Abstract
Nonsense-mediated decay (NMD) is a quality control mechanism that identifies and degrades aberrant mRNAs containing premature termination codons (PTC). NMD also regulates the expression of many wild-type genes. In plants, NMD identifies a stop codon as a PTC and initiates the rapid degradation of the transcript if the 3′untranslated region (UTR) is unusually long or if it harbors an intron. Approximately 20% of plant transcripts have an upstream ORF (uORF) in the 5′UTR. In theory, if a uORF is translated, the 3′UTR downstream of the uORF will be long and harbor introns, thus these transcripts might be degraded by NMD. Therefore, if uORFs can trigger NMD, uORF containing transcripts would be a major group of NMD regulated wild-type plant mRNAs. The aim of this study was to clarify whether plant uORFs could activate NMD. Here we demonstrate that plant uORFs induce NMD in a size-dependent manner, a 50 amino acid (aa) long uORF triggered NMD efficiently, whereas similar but shorter (31 and 15 aa long) uORFs failed to activate NMD response. We have found that only ~2% of annotated Arabidopsis genes contain a first uORF that is longer than 35 aa, thus we propose that NMD regulates only a small fraction of uORF containing transcripts. However, as mRNAs having uORF that is longer than the critical size are strongly overrepresented within the up-regulated transcripts of NMD deficient plants, it is likely that this subset of natural NMD targets induces NMD because of containing a relatively long translatable uORF.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Regulatory elements present in the 5′untranslated regions (5′UTR) play a crucial role in post-transcriptional gene regulation.
In eukaryotes, the 43S preinitiation complex interacts with the 5′ end of the mRNA and starts to scan the transcript for AUG codon. Start codon recognition allows joining of the large ribosomal subunit and the formation of the final 80S initiation complex (Kozak 2002).
Although this scanning model suggests that the eukaryotic translation initiates at the first AUG, many transcripts (20–40% of eukaryotic mRNAs) contain AUGs and/or short upstream ORFs (uORFs) in the 5′UTRs (Churbanov et al. 2005; Hayden and Jorgensen 2007; Hayden and Bosco 2008; Kawaguchi and Bailey-Serres 2005; Tran et al. 2008). uORFs can negatively regulate the expression of the major ORF by inhibiting its translation and/or by destabilizing the transcript. If uORFs are present in the 5′ leader sequence, the major ORF can be translated only if the scanning ribosomes do not initiate translation at first AUG instead they resume scanning and initiate translation at the start codon of the major ORF (leaky scanning). Alternatively, the major ORF could be expressed if translation is reinitiated after terminating the translation of uORF (Kozak 2005; Sachs and Geballe 2006). Both leaky scanning and reinitiation are supposed to be generally inefficient. Efficiency of the leaky scanning depends on the start codon context of uORF, while reinitiation efficacy depends on many factors including the length of uORF and the intercistronic region (the sequence between the stop of uORF and the start of major ORF). Reinitiation is less efficient after translation of longer ORFs compared with shorter ones. To reinitiate translation, the ribosome should reacquire initiation components while it is traversing the intercistronic region. Thus the longer intercistronic region allows more efficient reinitiation (Sachs and Geballe 2006). The transcript destabilization effects of uORFs are less understood. Protein products of uORFs might induce transcript degradation in cis or in trans. Translation of uORFs could also induce destabilization of mRNA by triggering nonsense-mediated mRNA decay (NMD; Lee and Schedl 2004; Mendell et al. 2004), a eukaryotic quality control mechanism that identifies and degrades mRNAs containing premature termination codons (PTC).
Recent “unified” model of NMD suggests that NMD recognizes a stop codon as a PTC and initiates the rapid decay of the transcript if the translation termination is inefficient (Muhlemann 2008; Neu-Yilik and Kulozik 2008; Rebbapragada and Lykke-Andersen 2009; Shyu et al. 2008; Silva and Romao 2009). This model proposes that efficient termination requires interaction of the eRF3 component of the terminating ribosome with the poly(A) binding protein (PABP). If the 3′UTR is unusually long, eRF3 fails to interact with PABP and instead it binds UPF1, which then recruits UPF2 and UPF3 NMD factors. The formation of the functional NMD complex triggers rapid degradation of the PTC containing transcript (Muhlemann 2008; Neu-Yilik and Kulozik 2008; Rebbapragada and Lykke-Andersen 2009; Shyu et al. 2008; Silva and Romao 2009). In mammals (but not in invertebrates or yeast), 3′UTR located introns can dramatically enhance the intensity of NMD. During splicing a protein complex called exon-junction complex (EJC) is deposited on the mRNA 20–25 nucleotides upstream of the exon–exon boundary (Le Hir et al. 2000). EJC serves as a binding platform for UPF3 and UPF2 NMD factors (Le Hir et al. 2001). The translating ribosomes remove EJC from the mRNA unless it locates in the 3′UTR. During mammalian translation termination a protein complex consisting of eRF1, eRF3, SMG-1 kinase and UPF1 NMD factor (called SURF complex) joins to the terminating ribosome. If an EJC is present on the 3′UTR, the UPF1 component of the SURF can interact with the EJC-bound UPF2. This interaction induces SMG-1 mediated phosphorylation of UPF1 leading to the formation of functional NMD complex and the rapid degradation of the transcript (Kashima et al. 2006).
The mechanistic details how NMD complex formation leads to the rapid degradation of the target transcript are not well understood. In yeast, NMD complex formation likely triggers deadenylation-independent decapping and accelerated deadenylation resulting in rapid mRNA decay (Cao and Parker 2003; Hagan et al. 1995; Mitchell and Tollervey 2003). In animals, UPF1 phosphorylation might be a key step that connects NMD complex formation and mRNA decay. SMG-5, -6 and -7, three related 14-3-3 like domain containing proteins can bind phosphorylated UPF1 and regulate the dephosphorylation of it by recruiting protein phosphatase 2A (PP2A). In addition, SMG-6 can cleave the NMD targeted mRNAs close to the PTC (Eberle et al. 2009; Gatfield and Izaurralde 2004; Huntzinger et al. 2008), while SMG-7 likely binds phospho-UPF1 and transports it with the associated PTC containing transcripts into the P-body, a specific compartment for mRNA decay (Fukuhara et al. 2005).
