13-day-old wild-type and rps6 mutants (top). As a loading control, we used a doublet of bands detected on a gel identically loaded and stained with Coomassie Blue (Bottom). b Level of RPS6 accumulation. The level of immunoreactive RPS6 was normalized and calculated by comparison to the RPS6 level in the WT. c Polysomes/monosomes profile from WT and rps6 plants. Extracts from 10-day-old seedlings were fractionated on sucrose gradients and absorbance at 260 nm was recorded along the gradients. The polysomes are at the bottom of the gradient whereas 80S monosomes and 60S and 40S ribosomal subunits are at the top. The arrowhead indicates the position of the 40S peak in the WT profile
Altogether, our results suggest a simple dosage-sensitive effector model which postulates that when there is 1 gene dose (i.e. in the WT) or 0.75 gene dose (i.e. in the simple heterozygotes) of total RPS6 the plant growth is normal but when there is only 0.5 gene dose (i.e. in simple mutants and in the double heterozygotes) the growth is delayed. In the gametophytic cells a quarter of a gene dose is enough for normal viability whereas the complete absence of RPS6 is lethal.
The RPS6A and RPS6B genes are expressed in the same tissues of the root tip
Our reciprocal complementation tests (see above) indicate that the two paralogous RPS6 genes are qualitatively equivalent. A corollary of this result is that they should be expressed in the same tissues. In order to test this idea, we took the root tip as a model system and we performed an in situ hybridization analysis. As shown in figure S4, both RPS6A and RPS6B transcripts are detected in the same tissues, in particular in the metabolically active cells of the meristem and in the elongating zone. This result is corroborated by the expression analysis displayed in the Genevestigator transcriptomic database (Zimmermann et al. 2004). The expression data from these DNA arrays also indicate that the RPS6A and RPS6B genes are expressed in all the other organs of the plant. Together, these expression data support the result from our reciprocal complementation tests, which reveals that RPS6A and RPS6B are functionally equivalent.
Analysis of rps6 polysomes
One highly likely explanation for the reduced growth of the Arabidopsis rps6 mutants is the lower general translation efficiency resulting from a reduced level of the functional 40S ribosome subunit. To test this idea, we analyzed the profiles of both polysomes and monosomes in mutant and WT lines. Cell extracts from 10-day-old seedlings (i.e. rapid growth phase) were separated on a sucrose gradient and detected at 260 nm. As shown in Fig. 6c, the overall profiles of the two rps6 mutants are roughly similar to that of the WT, with the exception that in the mutants there is a lack of free 40S subunits and a slight accumulation of free 60S subunits compared to WT. The 60S/80S ratio is 44.9 10−3 ± 22.10−3 for the WT, 102.6 10−3 ± 16.3 10−3 for the rps6a and 84.4 10−3 ± 33.10−3 for the rps6b.
These results show that the inactivation of one RPS6 paralogous gene slightly alters the stoichiometry of the free ribosomal subunits, a presumed consequence of the reduced 40S assembly or stability. However, the higher 60S/80S ratio in the mutants suggests a diminution of 80S assembly and/or more freely available 60S due to reduced level of the 40S subunits.
Discussion
The two RPS6 paralogs are functionally indistinguishable
In Arabidopsis, genes coding for basic cellular machinery, such as the ribosome, have survived in duplicate more often than would be predicted by chance (Blanc and Wolfe 2004), with most Arabidopsis RP encoded by three or four expressed genes (Barakat et al. 2001). Blanc and Wolfe (2004) proposed that the expression of several copies is necessary to achieve the cellular demand for RP. This idea has been further refined and the current explanation for the maintaining of duplicated RP genes is that ribosomal proteins are part of a multiprotein complex, sensitive to stoichiometric imbalance: the precise stoichiometry of all ribosomal proteins must been maintained to preserve ribosome activity (Veitia 2002). Our results indicate that this gene balance could be the evolutionary force that maintains two RPS6 genes in Arabidopsis. First, the rps6a and rps6b single mutants and the double heterozygote share the same, if not identical, mutant phenotype (reduced growth rate, altered leaf shape). Second, the published transcriptomic data available in Genevestigator indicate that both genes are expressed in all organs. Third, our whole mount detections of RPS6A and RPS6B mRNA show that both genes are expressed in the same tissues of the root tip. Fourth, our tetrad analysis shows that the RPS6A and RPS6B genes are functionally redundant in the gametophytes. Finally, each RPS6 gene is able to fully complement the rps6a/rps6a, rps6b/rps6b double mutant, demonstrating that the two RPS6 genes share identical functions. All of these results support the view that the two Arabidopsis RPS6 genes were conserved through evolution in order to maintain an appropriate level of RPS6. Although we have not tested if the rps6 mutations can reduce fitness, their altered growth and development suggest that this is the case (in support of the gene balance hypothesis).
