Abstract
The Arabidopsis thaliana Tonoplast Intrinsic Protein 1;1 (AtTIP1;1) is a member of the tonoplast aquaporin family. The tissue-specific expression pattern and intracellular localization of AtTIP1;1 were characterized using GUS and GFP fusion genes. Results indicate that AtTIP1;1 is expressed in almost all cell types with the notable exception of meristematic cells. The highest level of AtTIP1;1 expression was detected in vessel-flanking cells in vascular bundles. AtTIP1;1-GFP fusion protein labelled the tonoplast of the central vacuole and other smaller peripheral vacuoles. The fusion protein was not found evenly distributed along the tonoplast continuum but concentrated in contact zones of tonoplasts from adjacent vacuoles and in invaginations of the central vacuole. Such invaginations may result from partially engulfed small vacuoles. A knockout mutant was isolated and characterized to gain insight into AtTIP1;1 function. No phenotypic alteration was found under optimal growth conditions indicating that AtTIP1;1 function is not essential to the plant and that some members of the TIP family may act redundantly to facilitate water flow across the tonoplast. However, a conditional root phenotype was observed when mutant plants were grown on a glycerol-containing medium.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Water flux across cellular membranes has been shown to occur not only through the lipid bilayer, but also through proteinaceous water channels named aquaporins (AQPs) (Agre et al. 1993; Maurel 1997). Aquaporins are members of the Major Intrinsic Protein (MIP) family of integral membrane proteins. These proteins mediate the bidirectional flux of water and small solutes such as glycerol, urea, ammonia and carbon dioxide across membranes (Maurel 2007). They have been identified in almost all organisms but are especially abundant in plants. Plant MIPs are phylogenetically divided into four subfamilies: Plasma membrane Intrinsic Proteins (PIPs), Tonoplast Intrinsic Proteins (TIPs), Nodulin 26-like Intrinsic Proteins (NIPs), and Small basic Intrinsic Proteins (SIPs) (Johanson et al. 2001).
TIPs represent a subfamily of multifunctional MIPs which are named in accordance with their proven or putative subcellular localization, i.e. the tonoplast. In Arabidopsis, ten TIP isoforms have been identified and classified into five distinct groups (TIP1 to TIP5) (Johanson et al. 2001). Isoforms of the first three groups have been the most frequently investigated in other plant species and are known to reside in the tonoplast of different types of vacuoles inside the cell (Jauh et al. 1998; 1999; Jiang et al. 2000; Moriyasu et al. 2003).
Expression studies, as well as studies using AQP inhibitors such as mercurial compounds, have indicated that AQPs fulfil two main functions in plants, namely cell osmoregulation and control of transcellular water flow (Tyerman et al. 1999; Maurel et al. 2002). Investigations into the function of specific isoforms constitute a crucial goal for the molecular understanding of water relations in plants. Overexpression of a specific AQP or suppression of its expression are two means to unravel its functions in planta but they have been rarely used to investigate the implication of TIPs in plant growth and development. In tobacco suspension cells, overexpression of cauliflower TIP1;1 correlated with an increase in the cell size (Reisen et al. 2003). In Arabidopsis plants, overexpression of a tonoplast aquaporin from Panax ginseng (PgTIP1) increased growth rate (Lin et al. 2007). Transgenic plants had longer roots, larger leaves and leaf cells, and set larger seeds than wild-type plants. These two studies provide evidence that TIPs might be involved in water-driven cell enlargement by modulating the permeability of the tonoplast. Overexpression of PgTIP1 was also shown to alter salt tolerance, drought tolerance and cold acclimation ability (Peng et al. 2007). In Arabidopsis, the physiological role of AtTIP1;1, also known as γ-TIP (Höfte et al. 1992), was investigated using RNA interference (Ma et al. 2004). This work suggested that down-regulation of AtTIP1;1 leads to cell and plant death. Metabolite profiling in senescing plants and AtTIP1;1 subcellular localization in healthy plants led the authors to conclude that AtTIP1;1 is involved in vesicle-based metabolite routing between pre-vacuolar compartments and the central vacuole (Ma et al. 2004).
To gain new insights into AtTIP1;1 functions in plant physiology and development, we have characterized AtTIP1;1 expression and intracellular localization using transcriptional and translational gene fusions, and isolated and characterized an AtTIP1;1 gene knockout mutant. Results show that AtTIP1;1 is present in multiple vacuolar compartments inside differentiated cell types and is preferentially found in the apposing tonoplasts of adjacent vacuoles. When grown under optimal conditions, the knockout mutant displayed no phenotypic alteration and, in contrast to a previous report (Ma et al. 2004), AtTIP1;1 loss of function did not lead to early plant death. A conditional root phenotype was detected when mutant plants were grown on a glycerol-containing medium.
Materials and methods
Plant materials and growth conditions
The AtTIP1;1 mutants were obtained from the Arabidopsis Knockout Facility at the University of Wisconsin-Madison as a pool of seeds from 10 independent T-DNA lines. For mutant isolation and routine plant growth, seeds were sown on a 1:1 mixture of soil and compost. Plants were grown in a growth chamber under a 16-h-day (minimum photon flux density of 50 μmol m−2 s−1)/8-h-night and 24°C/20°C cycle. For root RNA extraction, seeds were surface-sterilized with 5% sodium hypochlorite containing 0.1% Triton X-100, washed four times with sterile distilled water, plated onto ABIS medium (5 mM KNO3, 2 mM MgSO4, 1 mM Ca(NO3)2, 2.5 mM K2HPO4 + KH2PO4, 50 μM Fe-EDTA, Murashige and Skoog microelements, 10 g l−1 sucrose, 7 g l−1 agar, 1 mM MES, pH 6.1) and cold-treated at 4°C for 3 days in the dark. Plants were then grown vertically in a growth chamber for 21 days under a 8-h-day (minimum photon flux density of 100 μmol m−2 s−1)/16-h-night and 22°C/18°C cycle. For glycerol treatment, seeds were allowed to germinate for 4 days under the same conditions, after which seedlings were transferred onto ABIS plates supplemented or not with 50 mM glycerol and grown vertically for an additional 8 days. Root systems were monitored using an Epson perfection 3170 photo scanner (Epson France, Levallois-Perret, France) and root lengths were measured using Optimas software 6.1 (Media Cybernetics, Silver Spring, MD, USA).
Plant vector construction
To construct the β-glucuronidase (GUS) reporter plasmid, the NcoI–XbaI restriction fragment of pNOM102 (Roberts et al. 1989) carrying the GUS coding region (uidA) was used to replace the NcoI–XbaI restriction fragment of pAVA393 (von Arnim et al. 1998) carrying the GFP coding region. In the resulting plasmid, named pAVA-uidA, the GUS coding region is placed under control of the CaMV 35S promoter and terminator. A sense primer 5′-TTC TCG AGT TAA CTA TTT TGG GTT G-3′ and an antisense primer 5′-TCG CCA TGG TCG GAC GGC TGA GAT TTA-3′ were used to amplify a ~3-kb upstream sequence of the AtTIP1;1 translational start codon with KOD hot start DNA polymerase (Novagen, Madison, WI) and genomic DNA as template. The PCR product was cut with XhoI and NcoI and used to replace the XhoI–NcoI restriction fragment carrying the 35S CaMV promoter in pAVA-GUS, placing the expression of GUS under the control of the AtTIP1;1 promoter. A XhoI–XmaI fragment carrying the fusion gene (AtTIP1;1 promoter:GUS coding region:35S transcription terminator) was then subcloned into the polylinker SalI and XmaI sites of the transformation vector pDE1001 (Denecke et al. 1992). To construct the GFP reporter plasmid, a sense primer 5′-TTC TCG AGT TAA CTA TTT TGG GTT G-3′ and an antisense primer 5′-TAC CCA TGG CGT AGT CTG TGG TTG GGA-3′ were used to amplify a ~3.8-kbp fragment upstream of the AtTIP1;1 stop codon. The PCR product was cut with XhoI and NcoI and used to replace the XhoI–NcoI restriction fragment of pAVA393 carrying the 35S CaMV promoter to fuse, in frame, the 3′ end of the AtTIP1;1 coding region with the 5′ end of the GFP coding region. The resulting fusion gene (AtTIP1;1 promoter and coding region:GFP coding region:35S transcription terminator) was then subcloned as a XhoI–XmaI fragment into the polylinker SalI and XmaI sites of pDE1001.