Little is known about plant NMD (reviewed in Belostotsky and Sieburth 2009; Chiba and Green 2009). Previously we and others have shown that in plants both long 3′UTR and 3′UTR located introns can trigger NMD efficiently (Hori and Watanabe 2007; Kertesz et al. 2006; Schwartz et al. 2006; Wu et al. 2007). The key NMD trans factors are conserved within eukaryotes, plant orthologs of UPF1, UPF2, UPF3 and SMG-7 (but not SMG-5, SMG-6 or SMG-1) can be found and these factors are required for plant NMD (Arciga-Reyes et al. 2006; Hori and Watanabe 2005; Kerenyi et al. 2008; Wu et al. 2007; Yoine et al. 2006). The basic mechanism of plant PTC identification and NMD complex formation could be similar to animal NMD systems (Kerenyi et al. 2008). The long 3′UTR-based plant NMD, like yeast and invertebrate NMD systems, measures the distance between the stop codon and PABP, indicating that efficient translation termination also requires interactions between PABP and terminating ribosomes in plants. Moreover, we have also provided evidence that intron-based plant NMD, like mammalian NMD, is mediated by an EJC-like complex (Kerenyi et al. 2008). The molecular mechanism of PTC containing plant transcript degradation is still not known.
In addition to eliminating PTC containing aberrant transcripts, plant NMD is also involved in the regulation of wild-type mRNAs, ~1 to 2% of transcripts are up-regulated in NMD deficient plants (Kurihara et al. 2009; Yoine et al. 2006). For instance, plant NMD is involved in the selective degradation of alternative splicing products, riboswitched transcripts or many noncoding mRNA-like RNAs (Hori and Watanabe 2005; Kurihara et al. 2009; Schoning et al. 2008; Wachter et al. 2007). Plant NMD is autoregulated, the 3′UTR of SMG-7 NMD factor is unusually long and contains two introns, thus it is directly targeted by NMD (Kerenyi et al. 2008).
NMD regulated genes can be grouped as indirect and direct targets. Indirect targets are genes, whose expression is controlled by an NMD targeted gene. Direct targets are genes, whose mRNAs contain NMD cis elements, hence their transcripts are degraded by NMD pathway. The two direct target groups of plant NMD are transcripts with long 3′UTR (longer than 3–400 nt) and mRNAs with 3′UTR located introns (Kertesz et al. 2006). We have postulated that uORF containing transcripts could be a third group of direct plant NMD targets. In theory, if a uORF is translated, the 3′UTR downstream of the uORF will be extremely long and harbor introns, thus it could be directly targeted by NMD. uORFs are present in ~20% of plant transcripts, therefore if plants uORFs can trigger NMD, NMD would play a very important role in the regulation of many wild-type plant mRNAs. However, it has not been experimentally tested if plant uORF can destabilize a transcript by triggering NMD. The aim of this study was to clarify whether plant NMD can be activated by uORFs.
Here we show that plant uORFs can trigger NMD in a size-dependent manner, 15 or 31 amino acid (aa) long uORFs were unable to trigger NMD, while a similar but longer (50 aa long) uORF induced NMD efficiently. Our data suggest that only small fraction of uORF containing plant transcripts are regulated by NMD.
Materials and methods
Plasmid constructs
For agroinfiltrations, genes were cloned into Bin61S binary vector or into the derivatives of Bin61S. P14, GFP, G-L and U1DN (previously referred to as UPF1DN) clones were previously described (Kerenyi et al. 2008; Kertesz et al. 2006). To create (5′U1-G, 5′U2-G, 5′U3-G, 5′U4-G, 5′U5-G) 5′UTR testing vectors, 5′UTR genomic segments were amplified from Arabidopsis thaliana DNA with primer pairs (1for1 + 1rev1, 2for1 + 2rev1, 3for1 + 3rev1, 4for1 + 4rev1, 5for1 + 5rev1) carrying KpnI sites (primer sequences are available upon request). These PCR fragments were cloned into GFP binary reporter vector upstream of GFP ORF.
NoATG-U2-G construct was generated by PCR mutagenesis from 5′U2-G. PCR fragments were generated with primer pairs mutating ATG (2for1 + uORFmutR and uORFmutF + 2rev1). These overlapping PCR products were annealed, primer extended, reamplified with 2for1 + 2rev1 primers and then cloned with KpnI into GFP reporter vector. Similar PCR mutagenesis was used to generate U2-noPstop-G construct (primers used: 2for1 + uO2-noProR and uO2no-ProF + 2rev1) from 5′U2-G. 31-U2-G (primers used: 2for1 + uORF31asR and uO2noProF + 2rev1) and 15-U2-G (primers used: 2for1 + uORF15asR and uO2noProF + 2rev1) constructs were generated from U2-noPstop-G. To test the effect of reduced reinitiation on NMD sensitivity reporter constructs with shortened intercistronic regions were generated by PCR mutagenesis. PCR fragments with short intercistronic sequences were generated with primer pair (2for1 + 2uORF2rovR) from 5′U2-G, 31-U2-G and 15-U2-G templates. These PCR fragments were digested with KpnI and cloned into GFP vector creating U2-s-G, 31-U2-s-G and 15-U2-s-G reporter constructs.
To generate reporter constructs lacking major ORF, a 439 nt PCR fragment was produced from 5′U1 region, which contained only a single ATG. This ATG was eliminated with PCR mutagenesis (primer used: 1uORFBamH1 F + NoATGR and NoATGF + 1uORFXba1 R), and then the obtained ATG-less PCR fragment was cloned and sequenced (noORF sequence). The noORF sequence as a BamHI-XbaI fragment was moved into Bin61s (Bin61SnoORF), and then the 5′UTR segments from 5′U2-G, 31-U2-G and 15-U2-G templates were cloned as KpnI fragments into KpnI cleaved Bin61SnoORF vector leading to generation of U2-noORF, 31-U2-noORF and 15-U2-noORF constructs.