Our work provides strong evidence that two paralogous plant RP genes can be functionally interchangeable, under standard growth conditions. This is in contrast with a recent study in the yeast S.cerevisiae which showed that functional specificity occurs in all duplicated ribosomal proteins, therefore including paralogous RP that have nearly identical sequences (Komili et al. 2007). These paralog-specific phenotypic effects suggest a “ribosome code” where the functional specificity of the ribosomes could be modulated by the set of RP paralogs which they contain (Komili et al. 2007). Interestingly, the composition of ribosomes in Arabidopsis is also extensively heterogeneous (regarding protein composition and their post-translational modifications) (Chang et al. 2005). Additionally, in Arabidopsis and in maize there are different phosphorylation isoforms of RPS6 proteins (Chang et al. 2005; Williams et al. 2003). In maize root tip, oxygen supply and temperature stress modulate phosphorylation of several serines and threonines located at the carboxy-terminal region of RPS6 proteins (Williams et al. 2003). Although the cellular role of these phosphorylations are not yet known for plants, in mammals they modulate cell growth. Interestingly, in Arabidopsis, two potentially phosphorylatable amino acids (S247 and T249, Figure 3S) are lacking in RPS6B (by comparison to RPS6A). However, in the event that some paralog-specific variations of Arabidopsis RPS6 have biological significances, we were unable to detect them in our growth conditions.
RPS6 is a limiting factor of growth
In eukaryotes as diverse as yeast, Drosophila, or mammals, the reduction of RP dosage or activity is associated with reduced cell size and growth rate (Meyuhas 2008). In plants, RPS6 phosphorylation depends upon growth conditions (Perez et al. 1990; Scharf and Nover 1982; Turck et al. 1998, 2004; Williams et al. 2003). In Drosophila, in which most RP genes are encoded by a single gene, or a small family with a main gene, a reduced dosage of many of the RP genes, including RPS6, results in decreased fitness (haplodeficiency) (Marygold et al. 2007). We have shown here that in Arabidopsis too the reduction of RPS6 dosage below a threshold level reduces or delays the growth of roots and shoots. The slower root growth rate of rps6 single mutants is correlated with a smaller root meristem. Thus the high rate of cell proliferation in root meristem requires at least three doses of the RPS6 gene (the single heterozygote does not display altered growth). By contrast, the final root cell size of rps6 mutants is not reduced (at least in the root epidermis), presumably because cell expansion does not require a high rate of protein synthesis. Therefore, it appears that an optimal level of RPS6 accumulation is more important for cell proliferation in root meristem cells than for the function of the gametophytes.