Plant transformation
Binary vectors carrying the fusion genes were introduced into A. tumefaciens strain C58C1 according to An et al. (1988). The resulting bacteria were then used for floral infiltration of A. thaliana plants (Clough and Bent 1998). Infiltrated seeds were surface sterilized and plated on a kanamycin (50 μg/ml) solid agar medium. Plates were incubated in the dark at 4°C for 2 days, then exposed to light under a 16-h-day (minimum photon flux density of 100 μmol m−2 s−1)/8-h-night and 24°C/20°C cycle. Kanamycin-resistant seedlings with green cotyledons were then transferred into soil.
RNA gel blot analysis
Total RNA was isolated from shoots and roots of in vitro-grown 21-day-old plants using the RNeasy Plant Mini Kit (Qiagen, Courtaboeuf, France). For RNA gel blot analysis, 20 μg of RNA was loaded per lane, electrophoresed on a 1% agarose formaldehyde denaturing gel and then transferred to a Hybond N+ membrane (Amersham Biosciences, Orsay, France) for hybridization. Gene-specific probes were generated by PCR amplification and labelled with the DIG High Prime DNA Labelling and Detection Starter Kit II (Roche Diagnostics, Meylan, France). The primers used for amplification were for AtTIP1;1: forward 5′-TGA TTG ACT AGT AAC CCT CAC TGA CTG AC-3′ and reverse 5′-CAG CTC CCA ACC ACA GAC TAC-3′, and for ACTIN2 (loading control): forward 5′-GTT AGC AAC TGG GAT GAT ATG G-3′ and reverse 5′-CAG CAC CAA TCG TGA TGA CTT GCC C-3′. DIG probe labelling, hybridization and detection were performed according to the manufacturer’s protocol.
Real-time qRT-PCR analysis
Total RNA from roots of in vitro-grown 21-day-old plants was isolated with the RNeasy Plant Mini Kit (Qiagen, Courtaboeuf, France) and treated with RQ1 DNaseI (Promega, Charbonnières, France) to eliminate genomic DNA. First-strand cDNA was produced from 1 μg of total RNA using the iScript Select cDNA Synthesis Kit (Bio-Rad, Marnes-la-Coquette, France). The product was diluted 1:5 with water, and 5 μl was used as template for RT-qPCR amplification with an iQ SYBR Green Supermix kit (Bio-Rad) on a MyiQ iCycler (Bio-Rad). The housekeeping gene UBQ10, whose expression has been shown to be stable in Arabidopsis (Czechowski et al. 2004), was used to normalize candidate gene transcripts. The primers used for amplification are listed in Supplemental Table S1. The 2−ΔΔCT method was used for relative gene expression analysis (Livak and Schmittgen 2001) and the Mann–Whitney U-test was used to compare ΔΔCt values between mutant and wild-type plants.
Screening for an AtTIP1;1 T-DNA knockout line
The Arabidopsis Knockout Facility (AKF) alpha and BASTA populations (Sussman et al. 2000) were screened by PCR, using a primer specific for the T-DNA left border (JL 202, 5′-CAT TTT ATA ATA ACG CTG CGG ACA TCT AC-3′) in combination with either a forward (GFM-790, 5′-TTG CGA TTA GAA TAA AGC GTA GTG ATT TG-3′) or a reverse AtTIP1;1-specific primer (GR-1433, 5′-TGA TTG ACT AGT AAC CCT CAC TGA CTG AC-3′). Identification of positive DNA pools was performed by Southern blot analysis of the PCR products and confirmed by sequencing of the amplified DNA. The probe used was a DIG-labelled AtTIP1;1-specific PCR product obtained with the primers pair GFM-790/GR-1433. Individual plants in positive pools were screened by PCR with the same oligonucleotides. The screening procedure yielded a single knockout mutant line, named J8, in the BASTA population.
Histochemical GUS staining
GUS staining was performed in 35 mM NaH2PO4/Na2HPO4 (pH 7.0), 0.5 mM potassium ferrocyanide, 10 mM EDTA, 0.1% Triton-X100 and 1 mM X-Gluc. Organs were incubated for 1–10 h at 37°C and then destained in 70% EtOH. GUS-stained organs were then fixed for 20 min in FAA (50% EtOH, 5% acetic acid, 3.7% formaldehyde) and observed with differential interference contrast microscopy.
Cytochalasin D treatment
Stock concentrations of 10 mM cytochalasin D (Sigma, Saint-Quentin-Fallavier, France) and 1 mM MitoTracker® Orange CM-H2TMRos (Invitrogen, Cergy-Pontoise, France) were made in dimethyl sulfoxide (DMSO). Stock solutions were then dissolved in water to the appropriate concentrations. Effectiveness of the cytochalasin D treatment was assessed through the inhibition of mitochondrial movement. To visualize mitochondria, Arabidopsis leaves were treated with 5 μM MitoTracker® Orange CM-H2TMRos for 30 min and were then rinsed three times with water before observation or treatment with cytochalasin D. To depolymerize actin filaments, leaves were treated with 100 μM Cytochalasin-D for 30 min and were rinsed three times with water before observation.
Confocal microscopy
Before observation, pieces of leaves, roots or flowers were mounted in water under a coverslip. Fluorescence observations were conducted with a Leica TCS-SP2-AOBS confocal microscope (Leica Microsystems, Wetzlar, Germany). Excitation wavelengths and emission filters were 488 nm/band-pass 506–538 nm for GFP, 488 nm/band-pass 664–696 nm for chlorophyll and 543 nm/band-pass 565–615 nm for MitoTracker® Orange CM-H2TMRos. Images were processed using Imaris 4.0.6 (Bitplane, Zurich, Switzerland) and Photoshop 6.0 (Adobe Systems, San Jose, CA). GFP fluorescence intensity of unprocessed TIFF images was quantified by using ImageJ 1.38× (NIH, Bethesda, MD). Fluorescence intensities were acquired by drawing a one-pixel wide line over the fluorescent membrane and averaged. These averaged intensities were used to compute the ratios of fluorescence between the highly fluorescent structures and the weakly fluorescent peripheral tonoplast.
Electron microscopy
Sepals from Arabidopsis thaliana were dissected in freshly prepared 1% (v/v) paraformaldehyde and 2% (v/v) glutaraldehyde in 0.1 M NaH2PO4/Na2HPO4 phosphate buffer pH 7.2, and immersed 4 h at room temperature in the same fixative mix. Samples were washed in the same buffer and post-fixed with 1% (v/v) OsO4 in phosphate buffer for 2 h at 4°C. After washing in phosphate buffer, sepals were incubated in 1% (w/v) tannic acid in phosphate buffer for 30 min at room temperature, in the dark. Samples were washed in distilled water, dehydrated through a graded ethanol series, followed by propylene oxide and embedded in an Araldite/Epon resin mixture. Sections (80 nm) were cut on an Ultracut E ultramicrotome (Reichert Jung, Leica, Heidelberg, Germany), mounted on copper grids, and stained in 2.5% uranyl acetate in 50% (v/v) methanol for 30 min, followed by lead citrate for 15 min. Sections were viewed in an H-7500 (Hitachi Ltd., Tokyo, Japan) operating at 80 kV and equipped with an AMT camera driven by AMT software (AMT, Danvers, USA).
Results
AtTIP1;1 is highly expressed in the root and the vascular system of the shoot
To monitor development-related AtTIP1;1 expression, a trancriptional fusion gene with the GUS coding sequence (AtTIP1;1-uidA) and a translational fusion gene with the GFP coding sequence (AtTIP1;1-gfp) were designed. Twenty-two transformed lines for the GUS construct were selected and 8 of them were assayed for GUS activity. The transgenic lines showed similar expression patterns with minor differences in expression levels. Twenty transformed lines for the GFP construct were also selected and 15 were analyzed by fluorescence microscopy. Among them, 4 lines showed no GFP fluorescence while all the others showed very similar expression patterns with slightly different levels of expression.
