Abstract
In higher plants, phosphate (Pi) deficiency induces the replacement of phospholipids with the nonphosphorous glycolipids digalactosyldiacylglycerol (DGDG) and sulfoquinovosyldiacylglycerol (SQDG). Genes involved in membrane lipid remodeling are coactivated in response to Pi starvation, but the mechanisms that guide this response are largely unknown. Previously, we reported the importance of auxin transport for DGDG accumulation during Pi starvation. To understand the role of auxin signaling in Arabidopsis membrane lipid remodeling, we analyzed slr-1, a gain-of-function mutant of IAA14 (a repressor of auxin signaling), and arf7arf19, a loss-of-function mutant of auxin response factors ARF7 and ARF19. In slr-1 and arf7arf19, Pi stress-induced accumulation of DGDG and SQDG was suppressed. Reduced upregulation of glycolipid synthase and phospholipase genes in these mutants under Pi-deficient conditions indicates that IAA14 and ARF7/19 affect membrane lipid remodeling at the level of transcription. Pi stress-dependent induction of a non-protein-coding gene, IPS1, was also lower in slr-1 and arf7arf19, whereas expression of At4 (another Pi stress-inducible non-protein-coding gene), anthocyanin accumulation, and phosphodiesterase induction were not reduced in the shoot. High free Pi content was observed in slr-1 and arf7arf19 even under Pi-deficient conditions, suggesting that Pi homeostasis during Pi starvation is altered in these mutants. These results demonstrate a requirement of auxin signaling mediated by IAA14 and ARF7/19 for low-Pi adaptation in Arabidopsis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phosphorus is an essential plant nutrient that has a pivotal role in metabolism. Although the total amount of phosphorus in the soil may be high, inorganic phosphate (Pi) is often limiting for plant growth because of its strong interactions with soil constituents (Holford 1997). Thus, plants have evolved various adaptation mechanisms to tolerate low Pi bioavailability (Ticconi and Abel 2004). One of these dynamic adaptations is that plants change their membrane lipid composition by substituting nonphosphorous glycolipids for phospholipids (Benning and Ohta 2005). In higher plants, the glycolipids monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG) and sulfoquinovosyldiacylglycerol (SQDG) constitute the bulk of membrane lipids in plastids and play pivotal roles in chloroplast function, whereas phospholipids are the main lipid class in other subcellular membranes under nutrient-sufficient conditions (Hölzl and Dörmann 2007). During Pi deficiency, however, the proportion of DGDG dramatically increases not only in plastidic membranes but also in extraplastidic membranes (Härtel et al. 2000) such as cell membranes (Andersson et al. 2003), tonoplasts (Andersson et al. 2005) and mitochondrial membranes (Jouhet et al. 2004). SQDG also increases during Pi starvation to compensate for reduced phosphatidylglycerol in chloroplasts (Essigmann et al. 1998; Yu et al. 2002).
Digalactosyldiacylglycerol accumulation upon Pi starvation is mainly achieved in Arabidopsis by the contribution of the MGDG synthases MGD2 and MGD3 and DGDG synthases DGD1 and DGD2 (Benning and Ohta 2005). In Arabidopsis, SQD1 and SQD2, whose gene expression is upregulated by Pi starvation, are required for SQDG synthesis (Essigmann et al. 1998; Yu et al. 2002). In addition to the increase in glycolipid biosynthesis, phospholipid degradation is also required for membrane lipid remodeling during Pi starvation. In Arabidopsis, the phospholipases PLDζ1 and PLDζ2 have a role in Pi starvation-induced lipid remodeling in the root (Cruz-Ramírez et al. 2006; Li et al. 2006a, b). The non-specific phospholipases NPC4 and NPC5 are also involved in lipid remodeling in Arabidopsis (Gaude et al. 2008; Nakamura et al. 2005). Several mutant studies in Arabidopsis have provided genetic evidence that membrane lipid remodeling is a potent mechanism for adapting to Pi deficiency (Cruz-Ramírez et al. 2006; Gaude et al. 2008; Härtel et al. 2000; Kobayashi et al. 2009; Li et al. 2006a, b; Yu et al. 2002).
As another adaptation to Pi starvation, plants change their root systems by promoting lateral root growth and root hair development to access a larger soil volume and enhance Pi uptake (López-Bucio et al. 2003). In contrast, shoot growth is severely repressed to economize phosphorus under these conditions, resulting in an increase in the root-to-shoot growth ratio (López-Bucio et al. 2003). Auxin is one of the central regulators of root development; auxin signals are involved in the alteration of root system architecture, especially for lateral root induction during Pi starvation (Jain et al. 2007; López-Bucio et al. 2002; Nacry et al. 2005). Moreover, in some auxin-insensitive mutants, the root phenotypes such as reduced growth of lateral roots and root hairs are rescued by Pi starvation (Bates and Lynch 1996; Jain et al. 2007; López-Bucio et al. 2002; Nacry et al. 2005).
Auxin signaling is mediated by auxin response factors (ARFs) and their inhibitors, auxin/indole-3-acetic acid (Aux/IAA) proteins. ARFs are transcription factors that regulate auxin-responsive gene expression, whereas Aux/IAA proteins inhibit the activity of ARFs by binding directly to them. Auxin receptors transport inhibitor response 1 (TIR1) and TIR1-like F-box proteins interact with Aux/IAA proteins in the presence of auxin and promote their degradation; the liberation of ARFs from inhibition by Aux/IAA can positively or negatively regulate the transcription of auxin-responsive genes (Guilfoyle and Hagen 2007). A recent report proposed a model in which the increased level of TIR1 in response to Pi starvation induces the degradation of Aux/IAA proteins and the liberation of ARFs, which then activate pericycle cell division and lateral root formation during Pi starvation (Pérez-Torres et al. 2008). Among various auxin signaling mutants with a reduced number of lateral roots and root hairs, slr-1 and arf7arf19 show a crucial defect in lateral root formation in Arabidopsis (Fukaki et al. 2002; Okushima et al. 2005; Wilmoth et al. 2005). In the gain-of-function slr-1 mutant, stabilized mutant IAA14 protein inactivates ARF proteins, including ARF7 and ARF19, thereby completely blocking lateral root initiation (Fukaki et al. 2005, 2006). In slr-1, root hair formation is also prevented, suggesting that IAA14 is involved in the development of lateral roots and root hairs (Fukaki et al. 2002, 2005). The involvement of IAA14 and ARF19 in enhanced lateral root formation during Pi starvation has also been reported (Pérez-Torres et al. 2008).