The genetics of the Arabidopsis rps6 mutations resembles that of the Drosophila Minute mutations. The Minute mutations are characterized by a set of similar dominant haploinsufficient phenotypic traits that include a small body size, prolonged development, shorter and thinner bristles, reduced fertility, and a recessive lethality; extreme Minute phenotypes include the arrest of gametogenesis. All the 66 known Minute mutations (with the exception of eIF2α) are in RP genes including RPS6 (most of them are encoded by a single gene or, in some few cases, by a small family with a main gene) (Marygold et al. 2007). The haploinsufficiency traits result from halving the copy number of RP which results in a reduced dosage of RP, and consequently a suboptimal concentration of functional ribosomes. Until now, the only known Arabidopsis RP gene displaying a Minute-like genetics behaviour is RPS5A. Similar to RPS6, there are two RPS5 genes, however RPS5A and RPS5B are differentially expressed. When homozygous, the disruption of RPS5A is embryonic lethal, and when heterozygous it confers semi-dominant growth retardation and developmental defects (Weijers et al. 2001). The phenotype of rps5b mutants has not been investigated yet. Our results show that the Arabidopsis RPS6A and RPS6B pair of paralogous genes behaves like a single Drosophila RP gene: halving the copy number of Arabidopsis RPS6 genes (i.e. in the rps6 single mutants or in the RPS6A/rps6a,RPS6B/rps6b double heterozygote) results in the Rps6 mutant phenotype. The non-allelic non-complementation between the rps6a and rps6b mutations substantiates the idea that what matters for Arabidopsis cell growth is the dosage of total RPS6, not the dosage of one particular RPS6 paralog. Taken together, these observations suggest that the pair of Arabidopsis RPS6 paralogs genetically behaves similar to the single copy Drosophila Minute gene. Recently, a similar result was found with RPL4A and RPL4D (Rosado et al. 2010).
One assumption of this model is that reduction of RPS6 accumulation impacts the efficiency of translation. However, the stoichiometry of ribosomal subunits in the rps6 mutants is modestly altered as shown by the slight increase of the 60S/40S ratio. By contrast, in the yeast deletion mutants ΔRPS6A and ΔRPS6B there is a dramatic accumulation of the 60S subunit as a consequence of the low level of 40S (Chiocchetti et al. 2007); a similar pattern has been observed under certain growth conditions in mice liver (Volarevic et al. 2000). It thus appears that lowering the RPS6 pool alters 40S biogenesis in Arabidopsis, although the effect on the stoichiometry of ribosomal subunits is not as intense as in yeast or mammals. Furthermore, as in yeast and mammals cells, the Arabidopsis rps6 mutations apparently do not modify the profile of polysomes. It is worth noting that the depletions of distinct RP can result in very different profiles of polysomes and monosomes in yeast (Chiocchetti et al. 2007) suggesting that RP deficiencies can alter the translational capacity of the yeast cells by different mechanisms.
Does leaf polarity need a specific ribosomal function?
The leaf shape results from the coordination of cell growth and differentiation in the primordia. Leaves display a “dorsoventral” polarity: the upper (adaxial) and lower (abaxial) side of the blade are distinct for trichomes, stomata or spongy mesophyll. This adaxial-abaxial polarity is controlled by a set of genes and small RNA (Husbands et al. 2009). Interestingly, loss-of-function mutations in the RPL5A, RPL5B, RPL9, RPL10B, RPL24B and RPL28A genes (Degenhardt and Bonham-Smith 2008; Pinon et al. 2008; Yao et al. 2008) enhance the phenotype of mutations that promote leaf abaxialisation such as1 and/or as2. Mutations in these RP genes also induce a pointed-leaves phenotype, as observed in rps6a and rps6b mutants. Other RP genes (RPS13A, RPS18A, RPL23aA, RPLA4, RPL4D) confer a pointed leaf phenotype when mutated or knocked down (Degenhardt and Bonham-Smith 2008; Pinon et al. 2008; Rosado et al. 2010). It is significant that among the seventeen Arabidopsis rp mutants that have been analyzed so far (including rps6a and rps6b), twelve have pointed leaves. However, none of the known rp mutants display leaf adaxial-abaxial polarity defects on their own, with the exception of rpl4a (Rosado et al. 2010), rpl5a, rpl5b, and the RPL5A/rpl5a, RPL5B/rpl5b double heterozygote (Yao et al. 2008). Therefore, does the regulation of the leaf adaxial-abaxial polarity require a conserved translational function of the ribosome (Yao et al. 2008)? How can a specific defect (leaf adaxial-abaxial polarity) be reconciled with its general function (ribosomal translation)? One possibility is that the balance between processes promoting adaxialisation and abaxialisation is sensitive to reduced translational activity, resulting in a shift toward abaxialisation in the case of a RP haplodeficiency.