No expression of any fusion genes was detected in embryos from imbibed mature seeds or from germinating seeds (Fig. 1a, data not shown for the AtTIP1;1-gfp transgenic embryos). Expression of the AtTIP1;1-uidA fusion gene was first seen in the root system 1 day after germination (Fig. 1b). In older seedlings, GUS and GFP labelling was detected all along the growing root with the exception of the meristematic region (Figs. 1b–d, f; 2a–b). In the proximal end of the elongation zone, fusion gene expression was mostly restricted to the endodermis (Figs. 1c, 2a, b). In the maturation zone, the fusion genes were expressed in all cell types but AtTIP1;1-GFP accumulation was higher in the endodermis (Figs. 1d, e; 2c, d). GUS activity was also detected in the aerial plant parts (cotyledons and leaves) when tissues were stained for a longer period of time (Fig. 1f), with the exception of the apical meristem (data not shown). This indicates that AtTIP1;1 promoter activity is higher in the root system, at least at the earlier stages of plant development. In cotyledons and leaves, AtTIP1;1 promoter activity was high in the veins (Fig. 1g, h) and, more specifically, in xylem vessels-flanking parenchyma cells (Fig. 1i), but it was also detected in pavement and mesophyll cells (Fig. 2e–h). In the inflorescence stem, cross-section analysis revealed that GUS activity was localized primarily in phloem cells (Fig. 1j). In flowers and developing siliques, GUS activity was also associated with vascular bundles of sepals (Fig. 1k), petals (Fig. 1l), stamen filaments (Fig. 1k), or silique valves (Fig. 1m), and with plant parts where tracheary elements occur such as in the style (Fig. 1n), stamen connective tissue (Fig. 1o), the receptacle of elongating siliques (Fig. 1p) and the funiculus-ovule connecting region (Fig. 1q). In sepals, the brightest GFP fluorescence was found in bundle sheath cells (Fig. 2i–k). Finally, high expression of both reporter genes was observed in a nonvascular cell type, the guard cells of anther stomata, before dehiscence (Figs. 1o, 2l–n). Interestingly the AtTIP1-1 promoter was not, or only very weakly, active in guard cells from other epidermal tissues (data not shown).
AtTIP1;1-uidA expression pattern in transgenic Arabidopsis plants. a Imbibed embryos a few hours before (left) or after germination (right). b Seedling 1 day after germination showing GUS expression in the root. c Primary root tip of a 1-week-old seedling. M: meristem, EZ: elongation zone, MZ: maturation zone. d, e Partial longitudinal view and cross-section of the maturation zone in the primary root of a 1-week-old seedling. All cell types express the fusion gene. f Two-week-old plant GUS-stained for a longer time period (10 h). g, h Cotyledon and cauline leaf showing GUS expression in vascular tissue. i Transverse section of a cauline leaf; GUS expression is high in small cells (arrows) flanking the xylem elements (asterisk). j Partial view of an inflorescence stem cross-section showing strong GUS expression in phloem cells (arrowhead). k Open flower. l Petal (10-h staining). m Silique valve. n Pistil. o Anther showing a strong GUS expression in the stomata and connective tissue. p Receptacle of an elongating silique. q Developing seeds. Scale bars = 100 μm in (a–d, n, and o), = 50 μm in (e, i, and j), = 1 mm in (f–h, and m), and = 500 μm in (k, l, p, and q)
AtTIP1;1-GFP expression pattern and subcellular localization in transgenic Arabidopsis plants. GFP fluorescence (green) is shown in (a–n), merged with chlorophyll fluorescence (red) in (h), with DIC images in (a, c, and e), or with chlorophyll fluorescence and DIC images in (g, i, j, l, and m). a, b In roots, the overall pattern of AtTIP1;1-gfp expression is similar to that observed with the AtTIP1;1-uidA fusion gene (Fig. 1). M: meristem, EZ: elongation zone, and MZ: maturation zone. c, d In the maturation zone, GFP fluorescence is clearly detected in all cell layers but is brighter in the endodermis (E). R: rhizoderm, C: cortical cell, and S: stele. e, f In pavement cells of a rosette leaf, GFP fluorescence highlights the outline of the cell as well as small internal spherical structures. g, h In cells from the palisade parenchyma, GFP fluorescence outlines the turgid central vacuole, maintaining the chloroplasts at the cell periphery. i, j In sepal tissues, highly fluorescent bundle sheath cells flank the tracheary elements. k In bundle sheath cells, the tonoplast of the central vacuole is labelled as well as multiple and multiform structures. l–n Highly fluorescent spherical structures are also found inside vacuoles of anther stomata. Scale bars = 100 μm in (a, b, i, and l), = 20 μm in (c, d, e–h, j, and k), and = 5 μm in (m, n)
Altogether these data indicate that although AtTIP1;1 is expressed at a basal level in most cell types, with the notable exception of meristematic cells, it is most strongly expressed in cells associated with vascular bundles, suggesting that this TIP is required in tissues dedicated to water and solute transport. No striking discrepancies were observed between data obtained with GFP and GUS reporter genes which points to transcriptional control as a primary, and probably the most important, determinant of AtTIP1;1 expression in most of the investigated cell types.
The AtTIP1;1-GFP fusion protein is unevenly distributed along the tonoplast continuum and AtTIP1;1-enriched macrodomains frequently define contact zones between adjacent vacuoles
By placing the AtTIP1;1-GFP fusion protein under the control of the native promoter, it can reasonably be assumed that the expression level of the fusion protein and its cell specificity in transformed plants are similar to those of the native protein in wild-type plants. Therefore, the AtTIP1;1-gfp transformed lines represent a suitable tool to address the question of the AtTIP1;1 subcellular localization. Observation of AtTIP1;1-GFP fluorescence by confocal laser scanning microscopy clearly showed that the labelling was not evenly distributed in the tonoplast of transformed cells (Fig. 2f, k, n). Highly fluorescent intravacuolar structures, more or less similar to those described as bulbs in cotyledon and root cells by Saito et al. (2002), were observed in numerous cell types, the most spectacular ones being in the bundle sheath cells of sepals (Fig. 2k). Because of their wide diversity, we decided to classify them into 5 types (I–V, Fig. 3). Type I structures represented nonmobile arc-shaped entities (Fig. 3a). A Z-series analysis indicated that these structures are contact zones between two vacuolar compartments of different size and quantification of fluorescence revealed that the contact zone is about 5 times more fluorescent than the peripheral tonoplast (Table 1). These data strongly suggest that the level of fluorescence is not simply a consequence of two tonoplasts being placed adjacently, but that the contact zone is definitely enriched in AtTIP1;1-GFP protein. Type II structures were brightly fluorescent membranes (Table 1) that extended across the longitudinal axis of a cell, from one side to the other (Fig. 3b). A Z-series analysis revealed that type II structures also separated two distinct vacuolar compartments. Type III structures corresponded most closely to the bulbs described by Saito et al. (2002). They seemed to be anchored to the cell periphery (Fig. 3c) by one or several stalks that linked the swollen part of the type III structure to the peripheral tonoplast. A Z-series analysis and orthogonal projections showed that the swollen part of the structure was an almost entirely enclosed compartment (Fig. 3c, upper and middle panels). The fluorescent labelling was intense (Table 1) and sometimes found inhomogeneously distributed all around the structure (Fig. 3c, lower panels). Type III structures were highly mobile inside the vacuole of most of the studied cell types with the notable exception of bundle sheath cells (movie 1 in supplemental data). Bulb immobilization was obtained when tissues were treated with 100 μM of the actin filament inhibitor cytochalasin D (movie 2 in supplemental data). Effectiveness of the cytochalasin D treatment was assessed through the inhibition of the cytoplasmic streaming of mitochondria. At 100 μM of cytochalasin D, cytoplasmic streaming was completely abolished while a few freely moving mitochondria were still visible inside the lumen of the central vacuole (movie 2 in supplemental data). We believe that this was the consequence of drug-induced disruption of some transvacuolar strands and the release of entrapped mitochondria into the vacuolar lumen. The cytochalasin D experiment suggests that type III bulbs are not freely moving entities inside the vacuolar lumen but that they are connected to the actin filament network of the peripheral cytoplasm, probably through their stalk(s). Type IV structures can be described as “open” type III structures with the opening being located at the opposite side of the stalk (Fig. 3d). Type V structures represented domains in the tonoplast of the central vacuole in which AtTIP1;1-GFP was particularly abundant (Fig. 3e). Such structures were frequently of sub-micron size but could sometimes be as large as the ones presented in Fig. 3e. Confocal analyses did not allow to ascertain whether such regions of the peripheral tonoplast were in close contact with a flattened vacuolar compartment or other cellular endomembranes. Their fluorescence intensity was slightly weaker than those found for the other structures (Table 1). Finally, nested highly fluorescent structures were frequently observed (Fig. 3f). Mobility of the nested compartment was consistently constrained by the nesting compartment suggesting that the latter probably has a closed conformation (movie 3 in supplemental data).