Recently, we reported that treatment with auxin transport inhibitors or the mutation of auxin transport genes greatly decreases MGD2/3 transcription in Arabidopsis during Pi starvation (Kobayashi et al. 2006). Moreover, auxin transport inhibition limits the increase in the proportion of DGDG in Pi-deficient plants, indicating the necessity of auxin for DGDG accumulation during Pi starvation. The auxin signaling pathway involved in Pi starvation-induced DGDG accumulation has not, however, been identified. Moreover, although SQDG synthesis is also important for the compensation of plastidic phosphatidylglycerol during Pi starvation (Essigmann et al. 1998; Yu et al. 2002), the mechanism(s) that regulates its synthesis under these conditions remains unknown. Here we describe the analysis of Pi starvation responses in auxin-signaling mutants and show that membrane lipid remodeling is impaired in the gain-of-function mutant slr-1 and the loss-of-function mutant arf7arf19. In these mutants, Pi stress-induced gene expression was substantially reduced for genes involved in glycolipid biosynthesis, phospholipid degradation, and other Pi starvation responses. Our results provide a better understanding of the involvement of auxin signaling mediated by IAA14 and ARF7/19 in membrane lipid remodeling and other Pi starvation responses.
Materials and methods
Plant material and growth conditions
For all experiments described here, Arabidopsis thaliana wild type (Columbia-0), arf7 (nph4-1), arf19 (arf19-1), arf7arf19 (npf4-1 arf19-1), slr-1 and pIAA14::mIAA14-GR, which are all in the Columbia background (Fukaki et al. 2002, 2005; Okushima et al. 2005), were grown in a cultivation chamber at 23°C under continuous white light. For fresh weight measurements (Fig. 1e), the lipid analysis (Fig. 2), qRT-PCR analysis (Figs. 3–5), phosphodiesterase assay (Fig. 6a) and free Pi measurements (Fig. 6c), plants were grown on solidified Murashige and Skoog (MS) medium containing 0.8% (w/v) agar and 1.0% (w/v) sucrose for 10 days and then were grown on Pi-replete (1.0 mM) or Pi-depleted (0 mM) medium prepared as described (Härtel et al. 2000) for another 10 days. For anthocyanin measurements (Fig. 6b), 5-day-old seedlings grown on MS medium were transferred to Pi-replete or Pi-depleted medium for another 7 days. Shoot samples of wild type and the mutants were then collected. For DEX treatments (Fig. 7), 10-day-old pIAA14::mIAA14-GR seedlings grown on MS medium were transferred to Pi-controlled medium with (5.0 μM) or without (0 μM) DEX. After incubation for 12 h on DEX-containing media, the root samples were collected for qRT-PCR analyses. Collected samples were frozen immediately in liquid nitrogen and used for the subsequent assays.
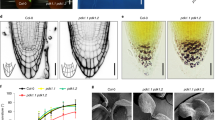
Growth phenotypes of wild type, slr-1 and arf7arf19 under Pi-sufficient or Pi-deficient conditions. a Five-day-old seedlings of wild type, slr-1 and arf7arf19 grown on MS medium were transferred to Pi-controlled medium and then grown for another 7 days. +Pi, Pi-sufficient (1.0 mM) conditions; −Pi, Pi-deficient (0 mM) conditions. Scale bars = 1.0 cm. b Root hairs of wild type, slr-1 and arf7arf19 grown as described in a. Light microscopic images approximately 1 cm from the primary root tips were captured in each plant. Scale bars = 500 μm. c,d Five-day-old seedlings of wild type, slr-1 and arf7arf19 grown on MS medium were transferred to Pi-controlled medium and then grown for the indicated number of additional days. Primary root length of the plants grown under Pi-sufficient conditions (c) or Pi-deficient conditions (d). e Ten-day-old seedlings of wild type, slr-1 and arf7arf19 grown on MS medium were transferred to Pi-controlled medium and then grown for another 10 days. Fresh weight of roots and shoots was measured separately. Values are the mean ± SD (n ≥ 15)
Lipid analysis of wild type and auxin mutants grown under Pi-controlled conditions. Membrane lipid composition of shoots (a) and roots (b) grown under Pi-sufficient conditions. Membrane lipid composition of shoots (c) and roots (d) grown under Pi-deficient conditions. MGDG, monogalactosyldiacylglycerol; DGDG, digalactosyldiacylglycerol; SQDG, sulfoquinovosyldiacylglycerol; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PC, phosphatidylcholine. SQDG in the root was undetectable in our experimental conditions. Values are the mean ± SD measured from three biologically independent samples. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with wild type (Student’s t-test)
Expression of glycolipid synthase genes in wild type, slr-1 and arf7arf19. Relative expression levels of MGD2 (a, b), MGD3 (c, d), DGD1 (e, f), DGD2 (g, h), SQD1 (i) and SQD2 (j) in the shoots (a, c, e, g, i, j) and the roots (b, d, f, h) of plants grown under Pi-sufficient (+Pi) or Pi-deficient (−Pi) conditions. Open, gray and black bars indicate wild type, slr-1 and arf7arf19, respectively. Gene expression levels were normalized to wild-type levels under +Pi conditions. Values are the mean ± SE measured from three biologically independent plant samples. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with wild type under −Pi conditions (Student’s t-test)
Root growth analyses
Five-day-old seedlings grown vertically on MS medium were transferred to vertically placed medium with (1.0 mM) or without (0 mM) Pi. Root growth of each plant was captured with a digital camera from day 0 to day 7 after the transplant to Pi-controlled media (Fig. 1c, d). Primary root length was digitally analyzed using image analyzing software (Scion Image, Scion, Frederick, MD). For Fig. 1b, images of root hairs approximately 1 cm from the primary root tips were captured with a digital microscope (VHX-600, Keyence, Osaka, Japan).