In conclusion, we have genetically demonstrated that: (1) the RPS6 function is indispensable for cell survival, (2) the two Arabidopsis RPS6 proteins are functionally indistinguishable, and (3) the pool of Arabidopsis RPS6 represents a limiting effector of plant growth. Finally, as for several other Arabidopsis rp mutants, leaf shape of rps6 plants is altered (pointed), possibly because leaf patterning is particularly sensitive to reduced levels of ribosomal activities.
Experimental procedures
Plant material and growth conditions
Unless otherwise indicated, Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild-type in this study. The SALK lines (Alonso et al. 2003) and the quartet1 mutant (Preuss et al. 1994) were provided by the Nottingham Arabidopsis Stock Centre. SALK_048825 (rps6a) and SALK_012147 (rps6b) are both in the Col-0 background. qrt1-1 (NASC number: N8050) is in Landsberg erecta (L.er).
For propagation of the lines and in vitro cultures, seedling and plant growth conditions were as in Léonard et al. (2003).
General PCR conditions and sequencing
The PCR were performed with ~20 ng total DNA, 125 μM of each deoxynucleotide, 0.5 μM of each primer, in 1× PCR buffer and 0.1 U μl−1 of Taq polymerase on a GeneAmp System (Perkin-Elmer model 9700, USA) or on a Mastercycler epgradient S (Eppendorf, France). The PCR products purification and sequencing were made as described in Léonard et al. (2003). For the SALK_048825 line, the flanking sequence has been amplified using primers LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) and S61-R1 (5′-TCGGTAAGCCCAGGAAGATCGTTCT-3′) in the following conditions: 94°C for 2 min followed by 35 cycles of 94°C for 15 s, 60°C for 30 s and 72°C for 1 min 10 s. For the SALK_012147 line, primers used were LBa1 and S6-F (5′- TTCAACGTCGCCAATCCGACCACCG-3′). The reaction was started by incubation for 2 min at 94°C followed by 35 cycles of 94°C for 15 s, 62°C for 30 s and 72°C for 1 min. The purified fragments have been sequenced with the primers used for amplifications.
Southern blots
Total DNA was extracted from leaves and inflorescences (Bouchez and Camilleri 1998). For the hydridization with the RPS6A probe, DNA of the control lines and mutants were digested with XbaI, whereas for the RPSB probe, they were digested with EcoRV or XhoI.
Probes were made with a PCR fragment amplified from genomic DNA by using the LBa1 and S61-R1 primers (or S61-F1: 5′-CGTTGAAATGACTTGGTTTGTAAGG-3′ and S61-R2: 5′- GGATTCGCAACGTTGAACTGAAACG-3′ in Fig. 4) for the RPS6A probe (1,360 bp), and primers S62-F (5′-AGTTAGCGGAGATGCTCTAGGCGAG-3′) and S62-R (5′-AACTTCTTGCCTGCACATGGAAGCC-3′) for the RPS6B probe (693 bp), under the cycling conditions described above. Nucleic acids were blotted on Hybond-N+ membranes (Amersham) and all the probes were labeled with digoxygenin (PCR DIG Probe Synthesis Kit; Roche Applied Science, Mannheim, Germany). The hybridization and detection procedures were performed as described (Creff et al. 2006).