Agroinfiltration assays
Agroinfiltrations and GFP detections were described (Silhavy et al. 2002). Leaves of wild-type plants or VIGS silenced N. benthamiana plants were agroinfiltrated with a mixture of cultures. The final OD of each culture was OD600 = 0.4 (except P14, which was OD = 0.2) in the mixture. The infiltrated leaves were studied at 3 days post inoculation (3 d.p.i.). Visual detection of GFP fluorescence was performed using a 100 W handhold long-wave ultraviolet lamp (UV products, Upland, CA 91786, Black Ray model B 100AP).
Virus-induced gene silencing
To trigger VIGS, leaves of ~21 days old greenhouse grown N. benthamiana plants were co-infiltrated with a mixture of three Agrobacterium cultures. One expressed P14, the second expressed TRV RNA1 (BINTRA6 vector; Ratcliff et al. 2001), while the third expressed TRV RNA2 (Liu et al. 2002; Valentine et al. 2004) containing a sequence from N. benthamiana PDS (PDS) or contained segments from N. benthamiana PDS and UPF1 (P-UPF1) or PDS and UPF2 (P-UPF2). PDS, P-UPF1 and P-UPF2 VIGS vectors were previously described (Kerenyi et al. 2008). VIGS induced (and/or agroinfiltrated) N. benthamiana plants were grown in a growth chamber under 16 h light (22°C)/8 h dark (18°C) cycles. At 9–10th days, when upper leaves of VIGS induced plants started to bleach (indicating that PDS silencing is efficient), leaves under the bleaching ones were agroinfiltrated with a mixture of cultures (see above).
RNA gel blot analysis
RNA methods and quantifications were described previously. PCR fragments labeled with random priming method were used for Northern analyses. Phosphorimagine measurements were used to quantify expression of mRNAs (Kertesz et al. 2006). RT-PCR was carried out with QIAGEN OneStep RT-PCR Kit.
Results
uORF containing 5′UTR segments could trigger plant NMD
Many uORF containing mRNAs are up-regulated in NMD deficient Arabidopsis mutant lines suggesting that these transcripts are negatively regulated by NMD (Kurihara et al. 2009). However, it is not known whether these transcripts are direct or indirect NMD targets. Moreover, even if they are direct NMD targets, it is possible that their 3′UTR segments instead of their uORFs induce NMD. To test whether uORFs can trigger NMD in plants, we have cloned the 5′UTRs of 5 uORF containing A. thaliana genes (5′UTR segment of At3g53400 is named 5′U1. Similarly, the 5′UTRs of At1g70780, At3g01060, At1g08730 and At3g18000 are named 5′U2, 5′U3, 5′U4 and 5′U5, respectively), which were overexpressed in NMD deficient mutants (Kurihara et al. 2009; Yoine et al. 2006). These 5′UTR segments were incorporated into a binary vector (GFP) between the 35S promoter and the GFP reporter gene (Fig. 1a) and then the effects of these leader regions on GFP expression were studied in agroinfiltration assays. Although all five genes encode at least one uORF, these uORF encoded proteins have not been experimentally detected. The deduced proteins of uORFs derived from At3g53400 (uORF1), from At1g70780 (uORF2) and from At3g18000 (uORF5) are well conserved between dicots and monocots. Therefore it is supposed that these uORFs are translated and that their protein products are functional (Hayden and Jorgensen 2007). By contrast, the putative protein products of the uORFs of At1g01060 (uORF3) and At3g08730 (uORF4) genes are not conserved suggesting that their protein products do not play evolutionally conserved physiological role.
Schematic representation of transcripts derived from the constructs used in this study. UTR segments are shown as lines, while ORFs are represented as rectangulars. Numbers under the lines shows the length of that segment in nucleotides. Dashed lines at noATG-U2-G and at different noORF constructs indicate that in these constructs the ORF was destroyed or was replaced by a start codon-less sequence. At VIGS constructs (P, P-UPF1 and P-UPF2) the lines represent TRV2 RNAs while the rectangular shows the plant genes from which segments were cloned into TRV2 VIGS vector. Note that the drawings are not proportional. For details of the constructs see the text
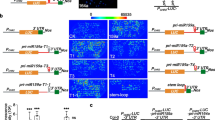
To test the effect of these different 5′UTR leaders on GFP expression, the reporter constructs were agroinfiltrated into N. benthaminana leaves (the construct carrying the 5′U1 region upstream of GFP is referred to as 5′U1-G. Similarly, 5′U2-G, 5′U3-G, 5′U4-G, 5′U5-G constructs contained 5′U2, 5′U3, 5′U4 and 5′U5 regions, respectively). The GFP construct was infiltrated as a control. As agroinfiltration induces RNA silencing response, P14 suppressor that inhibits silencing but does not interfere with NMD (Kertesz et al. 2006) was co-infiltrated with each construct (P14 was also co-infiltrated in all further experiments but it will not be mentioned later in the main text). GFP expression was strongly reduced in 5′U1-G, 5′U2-G, 5′U3-G and 5′U4-G infiltrated leaves compared to the GFP control leaves. By contrast, the green fluorescence of the 5′U5-G infiltrated leaf was only slightly weaker than the fluorescence of the GFP infiltrated control leaf (Fig. 2a, compare the right sides of the different leaves). These data suggest that the 5′U1, 5′U2, 5′U3 and 5′U4 can reduce the expression of the major gene, while 5′U5 might influence only slightly on it. 5′UTRs could inhibit the expression of the major gene by many different ways including transcript destabilization. To test whether the negative effect of the 5′UTRs on GFP expression is (at least partly) due to NMD, the reporter constructs were co-infiltrated with a dominant-negative mutant of UPF1 NMD factor (U1DN). Previously we have demonstrated that co-infiltration of U1DN leads to the inactivation of NMD in the infiltrated patch thereby enhancing the accumulation of co-expressed NMD target transcripts (Kertesz et al. 2006). If 5′UTR reporter constructs are targeted by NMD, their mRNA levels will be increased in U1DN co-infiltrated leaves. Co-infiltration of U1DN did not lead to significantly enhanced accumulation of 5′U3-G or 5′U5-G mRNAs suggesting that these UTRs were not down-regulated by NMD (Fig. 2b). By contrast, the mRNA levels of 5′U1-G, 5′U2-G and 5′U4-G were significantly increased in U1DN co-infiltrated leaves suggesting that 5′U1, 5′U2 and 5′U4 leader regions are negatively regulated by plant NMD (Fig. 2b).