Lipid analysis
Total lipids were extracted and separated by two-dimensional thin-layer chromatography as described (Kobayashi et al. 2007). Lipids isolated from silica gel plates were methylated, and fatty acid methyl esters were quantified by gas chromatography using myristic acid as an internal standard, according to Kobayashi et al. (2006).
qRT-PCR analysis
Total RNA was isolated from three independent plant samples using the SV Total RNA Isolation System (Promega, Madison, WI) according to the manufacturer’s instructions. Reverse transcription was performed using the PrimeScript RT reagent kit (TaKaRa Bio, Otsu, Japan). cDNA amplification was carried out using SYBR PreMix Ex Taq (TaKaRa Bio) and 200 nM of gene-specific primers as described in Table S1, in Li et al. (2006a) for PLDζ2, and in Bari et al. (2006) for IPS1, At4 and Pht1;1. Thermal cycling consisted of an initial step at 95°C for 10 s, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. Signal detection and quantification were performed in duplicate using the Thermal Cycler Dice Real Time System (TaKaRa Bio). The relative abundance of all transcripts amplified was normalized to the constitutive expression level of UBQ10 (Bari et al. 2006) according to the method of Pfaffl (2001).
Anthocyanin measurement
Anthocyanin measurements were determined according to Ticconi et al. (2001). Powdered samples in liquid nitrogen were homogenized in propanol/HCl/water (18:1:81) and boiled for 3 min. After centrifugation (25,000×g) for 10 min, the absorbance of the supernatant was measured at 535 and 650 nm. The absorbance corresponding to the amount of anthocyanins was calculated by A 535–A 650.
Phosphodiesterase assay
Phosphodiesterase activity in plant tissues was measured according to Ticconi et al. (2001) with minor modifications. Samples were powdered in liquid nitrogen using a pestle and homogenized with an extraction buffer consisting of 100 mM Tris–acetate, 100 mM potassium acetate, 250 mM sodium ascorbate, 2 mM EDTA, 5 mM DTT and 10% (w/v) glycerol (pH 8.0). The enzyme mixture was prepared by centrifugation (25,000×g) for 5 min at 4°C. Total protein of the enzyme mixture was quantified by the method of Bensadoun and Weinstein (1976) with bovine serum albumin as a standard. Phosphodiesterase activity was measured using the enzyme mixture corresponding to 10 μg of total protein and 5 mM bis(p-nitrophenyl) phosphate as the substrate in the reaction buffer (50 mM acetic acid, 5 mM MgCl2, adjusted to pH 6.0 with NaOH). After incubation at 23°C for 10 min, 800 μl of 1.0 M Na2CO3 was mixed with 200 μl of the reaction mixture to terminate the reaction. The activity was calculated using a molar extinction coefficient of 18,300 cm2 nmol−1 for p-nitrophenol at 405 nm. The assay was performed under conditions of demonstrated linearity with respect to time and protein amount.
Pi measurement
Pi concentration was analyzed according to Chiou et al. (2006) with minor modifications. Tissue samples (0.1 g fresh weight) were homogenized with 250 μl extraction buffer (10 mM Tris, 1.0 mM EDTA, 100 mM NaCl, and 1.0 mM β-mercaptoethanol, pH 8.0). After centrifugation (25,000×g) for 10 min, 100 μl of the supernatant was mixed with 900 μl of 1% glacial acetic acid and then incubated at 42°C for 30 min. After centrifugation (25,000×g) for 5 min, 300 μl of the supernatant was mixed with 700 μl assay solution (0.35% [w/v] NH4MoO4, 0.86 N H2SO4, 1.4% [w/v] ascorbic acid) and then incubated at 42°C for 30 min. Pi concentration was measured at A 820.
Results
Effects of Pi deficiency on root development in slr-1 and arf7arf19
To ascertain whether auxin signaling mediated by IAA14 and ARF7/ARF19 is involved in Pi stress-induced morphological changes, we analyzed the phenotypes of slr-1 and arf7arf19 mutants under Pi-controlled conditions (Fig. 1). As reported (Fukaki et al. 2002; Okushima et al. 2005; Wilmoth et al. 2005), lateral root formation is severely limited in slr-1 and arf7arf19 grown under Pi-sufficient conditions. In these mutants, lateral root formation was also strongly blocked under Pi-deficient conditions (Fig. 1a), showing that the auxin signaling mediated by IAA14 and ARF7/19 is important for lateral root formation even when Pi is limiting. These results are consistent with the report that IAA14 and ARF19 play an important role in lateral root formation during Pi starvation (Pérez-Torres et al. 2008). In slr-1, root hair formation was also severely disrupted under Pi-sufficient conditions (Fig. 1b). Under Pi-deficient conditions, however, a few root hairs were observed in slr-1, indicating that Pi starvation can partially rescue the defect of root hair formation in this mutant. We also examined primary root length of these plants under Pi-controlled conditions (Fig. 1c, d). In wild type, primary root growth was inhibited by Pi starvation, consistent with previous reports (Jain et al. 2007; López-Bucio et al. 2002). The strong inhibition of primary root growth by Pi starvation was also observed in slr-1 and arf7arf19, demonstrating that Pi stress-induced suppression of primary root growth is still functional or even enhanced in slr-1 and arf7arf19. In these mutants, fresh weight of the root was quite low as compared with wild type in both Pi conditions, probably owing to loss of lateral roots in these mutants (Fig. 1e). Fresh weight of the shoot was also lower in slr-1 and arf7arf19 than in wild type under Pi-sufficient conditions. Under Pi-deficient conditions, however, fresh weight of the shoot in slr-1 was not markedly different from that in wild type, indicating that control of the shoot/root ratio in response to Pi deficiency may be perturbed in slr-1.
