Abstract
Rhomboid proteins comprise a class of serine proteases that are conserved in all kingdoms of organisms. They contain six or seven transmembrane helices and control a wide range of cellular functions and developmental processes by intramembrane proteolysis. This paper provides experimental evidence for the existence of rhomboid proteases in plant mitochondria and chloroplasts. Among 15 putative rhomboid-like proteins in Arabidopsis thaliana, we selected five predicted as mitochondrially targeted. For these proteins we performed the GFP transient assay, and identified two homologues, AtRBL11 (At5g25752) and AtRBL12 (At1g18600) to be targeted into plastids and mitochondria, respectively. Phylogenetic analysis reveals that AtRBL12 or AtRBL11 have only one clear orthologue in plant species with completely sequenced genomes. Complementation of the yeast lacking a functional copy of mitochondrial rhomboid with AtRBL12 indicates that this plant protease, in contrast to the human orthologue, does not recognize the yeast substrates, cytochrome c peroxidase (Ccp1) or dynamin-like GTPase (Mgm1). In agreement with this, we did not observe processing of Mgm1 when labeled precursor of this protein was incubated in vitro with Arabidopsis mitochondrial extract. Our results imply that plant mitochondrial rhomboids function in a specific manner and thus differ from their yeast and mammal counterparts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Regulated intramembrane proteolysis (RIP) is a mechanism by which membrane-tethered substrates undergo site-specific proteolysis within a transmembrane domain (TMD), resulting in liberation of biologically active proteins (Wolfe and Kopan 2004). One of the protease families involved in RIP is rhomboids—polytopic integral membrane serine proteases that have been found in all kingdoms of living organisms (Urban 2006). Recently, several reports have brought to light new facts on the mechanism of action of bacterial rhomboid protease, GlpG, using a mutagenesis-based approach or crystallography (Wang et al. 2006; Wu et al. 2006; Ben-Shem et al. 2007; Baker et al. 2007; Wang and Ha 2007). Their results confirm the six conserved transmembrane helix topology of the protein and establish that the catalytic apparatus of enzyme is separated from the lipid phase by a hydrophilic cavity formed by a protein ring. Furthermore, their results have led to a conclusion of the important role of the fifth transmembrane domain (TMD5) and the fifth loop (L5) in the proteolysis. Despite relatively low sequence homology of rhomboid proteins, the overall structure and substrate specificity of these enzymes are thought to be conserved.
The eukaryotic active rhomboids split between two major subfamilies: large RHO (recently termed secretase) and smaller PARL (Koonin et al. 2003; Lemberg and Freeman 2007). The RHO and PARL subfamilies take their names from their founder members: Rhomboid-1 from Drosophila melanogaster and mammalian PARL (presenilin-associated rhomboid-like protein), respectively. Most of the experimentally characterized rhomboids belong to the RHO subfamily. The Drosophila Rhomboid-1 protein, the best characterized among the RHO subfamily, is located in the Golgi apparatus and regulates epidermal growth factor receptor (EGFR) signaling (Urban et al. 2001).
All mitochondrial rhomboids described so far belong to the PARL subfamily. Beside mammalian PARLs, mitochondrial rhomboids were experimentally proven to be present in Drosophila melanogaster (Rhomboid-7; McQuibban et al. 2006) and Saccharomyces cerevisiae (Pcp1p/Rbd1p/YGR101w; Esser et al. 2002). It was reported that yeast and Drosophila PARL-type rhomboids, are involved in processing species-specific dynamin-like GTPases, enzymes which take part in mitochondrial fusion and inner membrane remodeling (Cipolat et al. 2006; McQuibban et al. 2003; Herlan et al. 2003; McQuibban et al. 2006). Moreover, it was shown that the human PARL is able to cleave the yeast dynamin-like protein (Mgm1) indicating that proteolytic function against this substrate is conserved between yeast and mammals. In contrast, it was demonstrated that PARL does not cleave the human ortholog of Mgm1, OPA1 (Duvezin-Caubet et al. 2007).
The yeast rhomboid, Pcp1, was also shown to process another mitochondrial substrate—cytochrome c peroxidase precursor, Ccp1 (Esser et al. 2002). The mature Ccp1 has an antioxidant function and plays a role in oxidative stress response. Processing of both mitochondrial yeast rhomboid substrates, Mgm1 and Ccp1, involves two cleavages, from which the first cleavage is carried out by different proteases distinct for each substrate, and the second cleavage is mediated in both cases by Pcp1. Therefore, in the yeast cells lacking mitochondrial rhomboid protease, only the bands corresponding to the long isoform of Mgm1 and the intermediate of Ccp1 were observed. However, recent studies in yeast indicate that Ccp1 maturation by mitochondrial rhomboid does not depend on a preceding processing step but on the ATP-dependent membrane dislocation of Ccp1 (Tatsuta et al. 2007). A similar requirement was reported for Mgm1 processing by Pcp1 (Herlan et al. 2004).