Cloning of Arabidopsis 5′UTRs upstream of GFP could reduce the expression of the reporter gene. a The effects of different 5′UTRs on fluorescence of GFP reporter gene. The right sides of N. benthamiana leaves were co-infiltrated with P14 and with GFP control or with 5′U1-G, 5′U2-G, 5′U3-G, 5′U4-G and 5′U5-G test constructs containing the corresponding 5′UTR segment cloned upstream of GFP. The left sides of the leaves were co-infiltrated with P14, with U1DN (a mutant version of UPF1 NMD factor, which inhibits NMD in a dominant-negative manner) and with the control or test construct, which were used to infiltrate the right side of the same leaves. The U1DN co-infiltrated samples are referred to as U1DN treated samples (U1DN), while the samples at the right sides are referred to as non-treated samples (−). Note that P14 was co-infiltrated with each cultures but it is not shown on the photo. To evaluate the effect of 5′UTR on GFP expression, fluorescence of the GFP control and the different test constructs should be compared. UV pictures were taken at 3 days post inoculation (d.p.i.). b Inhibition of NMD leads to enhanced accumulation of certain 5′UTR containing GFP reporter mRNAs. RNA gel blot assays were carried out with samples isolated at 3 d.p.i. from leaves infiltrated with test constructs. Labeled GFP and P14 fragments were used as a probe. For each test constructs two samples are shown, the non-treated (−) and the U1DN treated (U1DN) pairs derived from the right and left sides of the same leaf. The lower bands of each RNA blot show P14 transcripts, while the upper bands represent the different test mRNAs. To quantify RNA samples, at each lane the signal intensity of the upper band was normalized to the corresponding P14 mRNA. To calculate the effect of U1DN co-infiltration, the normalized test mRNA levels of U1DN treated (U1DN) samples are compared to the levels of the corresponding non-treated samples. Enhanced accumulation of the test mRNA in U1DN treated samples indicates that the transcript is negatively regulated by NMD. Bold numbers show the fold change of normalized test transcript levels in the U1DN treated samples relative to the corresponding non-treated (−) samples. Mean values were calculated from three independent samples from which two are shown and the third was run on a separate gel. ± Indicates the standard deviation of the experiment
These data suggest that certain but not all plant uORF containing 5′UTR could induce NMD. Next we wanted to test what features of a 5′UTR define whether it can activate NMD or not. For further studies we selected the 5′UTR segment of At1g70780 (5′U2), because it triggered NMD very efficiently and because the structure of this leader region is as simple as possible, 5′U2 contains only a single uORF (referred to as uORF2), it has only one AUG (the start codon of the uORF2) and it does not harbor intron.
uORF2 translation activates NMD
5′U2-G mRNAs accumulated to increased levels in U1DN co-infiltrated leaves suggesting that 5′U2-G is negatively regulated by NMD. However, UPF1 is also involved in other RNA metabolic pathways including RNA silencing (Arciga-Reyes et al. 2006; Isken and Maquat 2008; Mango 2001), thus U1DN co-infiltration might enhance 5′U2-G mRNA levels because U1DN inhibited NMD or because it modified another RNA metabolic pathway. Therefore, we first wanted to confirm that 5′U2-G mRNA is a genuine NMD target. If 5′U2-G is down-regulated by NMD, it should overaccumulate in plants lacking either UPF1 or UPF2 NMD trans factors. To test this, UPF1 and UPF2 (Fig. 1d) were inactivated by virus-induced gene silencing (VIGS), and then G-L (Fig. 1a), a previously characterized NMD reporter construct, or 5′U2-G test construct was agroinfiltrated into the leaves of UPF1 or UPF2 silenced plants (P-UPF1 and P-UPF2, respectively). Previously we have shown that in P-UPF1 and P-UPF2 silenced leaves the UPF1 or UPF2 transcript levels are significantly reduced (25–50% of wild-type levels) and that the NMD system does not function in either P-UPF1 or P-UPF2 silenced leaves (Kerenyi et al. 2008). As a negative control, leaves of phytoene desaturase (P) silenced plants were infiltrated with G-L and 5′U2-G constructs. G-L was strongly overexpressed in P-UPF1 and P-UPF2 leaves relative to P control leaves (Fig. 3a), thus we concluded that both UPF1 and UPF2 silencing were effective. Importantly, 5′U2-G transcripts accumulated to significantly higher levels in UPF1 and UPF2 silenced leaves than in the PDS silenced control leaves strongly supporting that 5′U2-G is a genuine NMD target (Fig. 3a).