Transgenic complementation of rps6a and rps6b mutations
Each RPS6 gene was amplified with a High Fidelity Taq DNA Polymerase (Roche). For RPS6A, this includes 1,393 bp of promoter sequence, 13 bp of the 5′UTR and 234 bp of the 3′UTR and for RPS6B this includes 970 bp of promoter sequence, 87 bp of the 5′UTR and 230 bp of the 3′UTR. The PCR products were sub-cloned into the AscI/SalI site of the pGEM T-easy Vector (Promega) and sequenced before cloning into pGPTV Hygro binary vector (Becker et al. 1992). Each transgene was sequenced in its entirety with the following primers: RPS6A: REV 5′-CAGGAAACAGCTATGACC-3′; T7PROM 5′-TAATACGACTCACTATAGGG-3′; S61-F1; S61-R1; S61-GE 5′-GGCCGTGTTCGCCTTTTGCTT-3′. RPS6B : REV; T7; S62-F and S62-ATG 5′-TAAAAGCTGAGCCGCGTCGAGAGC-3′. The pGPTV-RPS6A and pGPTV-RPS6B vectors were used to transform the Agrobacterium tumefaciens strain GV3101 (pMP90). The transformations of Arabidopsis rps6 mutants were performed by floral dip as described by Clough and Bent (1998). For selecting transformants, seeds of the infiltrated plants were harvested and plated on a medium containing hygromycin B (50 μg ml−1).
RNA extraction and real time RT–PCR
Total RNA was extracted from 100 mg of 14-day-old frozen plantlets using the RNeasy Plant Mini Kit (Qiagen). In order to remove any residual genomic DNA, RNA was treated with RNAse-free DNAse (Ambion) according to the manufacturer’s instructions. Each extraction was performed in triplicate. Total RNA (1.5–2 μg) was used for first strand cDNA synthesis. Reverse transcription was performed as described in Creff et al. (2006). Specific primers were designed to the 3′ UTR region of each transcript:
-
S61-RTL 5′-CAGCGTGACAGGAGGAGTG-3′
-
S61-RTR 5′-GGTGACATCTTTGATTTGATTCTC-3′
-
S62-RTL 5′-CTTCTGCTCCTGCTAAACCC-3′
-
S62-RTR 5′-CGTTCTCATGTCTGTGGAGC-3′.
We carried out quantitative RT–PCR with 1:20 dilution cDNA using an ABI 7000 Sequence Detection System (Applied Biosystems) with SYBR Premix Ex Taq (Takara) as in Herbette et al. (2006).
Standard curves were generated by serial dilution of first-strand cDNA preparations and primer efficiency was determined. The following thermal profile was used : 2 min at 95°C and 40 cycles of 95°C for 10 s and 60°C for 35 s. GAPDH (Glyceraldehyde-3-Phosphate Dehydrogenase) was used as an internal control (Svistoonoff et al. 2007).
Northern blots
Total RNA of 14-day-old plantlets were isolated by phenol extraction and lithium chloride precipitation as in Verwoerd et al. (1989), with the exception that our phenol extraction was performed at room temperature. Gel-blots were performed with 10 μg total RNA. Specific probes were designed to the 3′-UTR region with following primers: S6A-F 5′-CAAGCCCTCTGTCACAGC-3′ and S6A-R 5′-GAGTTTAAAATGGTCTTC-3′ for a 200 bp RPS6A probe; S6B-F 5′-TAAACCCGTTGCTGCTTAAACTG-3′ and S6B-R 5′-TACTACTAAGAGGTTCAAAAC-3′ for a 186 bp RPS6B probe. The respective cycling conditions were: 2 min at 94°C and 35 cycles of 94°C for 15 s, 45°C for 30 s and 72°C for 20 s; and 2 min at 94°C and 35 cycles of 94°C for 30 s, 50°C for 45 s and 72°C for 20 s. The 25S probe (500 bp) was PCR amplified from an expressed sequence tag with primers PD 5′ ACGACGTTGTAAAACGACGGCCAG-3′ and PR 5′-CAGGAAACAGCTATGACCATGATTACG-3′ under the following conditions: 2 min at 94°C and 35 cycles of 94°C for 30 s, 65°C for 30 s and 72°C for 1 min.
Nucleic acids were blotted on Hybond-N+ (Amersham) membranes and all the probes were labelled with digoxygenin (PCR DIG Probe Synthesis Kit; Roche Applied Science, Mannheim, Germany). The hybridization and detection procedures were performed as previously described (Creff et al. 2006), with the exception that our northern blot hybridization was performed at 50°C.