Auxin signaling mediated by IAA14 and ARF7/19 is involved in Pi stress-induced membrane lipid remodeling
Previously we showed that DGDG accumulation in Pi-deficient Arabidopsis roots was diminished by the inhibition of polar auxin transport, suggesting that Pi stress-induced membrane lipid remodeling in the root requires auxin signals (Kobayashi et al. 2006). To investigate auxin signaling pathways involved in membrane lipid remodeling, we analyzed membrane lipid composition in slr-1, arf7, arf19, and the arf7arf19 double mutant under Pi-controlled conditions (Fig. 2). Under Pi-sufficient conditions, there were no notable differences in lipid composition of the shoots between wild type and these auxin mutants (Fig. 2a). In the root of slr-1, however, a significant increase in galactolipids was observed under Pi-sufficient conditions (Fig. 2b). A slight increase in MGDG was also detected in the root of Pi-deficient slr-1. The slr-1 roots accumulate high levels of chlorophyll (K. Kobayashi, S. Baba, E.-M. Aro, H. Ohta, H. Fukaki and T. Masuda, unpublished data), suggesting that photosynthetic membrane biogenesis is activated in slr-1 roots. Given that MGD1 is the major isoform responsible for photosynthetic membrane biogenesis (Kobayashi et al. 2007), the increase in galactolipids in slr-1 roots may be attributed to the activation of MGD1 pathway during root greening. Under Pi-deficient conditions, the amount of DGDG dramatically increased in wild-type shoots and roots as reported (Härtel et al. 2000). Under these conditions, the proportion of SQDG, which was undetectable in the root of any plants in our experimental conditions, also increased in wild-type shoots as shown by Essigmann et al. (1998). In Pi-deficient arf7arf19, however, the proportion of DGDG was significantly reduced by 15% in the shoot and by 29% in the root as compared with wild type (Fig. 2c, d). The amount of SQDG was also reduced by 21% in the shoot of Pi-deficient arf7arf19 (Fig. 2c). The reduced accumulation of DGDG was more pronounced in Pi-deficient slr-1; the amount of DGDG was reduced by 24% in the shoot and by 37% in the root as compared with wild type (Fig. 2c, d). The proportion of SQDG in the shoot was substantially reduced by 60% in Pi-deficient slr-1 as compared with wild type (Fig. 2c). In slr-1, the proportion of SQDG in the shoot under Pi-deficient conditions was almost equivalent to that under Pi-sufficient conditions, showing that Pi starvation-induced SQDG accumulation was severely impaired in slr-1 (Fig. 2c). These results suggest that auxin signaling mediated by IAA14 and ARF7/19 is one of the key regulatory systems involved in membrane lipid remodeling during Pi starvation. In arf7 and arf19, no significant reduction in the accumulation of DGDG or SQDG was observed during Pi starvation, indicating that ARF7 and ARF19 function redundantly (Fig. 2c, d).
Pi stress-dependent upregulation of glycolipid synthase genes is reduced in slr-1 and arf7arf19
Our lipid analysis implicated the involvement of IAA14 and ARF7/19 in membrane lipid remodeling during Pi starvation. To assess whether these factors are involved in the activation of glycolipid synthesis genes during Pi starvation, we examined the expression of Pi stress-inducible genes—MGD2, MGD3, DGD1, DGD2, SQD1 and SQD2—using real-time quantitative reverse transcription (qRT)-PCR analysis in slr-1 and arf7arf19 (Fig. 3). Consistent with previous reports on wild-type plant expression of the genes MGD2, MGD3 (Awai et al. 2001; Kobayashi et al. 2004), DGD1, DGD2 (Kelly et al. 2003), SQD1 (Essigmann et al. 1998) and SQD2 (Yu et al. 2002), these genes were highly upregulated in response to Pi deficiency. Although induction of these genes by Pi starvation was also observed in slr-1 and arf7arf19, the increased levels were generally lower than those in wild type. In slr-1, the MGD2 transcript level during Pi deficiency was 59% lower in the shoot and 80% lower in the root as compared with wild type (Fig. 3a, b). Reduced upregulation of MGD2 expression was also observed in Pi-deficient arf7arf19. Similar results were obtained for MGD3 expression in these mutants (Fig. 3c, d). Pi stress-induced upregulation of DGD1 and DGD2 was also lower in the roots of slr-1 and arf7arf19 (Fig. 3f, h); however, in the shoots of these mutants, the expression of these genes during Pi starvation was comparable to that in wild-type shoots (Fig. 3e, g). We also analyzed SQD1 and SQD2 transcript levels in the shoots of slr-1 and arf7arf19 (Fig. 3i, j). Under Pi-sufficient conditions, there was no considerable difference in expression of SQD1 or SQD2 between wild type and the auxin mutants. Under Pi-deficient conditions, however, the SQD1 transcript level in the shoot was reduced by 65% in slr-1 and by 46% in arf7arf19 as compared with wild type (Fig. 3i). In slr-1, Pi stress-induced upregulation of SQD2 was also significantly lower than that in wild type (Fig. 3j).
Pi stress-dependent upregulation of phospholipase genes is reduced in slr-1 and arf7arf19
Arabidopsis phospholipases NPC4 (Nakamura et al. 2005), NPC5 (Gaude et al. 2008) and PLDζ2 (Cruz-Ramírez et al. 2006; Li et al. 2006a, b) are induced by Pi starvation and are involved in Pi stress-dependent phospholipid degradation. To address the involvement of auxin signaling in the regulation of these phospholipase genes, we examined expression levels of NPC4, NPC5 and PLDζ2 in wild type, slr-1 and arf7arf19 (Fig. 4). Under Pi-sufficient conditions, only low expression levels of the phospholipase genes were observed in wild type and auxin mutants. In wild type, the expression of these genes strongly increased in both shoots and roots under Pi-deficient conditions, in agreement with previous reports (Cruz-Ramírez et al. 2006; Gaude et al. 2008; Li et al. 2006a, b; Nakamura et al. 2005). In slr-1, however, transcript levels of NPC4, NPC5 and PLDζ2 during Pi starvation were significantly reduced by 70, 76 and 47% in the shoot and by 73, 83 and 72% in the root, respectively, as compared with wild type. Reduced induction of these genes during Pi starvation was also observed in arf7arf19 although the reduction was more moderate and less significant than in slr-1. These results show that the mutations in slr-1 and arf7arf19 negatively affect Pi stress-dependent induction of phospholipase genes as well as glycolipid synthesis genes.