5′U2-G is a genuine NMD target. a 5′U2-G is overexpressed in plants lacking UPF1 or UPF2 NMD trans factor. 5′U2-G test construct and G-L NMD reporter construct, which has a 600 nt stuffer sequence cloned into the 3′UTR of GFP reporter construct, were co-infiltrated with P14 into PDS (P), PDS + UPF1 (P-UPF1) or PDS + UPF2 (P-UPF2) co-silenced leaves. The fluorescence of a construct in P control leaves and in P-UPF1 and P-UPF2 leaves should be compared. The strong fluorescence of G-L NMD reporter construct in P-UPF1 and P-UPF2 leaves indicate that the NMD factors were effectively silenced. RNA gel blot assay show that both G-L NMD reporter mRNAs and 5′U2-G test transcripts are overexpressed in both P-UPF1 (P-U1) and P-UPF2 (P-U2) silenced leaves relative to the PDS silenced control leaves. Labeled GFP and P14 fragments were used as a probe. RNA quantification and comparisons were described at Fig. 2. Bold numbers show the fold change of normalized test transcript levels in P-UPF1 or P-UPF2 silenced leaves relative to the corresponding transcript in P silenced control samples. b uORF2 translation is required for NMD mediated transcript degradation. 5′U2-G control and noATG-U2-G test constructs were co-infiltrated with P14 only (−) or were co-infiltrated with P14 and with U1DN (U1DN) into N. benthamiana leaves. Note that P14 was co-infiltrated but it is not shown on figure. RNA gel blot assay were conducted as described at Fig. 2. The right panel shows that proline-stop is not essential for NMD of 5′U2-G transcript as U2-noPstop-G construct is still efficiently targeted by NMD
5′U2-G can be a direct or an indirect NMD target. As NMD is a translation coupled RNA degradation system, if 5′U2-G is a direct target of NMD, the translation of the uORF should be required for NMD induction. To test this, we have removed the start codon of the uORF2 (noATG-U2-G, Fig. 1b), and then we studied the expression of the noATG-U2-G construct in agroinfiltration assay. The green fluorescence of the noATG-U2-G infiltrated leaf was very intense suggesting that the translation of the uORF2 is required to reduce the expression of the major gene. More importantly, co-infiltration of U1DN did not effect on the accumulation of noATG-U2-G mRNA levels indicating that noATG-U2-G was not targeted by NMD (Fig. 3b, middle panels). As the translation of uORF2 is required for NMD mediated down-regulation of 5′U2-G, we propose that 5′U2-G is a direct target of plant NMD. Next, we have asked what features of uORF2 are required for inducing NMD.
The specific feature of uORF2 is that it contains a proline-stop. In bacteria, if the last amino acid upstream of the stop codon is a proline (proline-stop), the translation termination will be ineffective leading to the rapid degradation of mRNA (Hayes et al. 2002). As the current NMD model suggests that inefficient termination could induce NMD in eukaryotes, we postulated that the proline-stop of uORF2 interferes with translation termination thereby triggering NMD. To test this hypothesis, we changed the last proline to a glutamine (U2-noPstop-G, Fig. 1b), and then studied in U1DN co-infiltration assays whether U2-noPstop-G is still NMD sensitive. As U2-noPstop-G was overexpressed in U1DN co-infiltrated leaves (Fig. 3b, right panels), we concluded that proline-stop did not play an essential role in NMD sensitivity of uORF2.
uORF2 activates NMD in a size-dependent manner
In mammals, mRNAs containing PTC close to the start codon frequently resistant to NMD (Silva et al. 2008). When the lengths of the predicted uORFs were compared, we found that the NMD activating 5′UTRs might encode longer protein than the non-NMD inducer 5′UTRs. For instance, uORF2 could express a 50 aa peptide, uORF1 might encode a 38 aa long peptide, whereas the predicted peptide product of uORF5 is only 29 aa long (Hayden and Jorgensen 2007). Therefore we postulated that in plants uORF could activate NMD in a size-dependent manner. To test this hypothesis, we created two constructs from 5′U2-G in which the length of uORF was reduced from 50 aa to 31 (31-U2-G, Fig. 1b) or to 15 aa (15-U2-G, Fig. 1b) by removing the middle region of the uORF. The sequences upstream of the uORFs and the intercistronic sequences were identical in the three constructs.
To study the effect of the length of uORF2 on gene expression, 31-U2-G and 15-U2-G test constructs and 5′U2-G control construct were agroinfiltrated. We found that the green fluorescences of the 31-U2-G and especially of the 15-U2-G infiltrated leaves were enhanced relative to 5′U2-G control leaves (Fig. 4a, compare the right sides of the leaves). To test the effect of NMD, we compared the expression of 5′U2-G, 31-U2-G and 15-U2-G constructs when they were infiltrated alone or were co-infiltrated with U1DN. As Fig. 4b shows, co-infiltration of U1DN resulted in strongly increased expression of 5′U2-G transcripts but failed to enhance the accumulation of either the 31-U2-G or the 15-U2-G mRNA (Fig. 4b, compare lanes 1 to 2, 7 to 8 and 13 to 14). These data suggest that, unlike 5′U2-G transcripts, the 31-U2-G or the 15-U2-G mRNAs were not targeted by NMD, they were NMD resistant. Therefore we concluded that uORFs induce NMD in a size-dependent manner in plants, short uORFs do not trigger NMD, whereas longer uORFs can induce NMD efficiently. Next we wanted to study why short uORF containing transcripts were resistant to NMD.
Plant uORFs trigger NMD in a size-dependent manner. a–c Short uORF containing transcripts are not protected from NMD by reinitiation. a, b 5′U2-G, 15-U2-G and 31-U2-G control constructs and their derivatives (U2-s-G, 15-U2-s-G and 31-U2-s-G) with shortened intercistronic segments (reduced reinitiation capacity) were co-infiltrated with P14 (−) only (a and the uneven lanes at b) or were co-infiltrated with P14 and with U1DN (U1DN) into N. benthamiana leaves (the even lanes at b). Note that 5′U2-G was used as a control on the RNA gel blots at (b) the middle and the right panels, therefore only one sample was run. Thus, in these experiments 5′U2-G quantifications were not possible. c uORF2 triggers NMD size-dependently even if the major ORF is eliminated. U2-noORF, 15-U2- noORF and 31-U2- noORF test constructs, in which the GFP major ORF was replaced with a sequence lacking start-codons, were co-infiltrated with P14 only (−) or were co-infiltrated with P14 and with U1DN (U1DN) into N. benthamiana leaves, and then RNA samples were isolated and gel blot assayed
Reinitiation is not required for NMD evasion of short uORF containing transcript
At least three different mechanisms can protect short, translatable uORF containing transcripts from NMD. In yeast, uORF harboring mRNAs having stabilizing elements in the 5′UTR downstream of the uORF are resistant to NMD (Ruiz-Echevarria and Peltz 2000). It has been shown that in mammals, efficient reinitiation of a downstream ORF can protect early PTC containing mRNAs from NMD (Zhang and Maquat 1997). Finally, it has also been proposed that mammalian mRNAs containing PTC close to the start codon are resistant to NMD because components of the translation initiation complex are only gradually lost during elongation, thus eIF4G and the interacting PABP could still be connected to the ribosome at the termination after translating a short ORF (Silva et al. 2008). Consequently, PABP can interact with the terminating ribosome thereby the translation termination will be efficient and the transcript is not subjected to NMD (referred to as efficient termination model).