Highly fluorescent structures in AtTIP1;1-GFP expressing cells. A triptych made of a single optical section and reconstructed orthogonal slices in the XZ and YZ planes is shown with (a, c) or without (b–e) an interpretative scheme. A projection image of a Z series is shown on the right in (b, middle panel of c, d and e). a A type I structure with a typical arc shape. Two different reconstructed orthogonal slices are shown in the XZ and YZ planes. In all planes the ends of the fluorescent arcs always join a line of weak fluorescence, indicating that the semi spherical entity is a closed compartment. The arrow indicates the peripheral tonoplasts of two adjacent cells that are visible as a single line of fluorescence. Note the difference in fluorescence intensity between this line and the arc. b Two type II structures split the vacuolar content of a single cell into 3 distinct compartments. c Type III structures with a typical bulb-like shape. A weakly fluorescent stalk connects the bulbs to the peripheral tonoplast (arrowheads). Three optical sections at 0.4-μm intervals show a type III structure with a non-uniform fluorescence (lower panels). d Type IV structure with a stalk (arrowhead). e Two type V stuctures define AtTIP1;1-enriched macrodomains within the peripheral tonoplast of the central vacuole. f Nested fluorescent structures. Scale bars = 5 μm
Adjacent or nested vacuolar compartments are observed in wild-type cells
In order to determine whether similar structures were also present in wild-type cells, an ultrastructural study of the vacuome of wild-type sepal cells was conducted. Electron micrographs presented in Fig. 4 are compared to confocal images obtained with AtTIP1;1-GFP-expressing cells. Figure 4a illustrates two distinct vacuoles, a small one (v1) with a clearer lumen protruding into a larger one (v2), the contact zone between the two tonoplasts displaying an arc shape similar to type I membranes. Figure 4b shows a large vacuole split into two compartments by two membranes. Such membranes could be interpreted as a type II structure or as the result of the longitudinal section of a transvacuolar strand. The fact that long and thin transvacuolar strands are usually fragile and disrupted during the sample preparation argues against the second interpretation. Figure 4c shows a circular compartment limited by a double membrane and engulfed into the central vacuole. Such a compartment is reminiscent of type III structures. Arrowheads in inset 1 show cytoplasmic bridges that connect the thin space delimited by the double membrane (inset 2) and the peripheral cytoplasm. Both luminal contents also appear slightly different suggesting that the 2 compartments are isolated from one another. Finally more sophisticated vacuolar inclusions were observed and the one presented in Fig. 4d is reminiscent of the nested bulbs found in confocal microscopy. The inclusion is characterized by two nested double-membrane compartments. The electron opacity of the space delimited by the double membranes is very similar to the one observed for the cytoplasm. Such electron microscope observations are clearly in line with the results obtained in confocal microscopy.
Ultrastructure of vacuolar compartments in Arabidopsis wild-type sepal cells. In all panels, confocal images of highly fluorescent structures from AtTIP1;1-GFP-expressing cells in sepals are presented on the right, and electron micrographs of very similar structures observed in wild-type cells are presented on the left. a Type I-like structure. The vacuolar lumens are different in the two compartments (v1 and v2). b Type II-like structure. A two-membrane structure splits the cell space into two compartments. c Type III-like structure. The electron micrograph shows a spherical vacuolar compartment (v1) engulfed in a larger vacuole (v2). Arrowheads indicate cytoplasmic bridges between the fringe of cytoplasm along the smaller vacuole and the cytoplasm of the cell periphery. d A four-membrane structure is visible in the electron micrograph and could represent nested vacuoles (v1, v2 and v3) surrounded by a thin layer of cytoplasm. Arrows point to single membranes. Scale bars = 2 μm in all confocal images, = 500 nm in (a, b) and left panel in (c, d), = 100 nm in central panel of (c, d)
Characterization of an AtTIP1;1 gene knockout
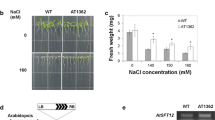
To gain insight into AtTIP1;1 function, knockout mutants were searched for in the Arabidopsis Knockout Facility (AKF) T-DNA mutant collections by reverse genetics. A single line was identified that contained two independent T-DNA insertions, one being in the first exon of AtTIP1;1. PCR-based genotype analysis of segregating populations led to isolation of a homozygous line for the AtTIP1;1 mutation, named J8. Molecular characterization of the mutant allele revealed that the inserted sequence was flanked by 2 T-DNA left borders (Fig. 5a), usually interpreted as the consequence of the insertion of two T-DNAs (or more) in inverted orientation. The AtTIP1;1 mutation was also characterized by a 303-bp deletion spanning the initiation codon. Altogether these data strongly suggested that the mutant allele was a null allele. Northern blot analysis of mRNA from wild-type and mutant plants confirmed the presence of AtTIP1;1 transcripts in the wild-type line but revealed their absence in the mutant line (Fig. 5b), thus validating it as a true knockout.
Molecular characterization of an AtTIP1;1 knockout mutant. a T-DNA insertion in the AtTIP1;1 gene led to the deletion of a 303-bp sequence that spans the translation initiation start. Dark boxes represent AtTIP1;1 protein coding regions and the black triangles represent T-DNA left borders. b Steady-state levels of AtTIP1;1 mRNAs in wild-type (WT) and mutant plants (J8). The left panel depicts the AtTIP1;1 transcripts probed with a 610 bp genomic DNA fragment 3′ to the last AtTIP1;1 exon and the right panel the actin2 (ACT2) transcripts as loading controls
Phenotypic characterization of the mutant failed to detect any macroscopic phenotype under optimal growth conditions. Germination, growth and development were normal and no sign of early senescence were detected (data not shown). Search for a conditional phenotype led us to test seed germination and seedling growth under stress conditions. Hyperosmotic, salt or desiccation stress did not differentiate between wild-type and mutant lines when applied continuously (data not shown). However, a subtle conditional phenotype was detected when seeds were grown on a glycerol-containing medium. High concentrations of exogenously applied glycerol are known to alter the plant cell metabolism and to inhibit root growth, cotyledon greening and development of true leaves (Aubert et al. 1994; Eastmond 2004). When grown on a 50 mM glycerol-containing medium, the primary root length of the mutant was shown to be 40% smaller than the wild-type one (Fig. 6a, b). This phenotypic alteration was reversed when the J8 mutant line was transformed with the AtTIP1;1-gfp fusion gene (Fig. 6a, c), confirming that it was truly the consequence of AtTIP1;1 gene knockout and indicating that the chimeric protein displays the function(s) of its TIP moiety.
Primary root elongation on glycerol-containing medium. a Root phenotypes of wild-type (WT), mutant (J8) and AtTIP1;1-GFP-expressing J8 (J8-38) seedlings on glycerol-containing medium. Primary root growth is more severely affected in the AtTIP1;1 mutant background. Seeds were allowed to germinate for 4 days on glycerol-free medium then transferred to medium supplemented with 50 mM glycerol. Scale bar = 5 mm. b Primary root lengths on normal and 50 mM glycerol-containing medium measured 8 days after transfer. J8 primary roots were significantly shorter than those of wild type plants on glycerol-containing medium (t test, P < 0.001). c Transformation of the J8 mutant with the AtTIP1;1-gfp fusion gene complements the conditional root phenotype. Primary roots of complemented seedlings (lines J8-4, J8-9 and J8-38) were significantly longer than those of J8 seedlings (t test, P < 0.001 for all three pairwise comparisons)
The lack of a visible phenotype in the J8 mutant under optimal growth conditions could be due to several reasons, one of them being a compensatory up-regulation of other members of the TIP gene subfamily. To test this hypothesis, the relative abundance of transcripts from the whole TIP subfamily was quantified in 21-day-old wild-type and mutant plants and normalized using the UBQ10 gene. Root tissues were chosen because the GUS reporter gene previously showed them to express high levels of the AtTIP1;1 aquaporin (Fig. 1). Relative levels of transcripts for 4 genes (AtTIP1;3, AtTIP3;1, AtTIP3;2 and AtTIP5;1) were near background and are not reported. The AtTIP1;1 mRNAs were the third most abundant transcripts in wild-type root cells and completely absent from mutant roots. No significant change in relative mRNA levels was detected in mutant root cells for the other members of the TIP gene subfamily (Fig. 7), suggesting that the mutated plant coped with the lack of AtTIP1;1 without up-regulating other TIP genes.
Relative expression of AtTIP genes in roots of wild type and AtTIP1;1 mutant J8 plants. Total RNAs were extracted from 21-day-old plants and used for qRT-PCR assays. The expression of each gene is given in arbitrary units relative to the expression of the ubiquitin10 (UBQ10) gene. Data are mean values and standard errors of 5 (WT plants) and 4 (J8 plants) independent experiments. The AtTIP1;3, AtTIP3;1, AtTIP3;2 and AtTIP5;1 gene expression levels were near background and are not reported here. The asterisk denotes a statistically significant difference between the two means at P < 0.05 using a Mann–Whitney U test
Discussion
AtTIP1;1 expression is correlated with water and solute transport
The first study of AtTIP1;1 expression using in situ mRNA hybridization and a promoter-GUS fusion construct led to the conclusion that AtTIP1;1 expression was primarily correlated with vacuolation and cell enlargement (Ludevid et al. 1992). The use of a transcriptional fusion construct with GUS and a translational fusion construct with GFP allowed us to revisit the cell specificity of AtTIP1;1 expression. This has revealed that while AtTIP1;1 expression is detected in most cell types, the highest levels of expression are found in cells associated with water and/or solute transport.