Expression of phospholipase genes in wild type, slr-1 and arf7arf19. Relative expression levels of NPC4 (a, b), NPC5 (c, d) and PLDζ2 (e, f) in the shoots (a, c, e) and the roots (b, d, f) of plants grown under Pi-sufficient (+Pi) or Pi-deficient (−Pi) conditions. Open, gray and black bars indicate wild type, slr-1 and arf7arf19, respectively. Gene expression levels were normalized to wild-type levels under +Pi conditions. Values are the mean ± SE measured from three biologically independent plant samples. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with wild type under −Pi conditions (Student’s t-test)
Involvement of IAA14 and ARF7/19 in transcription of other Pi stress-responsive genes
To address whether the mutations in slr-1 and arf7arf19 affect the transcription of other Pi starvation-responsive genes, expression of IPS1 and At4, which are non-protein-coding genes that are strongly and specifically induced by Pi starvation (Bari et al. 2006; Martín et al. 2000; Rubio et al. 2001), was analyzed in slr-1 and arf7arf19 (Fig. 5a–d). Although only very low IPS1 and At4 expression occurred in wild type in both shoots and roots under Pi-sufficient conditions, their expression dramatically increased in response to Pi deficiency. In slr-1, however, induction of IPS1 expression by Pi starvation was lower; the IPS1 transcript level during Pi starvation was reduced by 75% in the shoot and by 69% in the root as compared with wild type (Fig. 5a, b). In arf7arf19, IPS1 induction in response to Pi deficiency was also significantly reduced in the shoot, although the reduction was more moderate than that in slr-1. Unlike IPS1, the At4 transcript levels in the shoot of slr-1 and arf7arf19 were not statistically different from that in wild type, even under Pi-starved conditions (Fig. 5c), suggesting that IAA14 and ARF7/19 have little effect on At4 expression. We also investigated the effects of the mutations in slr-1 and arf7arf19 on the expression of a Pi transporter gene Pht1;1 (Fig. 5e, f), which has a significant role in phosphate acquisition from both low- and high-Pi environments (Shin et al. 2004). As reported (Bari et al. 2006; Karthikeyan et al. 2002; Mudge et al. 2002; Shin et al. 2004), in wild-type plants the transcription of Pht1;1 was strongly upregulated both in shoots and roots under Pi-deficient conditions. In the shoots of slr-1 and arf7arf19, the expression of Pht1;1 also increased to a level similar to that of wild type in response to Pi deficiency. In the root of slr-1, however, Pi stress-induced upregulation of Pht1;1 was strongly reduced as compared with wild type (Fig. 5f). The different expression patterns among IPS1, At4 and Pht1;1 in these mutants may reflect the presence of multiple signaling pathways for Pi starvation responses of these genes, at least in shoots.
Expression of Pi starvation-responsive genes in wild type, slr-1 and arf7arf19. Relative expression levels of IPS1 (a, b), At4 (c, d), Pht1;1 (e, f) in the shoots (a, c, e) and the roots (b, d, f) of plants grown under Pi-sufficient (+Pi) or Pi-deficient (−Pi) conditions. Open, gray and black bars indicate wild type, slr-1 and arf7arf19, respectively. Gene expression levels were normalized to wild-type levels under +Pi conditions. Values are the mean ± SE measured from three biologically independent plant samples. * P < 0.05, ** P < 0.01 compared with wild type under −Pi conditions (Student’s t-test)
slr-1 and arf7arf19 show normal anthocyanin accumulation and phosphodiesterase activation in response to Pi starvation despite excessive Pi accumulation
To assess whether impairment of auxin signaling in slr-1 and arf7arf19 affects other typical Pi stress-dependent responses, we examined phosphodiesterase activity in wild type and in the auxin mutants under Pi-controlled conditions (Fig. 6a). In wild type, phosphodiesterase activity increased in response to Pi starvation in the shoot, consistent with a previous study (Ticconi et al. 2001). In the shoots of slr-1 and arf7arf19, phosphodiesterase activity increased to a level similar to that of wild type in response to Pi deficiency. The phosphodiesterase activity in the root of these mutants was also not statistically different from that of wild type under either Pi condition (Fig. 6a). The data suggest that the mutations in slr-1 and arf7arf19 have no substantial effect on upregulation of phosphodiesterase activity in response to Pi deficiency.
Phosphodiesterase activity, anthocyanin content and free Pi concentration in wild type, slr-1 and arf7arf19. a Phosphodiesterase activity in shoots and roots under Pi-sufficient (+Pi) or Pi-deficient (−Pi) conditions. Production of p-nitrophenol from bis(p-nitrophenyl) phosphate was used to determine phosphodiesterase activity. There is no statistically significant difference between wild type and the mutants. Anthocyanin content (b) was measured in shoots and free Pi concentration (c) was measured in shoots and roots of plants grown under +Pi or −Pi conditions. In a–c, open, gray and black bars indicate wild type, slr-1 and arf7arf19, respectively. Values are the mean ± SD measured from three biologically independent samples. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with wild type under −Pi conditions (Student’s t-test)
Because anthocyanin accumulation is also a typical response to Pi starvation (Ticconi and Abel 2004), we measured anthocyanin content in the shoot of wild type and in the auxin mutants under Pi-controlled conditions. Results showed that Pi stress-induced accumulation of high levels of anthocyanin was observed not only in wild type but also in slr-1 and arf7arf19 (Fig. 6b), indicating that mutations in slr-1 and arf7arf19 do not inhibit anthocyanin accumulation during Pi starvation. Taken together, these results suggest that auxin signaling mediated by IAA14 and ARF7/19 has no essential role in the activation of phosphodiesterase or anthocyanin accumulation during Pi starvation. To ascertain whether the mutations in slr-1 and arf7arf19 affect internal Pi status, we examined free Pi content in shoots and roots (Fig. 6c). Under Pi-sufficient conditions, Pi content in slr-1 roots was significantly higher than in wild type and arf7arf19. In the shoot, however, a slight reduction of Pi content was detected in both slr-1 and arf7arf19 under Pi-sufficient conditions. Under Pi-deficient conditions, notable differences in free Pi content were observed between wild type and the auxin mutants. In wild type, the amount of free Pi was substantially reduced during Pi starvation. In slr-1 and arf7arf19, however, the reduction in free Pi during Pi starvation was attenuated in both the shoot and the root. In slr-1, particularly, Pi content in the shoot and root increased 5.4-fold and 2.7-fold, respectively, relative to wild type. Because we used medium without any Pi for Pi-deficient conditions in these experiments, it is unlikely that the increase in free Pi content in these mutants was caused by enhanced uptake of Pi from the medium. Our data thus suggest that internal Pi homeostasis during Pi starvation is altered in slr-1 and arf7arf19.
Auxin signaling mediated by IAA14 and ARF7/19 indirectly regulates expression of MGD3 and IPS1 during Pi starvation
The core sequence of auxin response elements (AuxREs), to which ARFs specifically bind, is TGTCTC (along with its variations, TGTCAC, TGTCCC and TGTCGC) in the auxin-responsive promoters of primary/early genes (Ulmasov et al. 1995, 1997). We looked for the core AuxRE sequences (TGTCnC), including the inverted version (GnGACA), in the promoter regions (1,000 bp upstream from the transcription start site) of the Pi stress-inducible genes analyzed here. MGD2, NPC4 and IPS1, whose expression in slr-1 was lower as compared with wild type under Pi-deficient conditions (Fig. 3a, b, 4a, b, 5a, b), did not have core AuxRE sequences in their promoters (data not shown), whereas these sequences were found in the promoters of all the other genes analyzed here.