It is unlikely that a stabilizing sequence protected 31-U2-G and 15-U2-G mRNAs from NMD because the intercistronic regions of these transcripts and the NMD sensitive 5′U2-G mRNAs are identical. By contrast, both reinitiation and efficient termination models can explain the NMD resistance of short uORF containing transcripts. Reinitiation efficiency depends on the length of uORF and the size of intercistronic region (Kozak 2002; Sachs and Geballe 2006). Generally the reinitiation is more efficient if the translated ORF is short. Therefore reinitiation could be more efficient for 31-U2-G and 15-U2-G than for 5′U2-G. Indeed, GFP expression was more intense in 31-U2-G and in 15-U2-G infiltrated leaves than in 5′U2-G infiltrated leaves. To test whether efficient reinitiation protects 31-U2-G and 15-U2-G mRNAs from NMD, similar constructs with reduced reinitiation efficiency have been created by shortening the intercistronic sequences from 106 to 20 nt (named as U2-s-G, 15-U2-s-G and 31-U2-s-G, Fig. 1c). These new constructs were agroinfiltrated alone or were co-infiltrated with U1DN. Shortening of the intercistronic region likely reduced reinitiation because GFP expression of U2-s-G, and 31-U2-s-G infiltrated leaves was weaker than the fluorescence of leaves, which were infiltrated with the same constructs having longer intercistronic segments (Fig. 4a, compare the two sides of the leaves). Importantly, while U1DN co-infiltration led to overexpression of U2-s-G transcripts, co-expression of U1DN did not enhance the expression of either 15-U2-s-G or 31-U2-s-G transcripts (Fig. 4b, compare lanes 3 to 4, 9 to 10 and 15 to 16). These data suggest that although reinitiation was inefficient when the intercistronic sequence was shortened (at least at the 31-U2-s-G transcripts), 31-U2-s-G (and 15-U2-s-G) mRNAs were still NMD resistant. Therefore it is unlikely that reinitiation protected short uORF containing transcripts from plant NMD. To further confirm that reinitiation is not required for NMD evasion, GFP regions of the 5′U2-G, 31-U2-G and 15-U2-G constructs were replaced with 439 nt long sequences lacking AUG (U2-noORF, 31-U2-noORF and 15-U2-noORF, Fig. 1c). As in these constructs, AUGs downstream of the uORFs are present only in the 3′UTR sequence derived from the 35S terminator region and because these AUGs are not present in favorable contexts, the reinitiation of these transcripts should be inefficient. To test whether these mRNAs are targeted by NMD, the U2-noORF, 31-U2-noORF and 15-U2-noORF reporter constructs were co-infiltrated with U1DN. We have found that U2-noORF mRNAs were NMD sensitive, while 31-U2-noORF and 15-U2-noORF transcripts were NMD resistant (Fig. 4c, compare lanes 1 to 2, 3 to 4 and 5 to 6).
Taken together, our data show that short uORFs (15–31 aa long) fail to trigger NMD independently whether the reinitiation is effective or inefficient, suggesting that reinitiation is not required for NMD resistance of short uORF containing transcripts. As neither stabilizing sequences nor efficient reinitiation can explain the NMD resistance of short uORF harboring mRNAs, we propose that in plants, like in mammals, the translation of short ORFs is terminated efficiently independently from the length of 3′UTR. Consequently, uORFs induce NMD in a size-dependent manner, short uORF do not trigger NMD, whereas longer uORFs induce NMD response efficiently.
Only a small set of uORF containing plant transcripts could be directly regulated by NMD
Approximately 20% of plant mRNAs contain at least one uORF suggesting that a large fraction of plant transcriptome could be regulated by NMD (Hayden and Jorgensen 2007; Kawaguchi and Bailey-Serres 2005). However, as we have found that uORFs can induce plant NMD only if the translated uORF is longer than ~30 to 40 aa (Figs. 2, 3; “Discussion”), we postulate that NMD could directly target only a subset of uORF containing plant mRNAs. To estimate the potential gene set, whose transcripts contain NMD inducing (“NMD critical”) uORF, we have analyzed a 5′UTR database (www.abc.hu/RNA/orf.1.html) that was created by using the annotation of the NCBI 2005 assembly of A. thaliana (Kertesz et al. 2006). We have postulated that an NMD critical uORF should be efficiently translated, thus it should be the first ORF on the transcript and its start codon should be at least 10 nt from the 5′ end. Moreover, the uORF should be longer than ~35 aa. From the 16,800 5′UTRs, 3,928 (23.4%) contained at least one uORF but only 679 (4%) harbored uORF that was longer than 35 aa. 736 genes (3.6% of the genes) contained uORF, which was longer than 35 aa and the start codon was more than 10 nt from the 5′ end. 331 (2%) genes harbored a first uORF that was longer than 35 aa and the start codon was at least 10 nt from the 5′ end. These data suggest that the strictest constrain is the size-limitation of the uORF.
Recently, protein coding genes that were up-regulated (97 genes) in both upf1 and upf3 mutant Arabidopsis lines have been identified (Kurihara et al. 2009). 80 out of these 97 genes were also represented in our 5′UTR database. Importantly, 12 out of the 80 (15%) contained a NMD critical uORF. As NMD critical uORFs are present only in 2% of genes in the whole database but are present in 15% of genes which are up-regulated in both NMD mutants, we concluded that genes containing NMD critical uORFs are strongly overrepresented (~7.5 times) within the NMD down-regulated gene set. If we have looked for the representation of first uORFs that are shorter than 20 aa but longer than 10 aa, with a start codon at least 10 nt from the 5′ end, we found relatively mild overrepresentation (2.6 times) within the NMD down-regulated genes. Similarly, Kurihara et al. (2009) has also reported that uORF containing transcripts were only slightly overrepresented (~2 times) within the NMD down-regulated genes.