Several lines of evidence indicate that AtTIP1;1 expression profiles along the longitudinal and radial root axes are correlated with the transcellular pathway for water movements. Firstly, AtTIP1;1 is expressed in all cell types along the root axis, with the notable exception of the meristem cells. Secondly, expression is detected in the elongation zone but is higher in the maturation zone where water uptake is maximal and cells lose their symplastic continuity (Duckett et al. 1994). Finally, expression is higher in the endodermis where water mostly flows along the transcellular path across the radial axis (Steudle 2000).
In this context, AtTIP1;1 is also highly expressed in the xylem parenchyma cells of leaf veins, which form the first cellular barrier for water on its way out of the xylem vessels. In the shoot, high expression levels were also found in the phloem strands of the stem, in the bundle sheath cells of sepals, which are thought to play a role in phloem loading (van Bel 1993), and at sites of phloem unloading such as the pedicel of the developing silique, the ovule stalk and the anther connective tissue. In these tissues, symplastic unloading of metabolites requires an enhanced capacity for osmotic equilibration between cytosol and vacuole that could be ensured in part by the water channel activity of AtTIP1;1.
Of particular interest is the high expression level of AtTIP1;1 detected in stomatal guard cells of anthers before dehiscence. Stomatal opening and closure result from the rapid osmotically-driven flow of water across the plasma membrane and the tonoplast, which is believed to be facilitated by PIPs and TIPs; in anthers, correctly timed dehydration is important for pollen development and dehiscence (Keijzer et al. 1987). Thus, AtTIP1;1 expression might participate in this process in allowing stomatal opening to promote transpiration and subsequent cell dehydration.
It is generally assumed that the high water permeability of the tonoplast allows the vacuole to buffer the cytoplasm in case of an intense water flow, thus providing an efficient means to dissipate any osmotic gradient across this membrane and to prevent any cellular damages caused by a sudden change in cytoplasmic volume (Barrieu et al. 1999; Maurel et al. 2002; Tyerman et al. 2002). It has been reported that a very efficient cytosol osmoregulation can occur as long as the tonoplast has a ≥5-fold higher osmotic water permeability than the plasma membrane (Maurel et al. 2002). Assuming that water balance is finely tuned, the high expression level of TIPs in a given tissue would reflect concomitantly a high expression level of PIPs. In situ hybridization of PIP RNAs in Arabidopsis has revealed the highest transcript levels in the root endodermal layer and in the leaf vascular bundles (Schäffner 1998). Similarly, in maize, transcripts of two of the three most highly expressed PIPs in roots have been found in abundance in the exodermis and the endodermis, two apoplastic barriers in the root (Hachez et al. 2006).
AtTIP1;1 expression pattern is reminiscent of that found for ZmTIP1;1, a highly expressed TIP of maize (Barrieu et al. 1998). One interesting difference is that ZmTIP1;1 expression has been detected in the root meristem. The presence of TIPs in meristematic cells has also been reported in several other plant species suggesting that they are needed for the biogenesis of new vacuoles (Marty-Mazars et al. 1995; Paris et al. 1996; Barrieu et al. 1998). With the exception of AtTIP3;1 for 3 days just after germination, no other TIPs have been detected in the Arabidopsis root tip (Hunter et al. 2007) suggesting that vacuole swelling in this plant might have different requirements. However, the expression patterns of all members of the AtTIP subfamily have yet to be characterized to conclude on this point.
Since the highest levels of AtTIP1;1 expression are observed in cells experiencing trans-cellular water flow and/or exposed to rapid changes in cytosolic metabolite concentrations, AtTIP1;1 should contribute to the osmotic equilibration of the cytosol that is needed in response to water and/or solute entry.
AtTIP1;1 is preferentially found in the apposing tonoplasts of adjacent vacuoles
In most cells, the AtTIP1;1-GFP fusion protein weakly labels the tonoplast of the central vacuole (named peripheral tonoplast), but strongly labels luminal membrane structures which were classified into 5 different types (I–V). Several reports have described the occurrence of fluorescent intravacuolar membrane structures when tonoplast-targeted GFP are constitutively expressed in Arabidopsis or tobacco cells (Hawes et al. 2001; Saito et al. 2002; Uemura et al. 2002; Escobar et al. 2003; Tian et al. 2004; Boursiac et al. 2005; Reisen et al. 2005). Most of these previously described intravacuolar structures are of type III and were named bulbs by Saito et al. (2002). In contrast to previous reports in which vacuolar bulbs have been observed only in vacuoles of young hypocotyl and cotyledon cells (Saito et al. 2002) or in vacuoles of cells exposed to water stress (Boursiac et al. 2005; Reisen et al. 2005), we found bulbs, as well as the other types of fluorescent structures, in most of the cells in which the fusion protein is expressed. The use of AtTIP1;1 promoter to drive the expression of AtTIP1;1-GFP assured that the cell specificity and level of expression of the fusion protein are similar to the ones of the native aquaporin. Therefore, bulb occurrence is not strictly correlated with cell expansion or water stress but rather exemplifies the structural and functional complexity of the vacuome in plant cells.
Confocal analyses of Z series of type I and type II structures indicate that they most probably correspond to apposing tonoplasts from distinct vacuoles. The fact that their fluorescence intensity is about five-fold higher than that of the peripheral tonoplast argues in favor of an AtTIP1;1-GFP accumulation in these tonoplast macrodomains. Type III structures could be interpreted as double membrane-bound inclusions resulting from the engulfment of a small vacuole into a large one. Alternatively, they could represent bulb-shaped invaginations of the peripheral tonoplast. The confocal microscope observations do not allow to distinguish between these two interpretations. Nevertheless, several circumstantial evidences point to the first hypothesis. Firstly, the fluorescence intensity of type III membranes is similar to that of type I and type II structures suggesting that type III fluorescence delineates, in a similar way, contact zones between adjacent tonoplasts enriched in AtTIP1;1-GFP. Secondly, double membrane-bound inclusions are often observed in EM investigation of wild-type vacuoles. In rare cases, we observed the continuity between the thin cytoplasmic space surrounding the engulfed vacuole and the peripheral cytoplasm. Thirdly, some type III bulbs display more than one stalk-like extension. In the engulfed vacuole hypothesis, such confocal images could be interpreted as small vacuoles moving along a branched network of transvacuolar strands. Their mobility under normal conditions and their immobilization following treatment with an actin polymerisation inhibitor provide support for this interpretation. Organelles such as amyloplasts can also be trapped within transvacuolar strands of endodermal cells in Arabidopsis (Morita et al. 2002; Yano et al. 2003). A mutation in the tonoplast marker AtVAM3 prevents amyloplasts to travel through the transvacuolar strands (Yano et al. 2003). This result led the authors to speculate that flexibility of the tonoplast is diminished in the mutant cells thus preventing the engulfment process. AtVAM3 accumulation in type III-like membranes of Arabidopsis and tobacco cells (Uemura et al. 2002; Foresti et al. 2006) is an indication that this protein might also contribute to the engulfment of small vacuoles by the central vacuole.
However, the invagination hypothesis cannot be ruled out since single membrane compartments are sometimes found within the lumen of the central vacuole in EM (data not shown). In yeast, tonoplast invaginations have previously been reported (Müller et al. 2000) but their tubular shape and their gradual depletion of transmembrane proteins towards their tip make them clearly different from type III structures.
Of all the brightly fluorescent structures in the GFP transformants, type IV and type V structures are the most puzzling. Type IV structures could be interpreted as type III structures with an uneven distribution of AtTIP1;1-GFP. Alternatively, they could correspond to open type III structures resulting from the fusion of apposing tonoplasts. Type IV structures would thus perfectly fit to the description of bulbs as tonoplast remnants of vacuolar fusion proposed by Saito et al. (2002). Type V structure fluorescence could correspond to the contact zone between a flattened vacuole and the central vacuole. Alternatively, it could represent a subregion of the peripheral tonoplast enriched in fusion proteins. Uneven distribution of aquaporins in the plasma membrane has already been reported for plant cells (Siefritz et al. 2001; Hachez et al. 2006).