To check whether auxin signaling regulates expression of Pi starvation-responsive genes directly, we used the pIAA14::mIAA14-GR line described in Fukaki et al. (2005). In this transgenic line, dexamethasone (DEX) treatment induces the accumulation of a stable mIAA14 protein resistant to auxin-mediated degradation, resulting in the inhibition of ARF-mediated auxin signaling. Ten-day-old pIAA14::mIAA14-GR seedlings were transferred to Pi-controlled medium with or without DEX. After incubation on DEX-containing medium for 12 h, the expression of MGD3 and IPS1 was analyzed in the root by qRT-PCR (Fig. 7a, b) because their expression during Pi starvation was substantially reduced in slr-1 relative to wild type. Transient induction of the stabilized mIAA14 protein by DEX treatment under Pi-deficient conditions did not suppress Pi starvation-induced expression of MGD3 and IPS1 within 12 h, suggesting that auxin signaling mediated by IAA14 and ARF7/19 does not directly regulate the expression of these genes. Although IPS1 expression in DEX-treated plants was significantly higher than that in the untreated control under Pi-deficient conditions, the mechanism is currently unclear.
Expression analysis using the dexamethasone (DEX)-mediated induction system of the stabilized mIAA14 protein. Relative expression levels of MGD3 (a) and IPS1 (b) in the roots of pIAA14::mIAA14-GR seedlings grown for 12 h with (+) or without (−) DEX were analyzed under Pi-sufficient (+Pi) or Pi-deficient (−Pi) conditions. Values are the mean ± SE measured from three biologically independent samples. * Significant difference from the −DEX control grown under −Pi conditions (P < 0.05, Student’s t-test)
Discussion
We previously showed that auxin transport from the shoot to the root is essential not only for the transcriptional activation of MGD2 and MGD3 but also for DGDG accumulation during Pi starvation (Kobayashi et al. 2006). Here we show that Pi stress-dependent accumulation of DGDG is considerably reduced both in the shoot and the root of slr-1 and arf7arf19 (Fig. 2c, d), demonstrating that auxin signaling mediated by IAA14 and ARF7/19 affects DGDG accumulation during Pi starvation. Moreover, Pi stress-dependent upregulation of the galactolipid synthase genes MGD2, MGD3, DGD1 and DGD2 was reduced in the root of slr-1 and arf7arf19 (Fig. 3b, d, f, h). In the shoot, however, induction of DGD1 or DGD2 expression by Pi starvation was not reduced in these mutants, whereas the upregulation of MGD2 and MGD3 expression was weakened (Fig. 3a, c, e, g). Pi stress-induced expression of DGD1 and DGD2 may be less dependent on auxin signaling mediated by IAA14 and ARF7/19 in the shoot. In slr-1 and arf7arf19, although morphological defects were mainly observed in the root systems (Fig. 1), inhibition of DGDG accumulation was detected not only in the root but also in the shoot under Pi-deficient conditions (Fig. 2c, d). Moreover, reduced induction of the SQDG synthase genes during Pi starvation was observed in the shoot of these mutants (Fig. 3i, j), where a reduction in SQDG accumulation occurred (Fig. 2c). Our data show that auxin signaling is important both in the shoot and in the root to induce glycolipid biosynthesis during Pi starvation. In addition to the glycolipid synthase genes, the mutations in slr-1 and arf7arf19 affected the Pi stress-dependent gene expression of the phospholipases NPC4, NPC5 and PLDζ2 (Fig. 4), which supply substrates for glycolipid synthases by catalyzing phospholipid degradation under low-Pi conditions (Cruz-Ramírez et al. 2006; Gaude et al. 2008; Li et al. 2006a, b; Nakamura et al. 2005). Impaired phospholipid degradation might also contribute to the reduction of Pi stress-induced accumulation of glycolipids in slr-1 and arf7arf19. The slr-1 mutant showed more severe defects in membrane lipid remodeling than arf7arf19 during Pi starvation. Moreover, at least in the genes analyzed here, the slr-1 mutation had more pronounced effects on Pi stress-dependent gene expression than the arf7arf19 mutations. These data indicate that other ARFs controlled by IAA14 may also affect the regulation of membrane lipid remodeling and other responses to Pi starvation.
In slr-1, Pi stress-dependent upregulation of IPS1 and Pht1;1 was mitigated in the roots, whereas the induction of Pht1;1 and At4 was not significantly reduced in the shoots (Fig. 5). These results suggest that although IAA14 and ARF7/19 are not involved in every aspect of transcriptional regulation of Pi stress-responsive genes, these factors have a broad impact on gene expression involved in low-Pi responses. Pi stress-dependent expression of IPS1 and At4 is under the control of a MYB transcription factor, PHR1 (Rubio et al. 2001), which is post-translationally regulated by SIZ1-mediated sumoylation (Miura et al. 2005). The reduction in Pi stress-induced IPS1 expression in slr-1 and arf7arf19 suggests that PHR1 may have been inactivated in these auxin mutants during Pi starvation. The expression of At4, another PHR1-regulated gene, was not, however, significantly affected, particularly in the shoots of these mutants. Furthermore, Pi stress-induced anthocyanin accumulation, which is also controlled by PHR1 (Rubio et al. 2001), was not reduced, even increased in slr-1 and arf7arf19 (Fig. 6b). Taken together, these results suggest that the reduction in Pi stress-dependent gene expression in these auxin mutants was not caused by inactivation of PHR1. Furthermore, Gaude et al. (2008) reported that PHR1 is not a major regulator involved in membrane lipid remodeling during Pi starvation.
A recent study in Arabidopsis by Lai et al. (2007) proposed that cell-cycle activity under low-Pi conditions determines the magnitude of Pi-starvation responses by altering Pi demand in growing organs. They showed that treatments that diminish root cell proliferation repress transcriptional activity of MGD3 during Pi starvation. In the root of slr-1, cell divisions of pericycle cells are strongly impaired, resulting in the complete elimination of lateral root initiation (Fukaki et al. 2002; Vanneste et al. 2005). A similar defective phenotype was observed in the root of arf7arf19 (Okushima et al. 2005, 2007). These data suggest that auxin signaling mediated by IAA14 and ARF7/19 affects the expression of Pi stress-inducible genes by regulating cell-cycle activity in the root. However, an inhibition of membrane lipid remodeling as well as a reduction in the expression of genes involved in the remodeling system was observed even in the shoot of Pi-deficient slr-1 (Figs. 2–4), in which no obvious difference in fresh weight and tissue development was observed as compared with Pi-deficient wild type (Fig. 1a, e). These results suggest that auxin signaling mediated by IAA14 and ARF7/19 may regulate certain phosphate starvation responses independently of cell-cycle activity, at least in the shoot.