Taken together, as our result suggest that only relatively long uORFs can trigger plant NMD and that only a small fraction of mRNAs harbor NMD critical uORFs, we concluded that only a small set of uORF containing genes could be directly targeted by NMD. However, our data also revealed that many plant transcripts that contain NMD critical uORFs are down-regulated by NMD.
Discussion
Previously we have shown that plant NMD targets two types of mRNAs, transcripts with long 3′UTR and mRNAs harboring 3′UTR located introns. Here we have shown that plant NMD can also efficiently degrade transcripts having a translatable uORF. Importantly, uORFs induce NMD in a size-dependent manner, short uORFs failed to trigger NMD whereas a similar but 50 aa long uORF induced NMD efficiently.
Size-dependent uORF induced NMD
Many lines of evidence support that plant NMD can target uORF containing mRNAs. Genes having uORF were significantly overrepresented within the up-regulated transcripts in NMD deficient plants (Kurihara et al. 2009). Here we have provided different experimental evidence that translation of uORF can subject plant transcripts to NMD RNA degradation pathway (Figs. 2, 3).
Importantly, we have found that plant uORFs are size-dependent NMD cis elements as length reduction of the uORF2 from 50 to 31 aa abolished the capacity of uORF2 to trigger NMD. Although it has been reported that efficient reinitiation can rescue PTC containing transcripts in mammals (Zhang and Maquat 1997), impairment of reinitiation either by shortening the intercistronic region or by replacing GFP with a sequence lacking start codon did not alter the NMD resistance of the short uORF containing mRNAs in plants. Thus it is unlikely that reinitiation protects short uORF containing mRNAs from plant NMD. Instead we propose that short uORFs do not trigger NMD because their translation termination is efficient, while longer uORFs induce NMD because their translation termination is inefficient. In mammals, ß-globin reporter transcripts are NMD resistant when the PTC is introduced at codon 15, whereas ß-globin mRNA with a PTC at codon 39 is NMD sensitive. It is proposed that in mammals early PTCs do not trigger NMD because eIF4G and the bound PABP are released from the ribosome only gradually during elongation, therefore PABP can interact with the eRF3 component of the terminating ribosome if a short ORF but not if a longer ORF is translated (Silva et al. 2006, 2008). Mammalian NMD occurs only on transcripts associated with CBP20-80 cap binding complex at the pioneer round of translation (Hosoda et al. 2005; Ishigaki et al. 2001), hence eIF4G and the associated PABP might cooperate with CBP complex to facilitate translation termination of uORF containing mRNAs. We postulate that in plants, like in mammals, the translation is terminated efficiently if the ORF is very short because PABP could still interact with terminating ribosome, thus it facilitates termination preventing the formation of NMD complex. However, CBP complex is not required for plant NMD (Dzikiewicz-Krawczyk et al. 2008), therefore it is likely that in plants, eIF4E-bound eIF4G and the associated PABP can provide efficient termination for the short uORF. The critical length for efficient translation termination in plants could be around 30–40 aa because 31 aa long uORF failed to induce NMD, whereas the 5′U1 leader that encoded a 38 aa long predicted peptide (Hayden and Jorgensen 2007) likely triggered NMD. As a rough estimation we use 35 aa as the critical length of an uORF to trigger NMD. In mammals it has been shown that the time required for the translation of the ORF instead of the length of uORF is critical, shorter ORFs with stronger secondary structure were still NMD sensitive (Silva et al. 2008). This suggests that in certain cases shorter uORFs could activate NMD and in other cases longer uORF containing transcript could be NMD resistant.
Our result that short uORFs fail to activate plant NMD also predicts that plant transcripts with very early PTCs are not targeted by NMD. Indeed, it is reported that very early PTCs do not subject plant mRNAs to NMD (Hori and Watanabe 2007; Wu et al. 2007). However, PTC at the third codon of the GFP reporter gene could activate plant NMD (Schwartz et al. 2006). It is possible, that in the latter case the reinitiation is very efficient after translation of such an extremely short ORF leading to the efficient translation of another longer, NMD inducer ORF within the GFP reporter gene.
The NMD regulated uORF containing transcripts
Based on our experimental data, we have roughly defined the requirements for an NMD critical uORF as an at least 35 aa long first ORF whose start codon is more than 10 nt from the 5′ end. Bioinformatical analyses revealed that relatively few genes (2%) harbor NMD critical uORF indicating that only a small set of uORF containing transcripts are directly regulated by NMD. Moreover, it is likely that many of these NMD critical uORFs are translated inefficiently. These data explain the apparently conflicting findings that ~20% of the plant genes contain uORF but only 1–2% of the genes are up-regulated in NMD mutants. However, as genes having NMD critical uORF are strongly overrepresented (7.5 times) within the up-regulated genes of NMD mutants, we propose that plant NMD directly targets a number of mRNAs because they contain NMD critical uORF. Moreover, it is likely that certain genes, which are annotated not to contain uORF, could generate alternative transcript isoforms harboring NMD critical uORFs. For instance, alternative transcript isoforms with an NMD inducing uORF could be generated by alternative transcriptional start site usage, by alternative splicing within the 5′UTR or by translating a uORF from non-AUG initiation codon. However, the frequency and biological importance of these potential NMD target alternative mRNA isoforms cannot be estimated yet.