The diversity of fluorescent structures within some Arabidopsis cells raises the question of their putative ontogenic relationships. Since no transformation of one fluorescent type into another has been observed, this can only be speculated. Figure 8 depicts a plausible scenario (solid arrow) in which two vacuolar compartments fuse to form a single one. In a first step, a vacuole is engulfed into another one (type I/II–type III transition), then fusion of the two tonoplasts (type III–type IV transition) leads to the release of contents of the engulfed vacuole into the lumen of the phagocytizing vacuole. In a final step, unfolding of the invaginated tonoplast (type IV–type V transition) gives rise to a compartment with a smoother contour. Alternatively, the reverse scenario—in which a single vacuole gives rise to two vacuolar compartments (Fig. 8, dashed arrow)—could also explain the occurrence of the highly fluorescent membranes.
While it is generally assumed that plant cells contain functionally distinct types of vacuoles associated with specific tonoplast aquaporins (Paris et al. 1996; Neuhaus and Rogers 1998; Neuhaus and Paris 2006), two reports have recently challenged this view (Hunter et al. 2007; Olbrich et al. 2007). Using several fluorescent reporter proteins for the tonoplast and the vacuolar lumen, Hunter et al. (2007) identified a single vacuolar compartment in Arabidopsis cells. The three fluorescent AtTIPs used in the study (including AtTIP1;1) labelled membranous structures very similar to the ones described here but they were not finely characterized. Our study shows the presence of AtTIP1;1 in separate vacuolar compartments which coexist inside differentiated Arabidopsis cells. What is not known however is whether these compartments are functionally different. The accumulation of other TIPs in contact zones between vacuoles (Hunter et al. 2007) indicates that it is not a property that is specific to AtTIP1;1 but that it is shared with other members of the TIP subfamily in Arabidopsis. This raises the question of why do TIPs accumulate in such contact zones?
The most likely explanation is that aquaporins facilitate the flow of water across the two apposing membranes to dissipate any osmotic gradient that could result from the specific influx or efflux of solutes in one of the two vacuoles. Dynamic water exchange between two functionally different vacuoles has indeed been shown to occur in mesophyll cells of the common ice plant (Epimashko et al. 2004). Another putative role is suggested for animal aquaporins in cell adhesion where the term adhenel has been proposed to designate a proteinacious channel with adhesive properties (Gonen et al. 2004; Hiroaki et al. 2006). Do plant aquaporins function in a similar way to promote vacuole adhesion? Whilst regions of tight apposition between tonoplasts would facilitate water flow, such regions were rarely observed in EM studies of wild-type cells suggesting that turgescence might be sufficient to maintain vacuoles in close contact.
Is AtTIP1;1 a dispensable tonoplast aquaporin?
The knockout mutant for AtTIP1;1 identified in the T-DNA mutant collection of the Arabidopsis Knockout Facility did not display any visible phenotype under optimal growth conditions. This is in contrast with the results of Ma et al. (2004), who reported an early senescence phenotype associated with the knockdown of AtTIP1;1 using an RNAi approach. What could explain such a discrepancy? Among the major problems of using RNAi are off-target effects, whereby partial complementarity of an RNAi trigger to non-targeted transcripts causes unwanted silencing effects (Jackson et al. 2003). Ma et al. (2004) showed that the severity of the senescence phenotype is correlated with the level of RNAi trigger but not with the level of targeted mRNA (below 10% of the wild-type level for all phenotype classes). Such data are an indication that the mutant phenotype could be the consequence of an off-target effect. Unwanted silencing usually affects members from the same gene family. A 50–70% reduction in AtTIP1;2 steady-state transcript levels and a ≥85% reduction in AtTIP1;3 steady-state transcript levels were detected by Ma et al. (2004) in mutants with a moderate phenotype. Two sets of data rule out the hypothesis that AtTIP1;2 partial silencing alone is responsible for the observed phenotype. Firstly, there is no correlation between the severity of the mutant phenotype and the steady-state level of AtTIP1;2 transcripts. Secondly, AtTIP1;2 RNAi lines with mRNA levels reduced to 4% of wild type displayed no growth alteration (Ma et al. 2004). Very little is known about the AtTIP1;3 gene, nevertheless its stamen-restricted expression pattern makes AtTIP1;3 gene silencing an unlikely hypothesis for the mutant phenotype (Neuhaus and Paris 2006). In conclusion, one possibility is that the down-regulation of all AtTIP1 subgroup genes could be responsible for the RNAi phenotype. Alternatively, the mutant phenotype could be the consequence of the down-regulation of an unrelated gene. Global expression profiling in AtTIP1;1 RNAi lines revealed that eleven other genes were downregulated (Ma et al. 2004), one of which might have been an unexpected target for the RNAi trigger.
Complete inactivation of AtTIP1;1 alone has no apparent effect on the phenotype of Arabidopsis. The natural function of this gene may well be assumed by another gene or genes from the TIP subfamily. Investigation of AtTIP gene expression in the mutant background did not reveal any change in levels indicating that no compensation occurred through the up-regulation of another AtTIP gene. Such a result suggests that either the Arabidopsis tonoplast is sufficiently permeable to cope with the absence of AtTIP1;1 or an unknown mechanism is activated to compensate for the loss of the water channel. In the present and previous studies 5 genes have been reported to be expressed in Arabidopsis roots (Alexandersson et al. 2005; Boursiac et al. 2005). A more extensive functional analysis of the TIP gene family implying the recovery of multiple mutant plants should shed more light on the role of TIPs in plant development and physiology. It is interesting to note that Caenorhabditis elegans possesses 8 aquaporin genes, and that quadruple aquaporin mutant animals were needed to obtain a conditional phenotype (Huang et al. 2007).
The search for a conditional phenotype showed that the AtTIP1;1 knockout mutant of Arabidopsis displayed a higher sensitivity to the toxic effect of glycerol. It has been suggested that glycerol could enter root cells through plasma membrane glyceroporins or aquaglyceroporins (Eastmond 2004) and members of the AtNIP subfamily have been shown to transport glycerol in heterologous expression systems such as yeast and Xenopus oocytes (Weig and Jakob 2000; Wallace and Roberts 2005). Glycerol catabolism is known to affect plant metabolism and root development (Leegood 1988; Aubert et al. 1994; Eastmond 2004). It is tempting to speculate that AtTIP1;1 contributes to glycerol detoxication by facilitating vacuolar sequestration. However, AtTIP1;1 does not transport glycerol in Xenopus oocytes (Maurel et al. 1993) despite an unusual hydrophobic selectivity filter for a water channel (Wallace and Roberts 2004). One possibility is that AtTIP1;1 promotes glycerol permease activity of another TIP through heteromerization. Plant MIPs form tetramers in the membranes (Daniels et al. 1999) and heteromerization of maize and mimosa PIPs modulates their water channel activity (Fetter et al. 2004; Temmei et al. 2005). Moreover, the tonoplast intrinsic protein from tobacco NtTIPa is permeable to glycerol (Gerbeau et al. 1999) and TIPs can form heteromers in membranes (Harvengt et al. 2000). However, the physiological significance of glycerol permeation through plant MIPs is questionable. Glycerol concentrations are generally very low in plants (Gerber et al. 1988) and glycerol channelling could simply be the consequence of the structural similarity of glycerol to metalloids (Bienert et al. 2008). Transient glycerol accumulation inside the vacuolar lumen could result from phospholipid catabolism after autophagic recycling of membranes, as has been shown to occur during starvation-induced autophagy (Aubert et al. 1996). The characterization of a glyceryl-phosphodiester phosphodiesterase activity in the vacuolar sap of carrot cells is in line with the involvement of vacuoles in the degradative pathway of membrane lipids (van der Rest et al. 2002). Glycerol recycling into cytosolic biosynthetic pathways would thus be facilitated by permeation through glyceroporins. The AtTIP1;1 mutant could be impaired in this recycling pathway.
Whatever its permeation selectivity or hypothetical interaction with other TIPs, AtTIP1;1 appears to be a dispensable tonoplast intrinsic protein, at least under normal growth conditions. However, AtTIP1;1 may be essential for the response to a so far unidentified environmental stress so contributing to the fitness of Arabidopsis in the natural environment.
Note added in proof
During the review of this manuscript, an article was published describing another knockout mutant for the AtTIP1;1 gene (Schüssler et al. 2008). This mutant did not display any visible phenotype, which is consistent with our results.