Western blots
Western blotting of soluble proteins extracted from 100 mg of 13-day-old frozen plantlets (extraction buffer: 50 mM Tris–HCl pH8.5 mM EDTA, 150 mM NaCl, 2 mM dithiothreitol and 1% of a plant protease inhibitor cocktail (Sigma)) was as described (Witte et al. 2004). Blots were subsequently probed with an anti-RPS6 primary antibody (dilution 1:5000) from maize (Williams et al. 2003) and visualized with 1:10000 diluted Alexa Fluor 680 goat anti-rabbit antibody (Invitrogen, France). Detection was enabled by an Odyssey Infrared Imaging System (LI-COR Biosciences, Nebraska). Relative amount of proteins was normalized with a doublet of bands detected on the Coomassie stained gel.
Polysomes preparation
Polysomes were prepared as in Sormani et al. (2007) with 300 mg of 10-day-old seedlings.
Whole-mount in situ hybridization
In situ hybridization on 4-day-old seedlings was performed as described (Friml et al. 2003). To prepare RPS6A and RPS6B-specific probes, the full length genomic DNA of RPS6A and RPS6B were sub-cloned into pGEM-T Easy vector. A 577 bp fragment of the 3′ UTR region of AtRPS6A was PCR-amplified (S6A-F and T7PROM). A 347 bp specific AtRPS6B probe was obtained with primers S6B-F and SP6 5′-ATTTAGGTGACACTATAGAATACT-3′. PCR products were purified and used as templates to synthesize labelled riboprobes. Digoxigenin-labelled antisense RNA probes were synthesized using T7 or SP6 RNA-polymerase by in vitro transcription according to the manufacturer’s instruction (Roche). For hybridization we used 3 μg probe per ml hybridization volume. Observations were performed with a Leica DMRXA microscope.
Root length and meristem size analysis
Root length was measured as described in Svistoonoff et al. (2007).
Measurement of root meristem size was performed as in Dello Ioio et al. (2007) by using a Leica SP2 AOBS inverted confocal microscope (Leica Microsystems, Germany) with a 20× HC PL APO dry objective (NA = 0.70, Leica). Briefly, 7-day-old seedlings (n = ~30) were incubated in propidium iodide solution (20 μg ml−1) for 3 to 5 min and rinsed three times in water before observation. Propidium iodide was excited by 488 nm light produced by an Argon laser and observed using a window from 600 to 700 nm. The experiment was repeated twice, with similar results.
Tetrad analysis
A plant heterozygous for both the rps6a and rps6b mutations was crossed with the qrt1/qrt1 mutant. In the F1 progeny, one plant heterozygous for the rps6a mutation and another heterozygous for rps6b were screened by PCR with the LBa1 and S61-R1 primers (for rps6a), and with the S6-F and LBa1 primers (for rps6b). These two plants (rps6/+, qrt1/+) were crossed to each other and in the resulting F1 plants, we selected those that were homozygous for the qrt1 mutation by examining anthers under a stereo microscope. Among these plants we PCR-selected those that were heterozygous for both the rps6a and rps6b mutations. The presumed genotype (rps6a/+, rps6b/+) of these plants was then verified by southern blot, with RPS6A and RPS6B-specific probes (data not shown).
For pollen tetrad analysis, anthers of several flowers were harvested and pollen stained 15 min with the Alexander solution (Alexander 1969). The aborted (unstained) and alive (pink) pollen grains in tetrads were counted under a DMRXA microscope (Leica). The tetrad analysis was performed according to Fincham et al. (1979).