In slr-1 and arf7arf19, accumulation of high levels of free Pi was observed in both shoots and roots, even under Pi-deficient conditions (Fig. 6c). Excessive Pi accumulation under low-Pi conditions also occurs in growth-inhibited roots treated with cytokinin, high nitrogen, or a non-metabolizable glucose analog, 3-O-methyl glucose, suggesting that reduced Pi demand in the growth-inhibited roots causes the increase in free Pi pools during Pi starvation (Lai et al. 2007). The accumulation of large amounts of free Pi in slr-1 and arf7arf19, in which auxin-dependent organ development is strongly arrested, might be explained by reduced Pi demand in growing tissues during Pi starvation. However, because the excess Pi accumulation was also observed in the shoot of slr-1, in which no severe growth defects were observed relative to wild type during Pi starvation (Fig. 1e), we cannot presently exclude the possibility that IAA14 and ARF7/ARF19 are involved in the regulation of free Pi metabolism during Pi starvation by a signaling cascade. Indeed, Pi stress-induced expression of IPS1, which is involved in the regulation of Pi homeostasis via the miR399 and PHO2 signaling pathways (Franco-Zorrilla et al. 2007), was reduced in slr-1 and arf7arf19 (Fig. 5a, b). Auxin signaling mediated by IAA14 and ARF7/19 may affect Pi homeostasis by altering the IPS1 expression level during Pi starvation. Wang et al. (2006) proposed that excessive Pi accumulation after cytokinin treatment is one of the mechanisms for a global reduction in Pi-starvation signaling in rice (Oryza sativa). It is possible that high Pi accumulation in slr-1 and arf7arf19 during Pi starvation affects the gene expression involved in low-Pi responses such as membrane lipid remodeling. Anthocyanin accumulation and an increase in phosphodiesterase activity were not, however, diminished in the shoots of these mutants, despite the increase in free Pi content in the tissues (Fig. 6). Moreover, in the shoots of these mutants, the increase in free Pi concentration did not repress the expression of DGD1, DGD2, At4, or Pht1;1 during Pi starvation (Figs. 3e, g, 5c, e), indicating that the increase in free Pi content is independent of the Pi starvation responsiveness in slr-1 and arf7arf19, at least in the shoot.
The analysis using the DEX-mediated induction system of the stabilized mIAA14 protein showed an indirect involvement of auxin signaling by IAA14 and ARF7/19 in the upregulation of MGD3 and IPS1 expression during Pi starvation (Fig. 7). These data are consistent with the fact that core AuxRE sequences are not fully conserved in the promoter regions of the Pi stress-inducible genes analyzed here (data not shown). Together with recent studies of auxin signaling pathways (Guilfoyle and Hagen 2007), our data suggest a possible regulatory mechanism for Pi stress-induced membrane lipid remodeling. First, auxin transported from production sites or storage pools induces the degradation of IAA14 protein via SCFTIR1-mediated ubiquitination. Recent studies reported that an increased expression of TIR1 in response to Pi starvation accelerates the degradation of AUX/IAA proteins (Pérez-Torres et al. 2008), supporting this idea. Then, as ARF7 and ARF19 are released from IAA14 inhibition, they can modify the cell-cycle activities and other cellular responses, which may lead to the indirect induction of gene expression involved in membrane lipid remodeling under Pi-deficient conditions.
We also evaluated the involvement of IAA14 and ARF7/19 in Pi stress-dependent changes in root morphology (Fig. 1). Auxin signaling mediated by IAA14 and ARF7/19 was not necessary for primary root inhibition by Pi starvation but was required for lateral root formation. A similar phenotype was observed in the iaa28-1 mutant, which has a gain-of-function mutation in IAA28 (López-Bucio et al. 2002), indicating that IAA14 and IAA28 may have redundant roles in root development. The involvement of auxin signaling in lateral root growth during Pi starvation has been proposed in Arabidopsis (Jain et al. 2007; López-Bucio et al. 2002; Nacry et al. 2005). These studies have shown that Pi deficiency increases auxin sensitivity in the root, which then promotes lateral root growth. It has been demonstrated that auxin sensitivity in the root is enhanced by the elevated expression of TIR1, which increases the degradation of AUX/IAA and the transcriptional activities of ARFs (Pérez-Torres et al. 2008). By contrast, auxin signaling mediated by IAA14 and ARF7/19 was not necessary for the inhibition of primary root growth caused by Pi starvation (Fig. 1c, d). Localized Pi deficiency triggers the progressive loss of meristematic cells in the primary root, thereby causing determinate growth (Sánchez-Calderón et al. 2005). Our results suggest that auxin signaling mediated by IAA14 and ARF7/19 has no important role in the shift to determinate growth of the primary root during Pi starvation, consistent with the report by Jain et al. (2007) that auxin is unlikely to be involved in determinate growth of the primary root exposed to localized Pi deficiency.