Abbreviations
- AQP:
-
Aquaporin
- GFP:
-
Green fluorescent protein
- GUS:
-
β-Glucuronidase
- MIP:
-
Membrane intrinsic protein
- NIP:
-
Nodulin-like intrinsic protein
- PIP:
-
Plasma membrane intrinsic protein
- TIP:
-
Tonoplast intrinsic protein
References
Agre P, Preston GM, Smith BL, Jung JS, Raina S, Moon C, Guggino WB, Nielsen S (1993) Aquaporin CHIP: the archetypal molecular water channel. Am J Physiol Renal Physiol 265:F463–F476
Alexandersson E, Fraysse L, Sjovall-Larsen S, Gustavsson S, Fellert M, Karlsson M, Johanson U, Kjellbom P (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol 59:469–484. doi:10.1007/s11103-005-0352-1
An G, Costa MA, Mitra A, Ha S-B, Márton L (1988) Organ-specific and developmental regulation of the nopaline synthase promoter in transgenic tobacco plants. Plant Physiol 88:547–552
Aubert S, Gout E, Bligny R, Douce R (1994) Multiple effects of glycerol on plant cell metabolism. Phosphorus-31 nuclear magnetic resonance studies. J Biol Chem 269:21420–21427
Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R (1996) Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol 133:1251–1263. doi:10.1083/jcb.133.6.1251
Barrieu F, Chaumont F, Chrispeels MJ (1998) High expression of the tonoplast aquaporin ZmTIP1 in epidermal and conducting tissues of maize. Plant Physiol 117:1153–1163. doi:10.1104/pp.117.4.1153
Barrieu F, Marty-Mazars D, Thomas D, Chaumont F, Charbonnier M, Marty F (1999) Desiccation and osmotic stress increase the abundance of mRNA of the tonoplast aquaporin BobTIP26-1 in cauliflower cells. Planta 209:77–86. doi:10.1007/s004250050608
Bienert GP, Schüssler MD, Jahn TP (2008) Metalloids: essential, beneficial or toxic? Major intrinsic proteins sort it out. Trends Biochem Sci 33:20–26. doi:10.1016/j.tibs.2007.10.004
Boursiac Y, Chen S, Luu DT, Sorieul M, van den Dries N, Maurel C (2005) Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiol 139:790–805. doi:10.1104/pp.105.065029
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. doi:10.1046/j.1365-313x.1998.00343.x
Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38:366–379. doi:10.1111/j.1365-313X.2004.02051.x
Daniels MJ, Chrispeels MJ, Yeager M (1999) Projection structure of a plant vacuole membrane aquaporin by electron cryo-crystallography. J Mol Biol 294:1337–1349. doi:10.1006/jmbi.1999.3293
Denecke J, De Rycke R, Botterman J (1992) Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. EMBO J 11:2345–2355
Duckett CM, Oparka KJ, Prior DAM, Dolan L, Roberts K (1994) Dye-coupling in the root epidermis of Arabidopsis is progressively reduced during development. Development 120:3247–3255
Eastmond PJ (2004) Glycerol-insensitive Arabidopsis mutants: gli1 seedlings lack glycerol kinase, accumulate glycerol and are more resistant to abiotic stress. Plant J 37:617–625. doi:10.1111/j.1365-313X.2003.01989.x
Epimashko S, Meckel T, Fischer-Schliebs E, Lüttge U, Thiel G (2004) Two functionally different vacuoles for static and dynamic purposes in one plant mesophyll leaf cell. Plant J 37:294–300
Escobar NM, Haupt S, Thow G, Boevink P, Chapman S, Oparka K (2003) High-throughput viral expression of cDNA-green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell 15:1507–1523. doi:10.1105/tpc.013284
Fetter K, Van Wilder V, Moshelion M, Chaumont F (2004) Interactions between plasma membrane aquaporins modulate their water channel activity. Plant Cell 16:215–228. doi:10.1105/tpc.017194
Foresti O, daSilva LL, Denecke J (2006) Overexpression of the Arabidopsis syntaxin PEP12/SYP21 inhibits transport from the prevacuolar compartment to the lytic vacuole in vivo. Plant Cell 18:2275–2293. doi:10.1105/tpc.105.040279
Gerbeau P, Güçlü J, Ripoche P, Maurel C (1999) Aquaporin Nt-TIPa can account for the high permeability of tobacco cell vacuolar membrane to small neutral solutes. Plant J 18:577–587. doi:10.1046/j.1365-313x.1999.00481.x
Gerber DW, Byerrum RU, Gee RW, Tolbert NE (1988) Glycerol concentrations in crop plants. Plant Sci 56:31–38. doi:10.1016/0168-9452(88)90182-3
Gonen T, Sliz P, Kistler J, Cheng Y, Walz T (2004) Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature 429:193–197. doi:10.1038/nature02503
Hachez C, Moshelion M, Zelazny E, Cavez D, Chaumont F (2006) Localization and quantification of plasma membrane aquaporin expression in maize primary root: a clue to understanding their role as cellular plumbers. Plant Mol Biol 62:305–323. doi:10.1007/s11103-006-9022-1
Harvengt P, Vlerick A, Fuks B, Wattiez R, Ruysschaert JM, Homble F (2000) Lentil seed aquaporins form a hetero-oligomer which is phosphorylated by a Mg(2+)-dependent and Ca(2+)-regulated kinase. Biochem J 352:183–190. doi:10.1042/0264-6021:3520183
Hawes C, Saint-Jore CM, Brandizzi F, Zheng H, Andreeva AV, Boevink P (2001) Cytoplasmic illuminations: in planta targeting of fluorescent proteins to cellular organelles. Protoplasma 215:77–88. doi:10.1007/BF01280305
Hiroaki Y, Tani K, Kamegawa A, Gyobu N, Nishikawa K, Suzuki H, Walz T, Sasaki S, Mitsuoka K, Kimura K, Mizoguchi A, Fujiyoshi Y (2006) Implications of the aquaporin-4 structure on array formation and cell adhesion. J Mol Biol 355:628–639. doi:10.1016/j.jmb.2005.10.081
Höfte H, Hubbard L, Reizer J, Ludevid D, Herman EM, Chrispeels MJ (1992) Vegetative and seed-specific forms of tonoplast intrinsic protein in the vacuolar membrane of Arabidopsis thaliana. Plant Physiol 99:561–570
Huang CG, Lamitina T, Agre P, Strange K (2007) Functional analysis of the aquaporin gene family in Caenorhabditis elegans. Am J Physiol Cell Physiol 292:C1867–C1873. doi:10.1152/ajpcell.00514.2006
Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L (2007) Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol 145:1371–1382. doi:10.1104/pp.107.103945
Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21:635–637. doi:10.1038/nbt831
Jauh GY, Fischer AM, Grimes HD, Ryan CA, Rogers JC (1998) delta-Tonoplast intrinsic protein defines unique plant vacuole functions. Proc Natl Acad Sci USA 95:12995–12999. doi:10.1073/pnas.95.22.12995
Jauh GY, Phillips TE, Rogers JC (1999) Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell 11:1867–1882. doi:10.1105/tpc.11.10.1867
Jiang L, Phillips TE, Rogers SW, Rogers JC (2000) Biogenesis of the protein storage vacuole crystalloid. J Cell Biol 150:755–770. doi:10.1083/jcb.150.4.755
Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjovall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol 126:1358–1369. doi:10.1104/pp.126.4.1358
Keijzer CJ, Hoek IHS, Willemse MTM (1987) The processes of anther dehiscence and pollen dispersal. 3. The dehydration of the filament tip and the anther in 3 monocotyledonous species. New Phytol 106:281–287. doi:10.1111/j.1469-8137.1987.tb00143.x
Leegood RC (1988) Phosphate sequestration by glycerol and its effects on photosynthetic carbon assimilation by leaves. Planta 176:117–126. doi:10.1007/BF00392487
Lin W, Peng Y, Li G, Arora R, Tang Z, Su W, Cai W (2007) Isolation and functional characterization of PgTIP1, a hormone-autotrophic cells-specific tonoplast aquaporin in ginseng. J Exp Bot 58:947–956. doi:10.1093/jxb/erl255
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Ludevid D, Höfte H, Himelblau E, Chrispeels MJ (1992) The expression pattern of the tonoplast intrinsic protein gamma-TIP in Arabidopsis thaliana is correlated with cell enlargement. Plant Physiol 100:1633–1639
Ma S, Quist TM, Ulanov A, Joly R, Bohnert HJ (2004) Loss of TIP1;1 aquaporin in Arabidopsis leads to cell and plant death. Plant J 40:845–859. doi:10.1111/j.1365-313X.2004.02265.x
Marty-Mazars D, Clemencet MC, Dozolme P, Marty F (1995) Antibodies to the tonoplast from the storage parenchyma cells of beetroot recognize a major intrinsic protein related to TIPs. Eur J Cell Biol 66:106–118