References
Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44:117–122
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genomewide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Câmara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller AM, Kissmehl R, Klotz C, Koll F, Le Mouël A, Lepère G, Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Serrano V, Zagulski M, Dessen P, Bétermier M, Weissenbach J, Scarpelli C, Schächter V, Sperling L, Meyer E, Cohen J, Wincker P (2006) whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444:171–178
Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20:1195–1197
Birchler JA, Veitia RA (2007) The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19:395–402
Birchler JA, Veitia RA (2009) The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol doi:10.1111/j.1469-8137.2009.03087.x
Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691
Bouchez D, Camilleri C (1998) High molecular weight DNA extraction from Arabidopsis. Methods Mol Biol 82:61–70
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438
Byrne ME (2009) A role for the ribosome in development. Trends Plant Sci 14:512–519
Carvalho CM, Santos AA, Pires SR, Rocha CS, Saraiva DI, Machado JP, Mattos EC, Fietto LG, Fontes EP (2008) Regulated nuclear trafficking of rpL10A mediated by NIK1 represents a defense strategy of plant cells against virus. PLoS Pathog 4:e1000247
Chang IF, Szick-Miranda K, Pan S, Bailey-Serres J (2005) Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol 137:848–862
Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, Heeren G, Oender K, Bauer J, Hintner H, Breitenbach M, Breitenbach-Koller L (2007) Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol 42:275–286
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 6:735–743
Conant GC, Wolfe KH (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 12:938–950
Copenhaver GP, Keith KC, Preuss D (2000) Tetrad analysis in higher plants. A budding technology. Plant Physiol 124:7–16
Creff A, Léonard B, Desnos T (2006) Targeted Ds-tagging strategy generates high allelic diversity at the Arabidopsis HY2 locus. Plant Mol Biol 61:603–613
De Bodt S, Maere S, Van de Peer Y (2005) Genome duplication and the origin of angiosperms. Trends Ecol Evol 20:591–597
Degenhardt RF, Bonham-Smith PC (2008) Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol 147:128–142
Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17:678–682
Deprost D, Truong HN, Robaglia C, Meyer C (2005) An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem Biophys Res Commun 326:844–850
Fincham JRS, Day PR, Radford A (1979) Fungal genetics. 4th ed. Botanical monographs-vol. 4. Blackwell Scientific Publications, Oxford
Freeling M (2009) Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60:433–453
Friml J, Benkova E, Mayer U, Palme K, Muster G (2003) Automated whole mount localisation techniques for plant seedlings. Plant J 34:115–124
Fujikura U, Horiguchi G, Ponce MR, Micol JL, Tsukaya H (2009) Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J 59:499–508
Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C (2007) Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6:95–110
Hay N, Sonenberg N (2004) Upstream and downstream of mTOR. Genes Dev 18:1926–1945
Herbette S, Taconnat L, Hugouvieux V, Piette L, Magniette ML, Cuine S, Auroy P, Richaud P, Forestier C, Bourguignon J, Renou JP, Vavasseur A, Leonhardt N (2006) Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 88:1751–1765
Husbands AY, Chitwood DH, Plavskin Y, Timmermans MC (2009) Signals and prepatterns: new insights into organ polarity in plants. Genes Dev 23:1986–1997
Imai A, Komura M, Kawano E, Kuwashiro Y, Takahashi T (2008) A semi-dominant mutation in the ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana. Plant J 56:881–890
Ito T, Kim GT, Shinozaki K (2000) Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J 22:257–264
Komili S, Farny NG, Roth FP, Silver PA (2007) Functional specificity among ribosomal proteins regulates gene expression. Cell 131:557–571
Léonard B, Creff A, Desnos T (2003) The HY2 gene as an efficient marker for transposon excision in Arabidopsis. Mol Gen Genomics 269:746–752
Maeda I, Kohara Y, Yamamoto M, Sugimoto A (2001) Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol 11:171–176
Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102:5454–5459
Mahfouz MM, Kim S, Delauney AJ, Verma DP (2006) Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18:477–490
Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC, Leevers SJ, Cook KR (2007) The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol 8:R216
McIntosh KB, Bonham-Smith PC (2006) Ribosomal protein gene regulation: what about plants? Can J Bot 84:342–362
Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99:6422–6427
Meyuhas O (2008) Physiological roles of ribosomal protein S6: one of its kind. Int Rev Cell Mol Biol 268:1–37