Abbreviations
- NMD:
-
Nonsense-mediated mRNA decay
- uORF:
-
Upstream ORF
- VIGS:
-
Virus-induced gene silencing
- UTR:
-
Untranslated region
- EJC:
-
Exon-junction complex
- eRF:
-
Eukaryotic releasing factor
References
Arciga-Reyes L, Wootton L, Kieffer M, Davies B (2006) UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J 47:480–489
Belostotsky DA, Sieburth LE (2009) Kill the messenger: mRNA decay and plant development. Curr Opin Plant Biol 12:96–102
Cao D, Parker R (2003) Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113:533–545
Chiba Y, Green P (2009) mRNA degradation machinery in plants. J Plant Biol 52:114–124
Churbanov A, Rogozin IB, Babenko VN, Ali H, Koonin EV (2005) Evolutionary conservation suggests a regulatory function of AUG triplets in 5′-UTRs of eukaryotic genes. Nucleic Acids Res 33:5512–5520
Dzikiewicz-Krawczyk A, Piontek P, Szweykowska-Kulinska Z, Jarmolowski A (2008) The nuclear cap-binding protein complex is not essential for nonsense-mediated mRNA decay (NMD) in plants. Acta Biochim Pol 55:825–828
Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH (2009) SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16:49–55
Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E (2005) SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17:537–547
Gatfield D, Izaurralde E (2004) Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429:575–578
Hagan KW, Ruiz-Echevarria MJ, Quan Y, Peltz SW (1995) Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol 15:809–823
Hayden CA, Bosco G (2008) Comparative genomic analysis of novel conserved peptide upstream open reading frames in Drosophila melanogaster and other dipteran species. BMC Genomics 9:61
Hayden CA, Jorgensen RA (2007) Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol 5:32
Hayes CS, Bose B, Sauer RT (2002) Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem 277:33825–33832
Hori K, Watanabe Y (2005) UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J 43:530–540
Hori K, Watanabe Y (2007) Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol 48:1072–1078
Hosoda N, Kim YK, Lejeune F, Maquat LE (2005) CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol 12:893–901
Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E (2008) SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. Rna 14:2609–2617
Ishigaki Y, Li X, Serin G, Maquat LE (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106:607–617
Isken O, Maquat LE (2008) The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet 9:699–712
Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20:355–367
Kawaguchi R, Bailey-Serres J (2005) mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res 33:955–965
Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D (2008) Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J 27:1585–1595
Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D (2006) Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res 34:6147–6157
Kozak M (2002) Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1–34
Kozak M (2005) Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13–37
Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, Toyoda T, Shinozaki K, Seki M (2009) Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc Natl Acad Sci USA 106:2453–2458
Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19:6860–6869
Le Hir H, Gatfield D, Izaurralde E, Moore MJ (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20:4987–4997
Lee MH, Schedl T (2004) Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev 18:1047–1059
Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31:777–786
Mango SE (2001) Stop making nonSense: the C. elegans smg genes. Trends Genet 17:646–653
Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36:1073–1078
Mitchell P, Tollervey D (2003) An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′–>5′ degradation. Mol Cell 11:1405–1413
Muhlemann O (2008) Recognition of nonsense mRNA: towards a unified model. Biochem Soc Trans 36:497–501
Neu-Yilik G, Kulozik AE (2008) NMD: multitasking between mRNA surveillance and modulation of gene expression. Adv Genet 62:185–243
Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Technical advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25:237–245
Rebbapragada I, Lykke-Andersen J (2009) Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol 21:394–402
Ruiz-Echevarria MJ, Peltz SW (2000) The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101:741–751
Sachs MS, Geballe AP (2006) Downstream control of upstream open reading frames. Genes Dev 20:915–921
Schoning JC, Streitner C, Meyer IM, Gao Y, Staiger D (2008) Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res 36:6977–6987
Schwartz AM, Komarova TV, Skulachev MV, Zvereva AS, Dorokhov Iu L, Atabekov JG (2006) Stability of plant mRNAs depends on the length of the 3′-untranslated region. Biochemistry (Mosc) 71:1377–1384
Shyu AB, Wilkinson MF, van Hoof A (2008) Messenger RNA regulation: to translate or to degrade. EMBO J 27:471–481
Silhavy D, Molnar A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyan J (2002) A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J 21:3070–3080
Silva AL, Romao L (2009) The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision? FEBS Lett 583:499–505
Silva AL, Pereira FJ, Morgado A, Kong J, Martins R, Faustino P, Liebhaber SA, Romao L (2006) The canonical UPF1-dependent nonsense-mediated mRNA decay is inhibited in transcripts carrying a short open reading frame independent of sequence context. Rna 12:2160–2170
Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L (2008) Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. Rna 14:563–576
Tran MK, Schultz CJ, Baumann U (2008) Conserved upstream open reading frames in higher plants. BMC Genomics 9:361
Valentine T, Shaw J, Blok VC, Phillips MS, Oparka KJ, Lacomme C (2004) Efficient virus-induced gene silencing in roots using a modified tobacco rattle virus vector. Plant Physiol 136:3999–4009
Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR (2007) Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19:3437–3450
Wu J, Kang JH, Hettenhausen C, Baldwin IT (2007) Nonsense-mediated mRNA decay (NMD) silences the accumulation of aberrant trypsin proteinase inhibitor mRNA in Nicotiana attenuata. Plant J 51:693–706
Yoine M, Ohto MA, Onai K, Mita S, Nakamura K (2006) The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J 46:49–62
Zhang J, Maquat LE (1997) Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J 16:826–833
Acknowledgments
We are grateful to C. Lacomme (University of Edinburgh), D. Baulcombe (University of Cambridge) and S. P. Dinesh-Kumar (Yale University) for TRV vectors. This research was supported by grants from the OTKA (K60102 and C77086) and from the National Office for Research and Technology (4/064/2004, “Búzakalász”). We thank E. Barta (University of Debrecen) for his help with bioinformatical studies. D. Silhavy was supported by the Bolyai János Scholarship. T. Nyikó and Zs. Mérai are graduate students of ELTE “Classical and Molecular Genetics” PhD program. A. Hangyáné Benkovics is a graduate student of Corvinus University of Budapest (“Program of Viticulture” at the Doctoral School of Horticulture).
Author information
Authors and Affiliations
Corresponding author
Additional information
Tünde Nyikó and Boglárka Sonkoly contributed equally to this work.
Rights and permissions
About this article
Cite this article
Nyikó, T., Sonkoly, B., Mérai, Z. et al. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol Biol 71, 367 (2009). https://doi.org/10.1007/s11103-009-9528-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-009-9528-4