References
Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS (2003) Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett 537:128–132. doi:10.1016/S0014-5793(03)00109-1
Andersson MX, Larsson KE, Tjellstrom H, Liljenberg C, Sandelius AS (2005) Phosphate-limited oat. The plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J Biol Chem 280:27578–27586. doi:10.1074/jbc.M503273200
Awai K, Maréchal E, Block MA, Brun D, Masuda T, Shimada H, Takamiya K, Ohta H, Joyard J (2001) Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc Natl Acad Sci USA 98:10960–10965. doi:10.1073/pnas.181331498
Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141:988–999. doi:10.1104/pp.106.079707
Bates TR, Lynch JP (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorous availability. Plant Cell Environ 19:529–538. doi:10.1111/j.1365-3040.1996.tb00386.x
Benning C, Ohta H (2005) Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem 280:2397–2400. doi:10.1074/jbc.R400032200
Bensadoun A, Weinstein D (1976) Assay of proteins in the presence of interfering materials. Anal Biochem 70:241–250. doi:10.1016/S0003-2697(76)80064-4
Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 18:412–421. doi:10.1105/tpc.105.038943
Cruz-Ramírez A, Oropeza-Aburto A, Razo-Hernández F, Ramírez-Chávez E, Herrera-Estrella L (2006) Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci USA 103:6765–6770. doi:10.1073/pnas.0600863103
Essigmann B, Güler S, Narang RA, Linke D, Benning C (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95:1950–1955
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037. doi:10.1038/ng2079
Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29:153–168. doi:10.1046/j.0960-7412.2001.01201.x
Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M (2005) Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44:382–395. doi:10.1111/j.1365-313X.2005.02537.x
Fukaki H, Taniguchi N, Tasaka M (2006) PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J 48:380–389. doi:10.1111/j.1365-313X.2006.02882.x
Gaude N, Nakamura Y, Scheible WR, Ohta H, Dörmann P (2008) Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J 56:28–39. doi:10.1111/j.1365-313X.2008.03582.x
Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10:453–460. doi:10.1016/j.pbi.2007.08.014
Härtel H, Dörmann P, Benning C (2000) DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA 97:10649–10654. doi:10.1073/pnas.180320497
Holford ICR (1997) Soil phosphorus: its measurements and its uptake by plants. Aust J Soil Res 35:227–239
Hölzl G, Dörmann P (2007) Structure and function of glycoglycerolipids in plants and bacteria. Prog Lipid Res 46:225–243. doi:10.1016/j.plipres.2007.05.001
Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, Murphy AS, Raghothama KG (2007) Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol 144:232–247. doi:10.1104/pp.106.092130
Jouhet J, Maréchal E, Baldan B, Bligny R, Joyard J, Block MA (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J Cell Biol 167:863–874. doi:10.1083/jcb.200407022
Karthikeyan AS, Varadarajan DK, Mukatira UT, D’Urzo MP, Damsz B, Raghothama KG (2002) Regulated expression of Arabidopsis phosphate transporters. Plant Physiol 130:221–233. doi:10.1104/pp.020007
Kelly AA, Froehlich JE, Dörmann P (2003) Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15:2694–2706. doi:10.1105/tpc.016675
Kobayashi K, Awai K, Takamiya K, Ohta H (2004) Arabidopsis type B monogalactosyldiacylglycerol synthase genes are expressed during pollen tube growth and induced by phosphate starvation. Plant Physiol 134:640–648. doi:10.1104/pp.103.032656
Kobayashi K, Masuda T, Takamiya K, Ohta H (2006) Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. Plant J 47:238–248. doi:10.1111/j.1365-313X.2006.02778.x
Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H (2007) Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc Natl Acad Sci USA 104:17216–17221. doi:10.1073/pnas.0704680104
Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H (2009) Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J 57:322–331. doi:10.1111/j.1365-313X.2008.03692.x
Lai F, Thacker J, Li Y, Doerner P (2007) Cell division activity determines the magnitude of phosphate starvation responses in Arabidopsis. Plant J 50:545–556. doi:10.1111/j.1365-313X.2007.03070.x
Li M, Qin C, Welti R, Wang X (2006a) Double knockouts of phospholipases Dzeta1 and Dzeta2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol 140:761–770. doi:10.1104/pp.105.070995
Li M, Welti R, Wang X (2006b) Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases D zeta1 and D zeta2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol 142:750–761. doi:10.1104/pp.106.085647
López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129:244–256. doi:10.1104/pp.010934
López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6:280–287. doi:10.1016/S1369-5266(03)00035-9
Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24:559–567. doi:10.1111/j.1365-313X.2000.00893.x
Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ, Hasegawa PM (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102:7760–7765. doi:10.1073/pnas.0500778102
Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31:341–353. doi:10.1046/j.1365-313X.2002.01356.x
Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P (2005) A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 138:2061–2074. doi:10.1104/pp.105.060061
Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya K, Ohta H (2005) A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J Biol Chem 280:7469–7476. doi:10.1074/jbc.M408799200
Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17:444–463. doi:10.1105/tpc.104.028316
Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19:118–130. doi:10.1105/tpc.106.047761
Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20:3258–3272. doi:10.1105/tpc.108.058719
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15:2122–2133. doi:10.1101/gad.204401
Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46:174–184. doi:10.1093/pcp/pci011
Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39:629–642. doi:10.1111/j.1365-313X.2004.02161.x
Ticconi CA, Abel S (2004) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9:548–555. doi:10.1016/j.tplants.2004.09.003
Ticconi CA, Delatorre CA, Abel S (2001) Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol 127:963–972. doi:10.1104/pp.127.3.963
Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995) Composite structure of auxin response elements. Plant Cell 7:1611–1623. doi:10.1105/tpc.7.10.1611
Ulmasov T, Hagen G, Guilfoyle TJ (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276:1865–1868. doi:10.1126/science.276.5320.1865
Vanneste S, De Rybel B, Beemster GT, Ljung K, De Smet I, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M, Inzé D, Fukaki H, Beeckman T (2005) Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17:3035–3050. doi:10.1105/tpc.105.035493
Wang X, Yi K, Tao Y, Wang F, Wu Z, Jiang D, Chen X, Zhu L, Wu P (2006) Cytokinin represses phosphate-starvation response through increasing of intracellular phosphate level. Plant Cell Environ 29:1924–1935. doi:10.1111/j.1365-3040.2006.01568.x
Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43:118–130. doi:10.1111/j.1365-313X.2005.02432.x
Yu B, Xu C, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99:5732–5737. doi:10.1073/pnas.082696499
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (No. 18056007 and No. 20053005) from the Ministry of Education, Sports, Science and Culture in Japan. K.K. was supported by a research fellowship of the Japan Society for the Promotion of Science for young scientists. T.N. was supported by the Grant of the 21st Century COE Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Takafumi Narise and Koichi Kobayashi contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2009_9589_MOESM1_ESM.pdf
Table S1. Primers used for quantitative real-time PCR analysis. Primers for MGD1, MGD2, DGD1, DGD2, SQD1, SQD2, NPC4 and NPC5 were designed to avoid non-specific amplifications and primer dimerization. (PDF 27 kb)
Rights and permissions
About this article
Cite this article
Narise, T., Kobayashi, K., Baba, S. et al. Involvement of auxin signaling mediated by IAA14 and ARF7/19 in membrane lipid remodeling during phosphate starvation. Plant Mol Biol 72, 533–544 (2010). https://doi.org/10.1007/s11103-009-9589-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9589-4