Morimoto T, Suzuki Y, Yamaguchi I (2002) Effects of partial suppression of ribosomal protein S6 on organ formation in Arabidopsis thaliana. Biosci Biotechnol Biochem 66:2437–2443
Nishimura T, Wada T, Okada KA (2004) Key factor of translation reinitiation, ribosomal protein L24, is involved in gynoecium development in Arabidopsis. Biochem Soc Trans 32:611–613
Nishimura T, Wada T, Yamamoto KT, Okada K (2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17:2940–2953
Panić L, Tamarut S, Sticker-Jantscheff M, Barkić M, Solter D, Uzelac M, Grabusić K, Volarević S (2006) Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol 26:8880–8891
Papp B, Pál C, Hurst LD (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424:194–197
Perez L, Aguilar R, Mendez A, Sanchez de Jimenez E (1990) Phosphorylation of ribosomal proteins induced by auxins in maize embryonic tissues. Plant Physiol 94:1270–1275
Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME (2008) Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135:1315–1324
Preuss D, Rhee SY, Davis RW (1994) Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264:1458–1460
Rocha CS, Santos AA, Machado JP, Fontes EP (2008) The ribosomal protein L10/QM-like protein is a component of the NIK-mediated antiviral signaling. Virology 380:165–169
Rosado A, Sohn EJ, Drakakaki G, Pan S, Swidergal A, Xiong Y, Kang BH, Bressan RA, Raikhel NV (2010) Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell 22:143–158
Scharf KD, Nover L (1982) Heat-shock-induced alterations of ribosomal protein phosphorylation in plant cell cultures. Cell 30:427–437
Seoighe C, Gehring C (2004) Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet 20:461–464
Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99:13627–13632
Sormani R, Yao L, Menand B, Ennar N, Lecampion C, Meyer C, Robaglia C (2007) Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol 7:26
Stewart MJ, Denell R (1993) Mutations in the Drosophila gene encoding ribosomal protein S6 cause tissue overgrowth. Mol Cell Biol 13:2524–2535
Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, Volarevic S (2005) Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev 19:3070–3082
Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39:792–796
Turck F, Kozma SC, Thomas G, Nagy F (1998) A heat-sensitive Arabidopsis thaliana kinase substitute for human p70S6K function in vivo. Mol Cell Biol 18:2038–2044
Turck F, Zilbermann F, Kozma SC, Thomas G, Nagy F (2004) Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol 134:1527–1535
Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M (1994) An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J 13:3378–3388
Veitia RA (2002) Exploring the etiology of haploinsufficiency. Bioessays 24:175–184
Verwoerd TC, Dekker BMM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17:2362
Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G (2000) Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288:2045–2047
Warner JR, McIntosh KB (2009) How common are extraribosomal functions of ribosomal proteins? Mol Cell 34:3–11
Watson KL, Konrad KD, Woods DF, Bryant PJ (1992) Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc Natl Acad Sci USA 89:11302–11306
Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R (2001) An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128:4289–4299
Whittle CA, Krochko JE (2009) Transcript profiling provides evidence of functional divergence and expression networks among ribosomal protein gene paralogs in Brassica napus. Plant Cell 21:2203–2219
Williams ME, Sussex IM (1995) Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J 8:65–76
Williams AJ, Werner-Fraczek J, Chang IF, Bailey-Serres J (2003) Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol 132:2086–2097
Witte CP, Noël LD, Gielbert J, Parker JE, Romeis T (2004) Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol Biol 55:135–147
Yao Y, Ling Q, Wang H, Huang H (2008) Ribosomal proteins promote leaf adaxial identity. Development 135:1325–1334
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Acknowledgments
We gratefully acknowledge Julia Bailey-Serre for the RPS6maize antibody, the GRAP team for plant care, Elena Marin for comments on the manuscript, Brandon Loveall for editing the English, and Laurent Nussaume and Christophe Robaglia for their support. This work was supported by the Commissariat à l’Énergie Atomique to AC and TD. RS was supported by a doctoral grant from the ToxNuc program (Commissariat à l’Énergie Atomique).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Creff, A., Sormani, R. & Desnos, T. The two Arabidopsis RPS6 genes, encoding for cytoplasmic ribosomal proteins S6, are functionally equivalent. Plant Mol Biol 73, 533–546 (2010). https://doi.org/10.1007/s11103-010-9639-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-010-9639-y