Maurel C (1997) Aquaporins and water permeability of plant membranes. Annu Rev Plant Physiol Plant Mol Biol 48:399–429. doi:10.1146/annurev.arplant.48.1.399
Maurel C (2007) Plant aquaporins: novel functions and regulation properties. FEBS Lett 581:2227–2236. doi:10.1016/j.febslet.2007.03.021
Maurel C, Reizer J, Schroeder JI, Chrispeels MJ (1993) The vacuolar membrane protein gamma-TIP creates water specific channels in Xenopus oocytes. EMBO J 12:2241–2247
Maurel C, Javot H, Lauvergeat V, Gerbeau P, Tournaire C, Santoni V, Heyes J (2002) Molecular physiology of aquaporins in plants. Int Rev Cytol 215:105–148. doi:10.1016/S0074-7696(02)15007-8
Morita MT, Kato T, Nagafusa K, Saito C, Ueda T, Nakano A, Tasaka M (2002) Involvement of the vacuoles of the endodermis in the early process of shoot gravitropism in Arabidopsis. Plant Cell 14:47–56. doi:10.1105/tpc.010216
Moriyasu Y, Hattori M, Jauh GY, Rogers JC (2003) Alpha tonoplast intrinsic protein is specifically associated with vacuole membrane involved in an autophagic process. Plant Cell Physiol 44:795–802. doi:10.1093/pcp/pcg100
Müller O, Sattler T, Flötenmeyer M, Schwarz H, Plattner H, Mayer A (2000) Autophagic tubes: vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J Cell Biol 151:519–528. doi:10.1083/jcb.151.3.519
Neuhaus JM, Paris N (2006) Plant vacuoles: from biogenesis to function. In: Šamaj JBF, Menzel D (eds) Plant endocytosis. Springer Verlag, Berlin, pp 63–82
Neuhaus JM, Rogers JC (1998) Sorting of proteins to vacuoles in plant cells. Plant Mol Biol 38:127–144. doi:10.1023/A:1006032627036
Olbrich A, Hillmer S, Hinz G, Oliviusson P, Robinson DG (2007) Newly formed vacuoles in root meristems of barley and pea seedlings have characteristics of both protein storage and lytic vacuoles. Plant Physiol 145:1383–1394. doi:10.1104/pp.107.108985
Paris N, Stanley CM, Jones RL, Rogers JC (1996) Plant cells contain two functionally distinct vacuolar compartments. Cell 85:563–572. doi:10.1016/S0092-8674(00)81256-8
Peng Y, Lin W, Cai W, Arora R (2007) Overexpression of a Panax ginseng tonoplast aquaporin alters salt tolerance, drought tolerance and cold acclimation ability in transgenic Arabidopsis plants. Planta 226:729–740. doi:10.1007/s00425-007-0520-4
Reisen D, Leborgne-Castel N, Özalp C, Chaumont F, Marty F (2003) Expression of a cauliflower tonoplast aquaporin tagged with GFP in tobacco suspension cells correlates with an increase in cell size. Plant Mol Biol 52:387–400. doi:10.1023/A:1023961332391
Reisen D, Marty F, Leborgne-Castel N (2005) New insights into the tonoplast architecture of plant vacuoles and vacuolar dynamics during osmotic stress. BMC Plant Biol 5:13. doi:10.1186/1471-2229-5-13
Roberts IN, Oliver RP, Punt PJ, van den Hondel CA (1989) Expression of the Escherichia coli beta-glucuronidase gene in industrial and phytopathogenic filamentous fungi. Curr Genet 15:177–180. doi:10.1007/BF00435503
Saito C, Ueda T, Abe H, Wada Y, Kuroiwa T, Hisada A, Furuya M, Nakano A (2002) A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J 29:245–255. doi:10.1046/j.0960-7412.2001.01189.x
Schäffner AR (1998) Aquaporin function, structure, and expression: are there more surprises to surface in water relations? Planta 204:131–139. doi:10.1007/s004250050239
Schüssler MD, Alexandersson E, Bienert GP, Kichey T, Laursen KH, Johanson U, Kjellbom P, Schjoerring JK, Jahn TP (2008) The effects of the loss of TIP1;1 and TIP1;2 aquaporins in Arabidopsis thaliana. Plant J 56:756–767. doi:10.1111/j.1365-313X.2008.03632.x
Siefritz F, Biela A, Eckert M, Otto B, Uehlein N, Kaldenhoff R (2001) The tobacco plasma membrane aquaporin NtAQP1. J Exp Bot 52:1953–1957. doi:10.1093/jexbot/52.363.1953
Steudle E (2000) Water uptake by roots: effects of water deficit. J Exp Bot 51:1531–1542. doi:10.1093/jexbot/51.350.1531
Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124:1465–1467. doi:10.1104/pp.124.4.1465
Temmei Y, Uchida S, Hoshino D, Kanzawa N, Kuwahara M, Sasaki S, Tsuchiya T (2005) Water channel activities of Mimosa pudica plasma membrane intrinsic proteins are regulated by direct interaction and phosphorylation. FEBS Lett 579:4417–4422. doi:10.1016/j.febslet.2005.06.082
Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, Rhee SY, Raikhel NV, Citovsky V (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135:25–38. doi:10.1104/pp.104.040139
Tyerman SD, Bohnert HJ, Maurel C, Steudle E, Smith JAC (1999) Plant aquaporins: their molecular biology, biophysics and significance for plant water relations. J Exp Bot 50:1055–1071. doi:10.1093/jexbot/50.suppl_1.1055
Tyerman SD, Niemietz CM, Bramley H (2002) Plant aquaporins: multifunctional water and solute channels with expanding roles. Plant Cell Environ 25:173–194. doi:10.1046/j.0016-8025.2001.00791.x
Uemura T, Yoshimura SH, Takeyasu K, Sato MH (2002) Vacuolar membrane dynamics revealed by GFP-AtVam3 fusion protein. Genes Cells 7:743–753. doi:10.1046/j.1365-2443.2002.00550.x
van Bel AJE (1993) Strategies of phloem loading. Annu Rev Plant Physiol Plant Mol Biol 44:253–281. doi:10.1146/annurev.pp.44.060193.001345
van der Rest B, Boisson AM, Gout E, Bligny R, Douce R (2002) Glycerophosphocholine metabolism in higher plant cells. Evidence of a new glyceryl-phosphodiester phosphodiesterase. Plant Physiol 130:244–255. doi:10.1104/pp.003392
von Arnim AG, Deng XW, Stacey MG (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221:35–43. doi:10.1016/S0378-1119(98)00433-8
Wallace IS, Roberts DM (2004) Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Physiol 135:1059–1068. doi:10.1104/pp.103.033415
Wallace IS, Roberts DM (2005) Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry 44:16826–16834. doi:10.1021/bi0511888
Weig AR, Jakob C (2000) Functional identification of the glycerol permease activity of Arabidopsis thaliana NLM1 and NLM2 proteins by heterologous expression in Saccharomyces cerevisiae. FEBS Lett 481:293–298. doi:10.1016/S0014-5793(00)02027-5
Yano D, Sato M, Saito C, Sato MH, Morita MT, Tasaka M (2003) A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc Natl Acad Sci USA 100:8589–8594. doi:10.1073/pnas.1430749100
Acknowledgments
Azeez Beebo gratefully acknowledges the support of a doctoral fellowship from the French Ministère des Affaires Etrangères et Européennes and the Institut Kurde de Paris. This work was also supported by grants from the French Ministère de l’Enseignement Supérieur et de la Recherche, INRA and CNRS. The authors thank the Arabidopsis Knockout Facility of the University of Wisconsin Biotech Center for providing the pools of T-DNA mutants. We are grateful to Dr. Jeannine Lherminier for her help with electron microscopy. We also wish to thank Dr. Vivienne Gianinazzi-Pearson for her helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Azeez Beebo and Dominique Thomas have contributed equally to this work.
Accession number
The Arabidopsis Genome Initiative locus identifier for the AtTIP1;1 (γ-TIP) gene is At2g36830.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Movie 1
Motion of highly fluorescent vacuolar membranes in leaf epidermal cells (MPG 2741 kb)
Movie 2
Cytochalasin D-induced immobilization of highly fluorescent vacuolar membranes in leaf epidermal cells (MPG 1326 kb)
Movie 3
Constrained motion of nested vacuolar membranes in cortical parenchyma cells of stamen filament (MPG 425 kb)
Rights and permissions
About this article
Cite this article
Beebo, A., Thomas, D., Der, C. et al. Life with and without AtTIP1;1, an Arabidopsis aquaporin preferentially localized in the apposing tonoplasts of adjacent vacuoles. Plant Mol Biol 70, 193–209 (2009). https://doi.org/10.1007/s11103-009-9465-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9465-2