To date, information about plant rhomboids is rather limited. Searches of the sequence database reveal the existence of genes coding for rhomboid-like proteins (RBLs) in Arabidopsis thaliana (Koonin et al. 2003), Oryza sativa (Tripathi and Souwdhamini 2006), Populus trichocarpa (Garcia-Lorenzo et al. 2006) and Physcomitrella patens (within this paper). A link between organellar rhomboid proteases and a plastid translocon component was suggested using a yeast mitochondrial-based approach (Karakasis et al. 2007). Recently, A. thaliana RBLs were analyzed with respect to their similarity to Rhomboid-1 (Kanaoka et al. 2005). The closest Rhomboid-1 homologues, AtRBL1 and AtRBL2, were localized to the Golgi apparatus, however only AtRBL2 was able to cleave the Drosophila EGFR ligands (Kanaoka et al. 2005). These results suggest that at least some Drosophila and plant members of the RHO subfamily have similar substrate specificity. This is particularly interesting, since Arabidopsis genome miss genes coding for respective players of the EGF signaling pathway, which suggest that this mode of signaling does not operate in plants. The aim of this paper was to identify mitochondrially localized rhomboids in higher plants and investigate their substrate specificity compared to the yeast Pcp1 rhomboid.
Material and methods
Biological material and growth condition
Arabidopsis thaliana (ecotype Columbia) and Pisum sativum plants were grown in soil under short-day (8-h light/16-h dark) conditions at 23/21°C with a light intensity of 100 μmol m−2 s−1. The Saccharomyces cerevisiae strains BY4741 (MATα, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) and BY4743 (MATa/MATα, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, met15Δ0/+, lys2Δ0/+, ura3Δ0/ura3Δ0) and the corresponding Δpcp1 knockout strains were obtained from EUROSCARF (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/, Winzeler et al. 1999). Yeast strains were grown on complete YPD (1% yeast extract, 2% peptone, 2% glucose) and YPG (1% yeast extract, 2% peptone, 2% glycerol) media or on minimal YNB medium (0.67% yeast nitrogen base, 2% glucose) supplemented with auxotrophic requirements (Kaiser et al. 1994). S. cerevisiae cells were transformed with plasmids by a modified lithium acetate method using PLATE mixture (Kaiser et al. 1994). Total yeast DNA was isolated according to the method of Rose et al. (1990).
cDNA synthesis
Total RNA from leaves was isolated from 21-day old seedlings of Arabidopsis thaliana using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany; http://www1.qiagen.com/). Total cDNA was synthesized using Termoscript reverse transcriptase and (dT)16 primer according to the manufacturer’s instruction (Invitrogen, San Diego, CA; http://www.invitrogen.com/).
DNA constructions
The AtRBLs sequences predicted at least by one algorithm to localize in mitochondria were cloned into the expression vector p35S-GFP (Sakamoto et al. 2002). Two sets of constructs were made for each of the selected AtRBL. The first set contained full-length coding sequences (FL), while the second set of constructs contained DNA sequences corresponding to the putative target peptides and ten additional amino acids (N; Table 2). In the case of AtRBL13 and AtRBL15, for which transit peptide could not be established by TargetP (Table 2), DNA sequences encoded N-terminal regions preceding the first TMD were cloned. Full-length and N-terminal sequences of AtRBLs were amplified from cDNA using gene specific primers with SalI/NcoI or XhoI/NcoI (see Supplemental Table 1 for details) recognition sites added to the ends. Respective fragments were inserted in frame into the p35S-GFP vector. In the case of AtRBL15, we could not clone the full-length coding sequence because cloning sites from the vector polylinker were also present within AtRBL15 (Table 2). All constructs were verified by DNA sequencing. All primers used for cloning are listed in Supplemental Table 1.
For the yeast complementation analysis, we created constructs using two vectors: YEp352 (Fig. 6) and pRS316 (Fig. 5). In both vectors, inserts were cloned under control of the PCP1-Promoter and their structures are schematically presented in Fig. 3. Construct A contains the whole ORF of AtRBL12 (1–336 aa). Construct B has two versions. One of them used in the experiment shown in Fig. 6 contains the PCP1 signal sequence (1–88 aa) followed by the AtRBL12 sequence coding for transmembrane domains 1–6 (59–336 aa). The second version was used in the experiment illustrated in Fig. 5 and contains the PCP1 signal sequence (1–86 aa) followed by the AtRBL12 coding sequence lacking its own signal sequence (92–336 aa). Construct C contains the PCP1 signal sequence (1–88 aa) followed by the AtRBL12 sequence coding for transmembrane domains 1–3 (59–248 aa) and the PCP1 sequence coding for transmembrane domains 4–6 (244–346 aa). Construct D contains the PCP1 sequence coding for the signal sequence and transmembrane domains 1–3 (1–241 aa) and the AtRBL12 sequence for transmembrane domains 4–6 (242–336 aa). The PCP1 sequence for transmembrane domain 4 (242–272 aa) is replaced by the corresponding AtRBL12 sequence (242–276 aa) in construct E. Construct F contains the PCP1 signal sequence (1–88 aa) followed by the AtRBL12 sequence (59–328 aa) and the GFP gene.
For in vitro import assay insert B (the second version) was cloned into a TOPOII vector under T7 promoter (Invitrogen, San Diego, CA) according to the manufacturer’s instruction. For in vitro processing experiments sequences of Ccp1/YKR066C and Mgm1/YOR211C were amplified from yeast genome with gene-specific primers, and subsequently cloned into TOPOII vector (Invitrogen, San Diego, CA). All constructs were verified by sequencing. All primers used for cloning are listed in Supplemental Table 1.
GFP transient assay
A GFP transient assay was performed as described previously (Sakamoto et al. 2003) except that protoplasts for transfection were prepared from mature leaves of Arabidopsis ecotype Columbia. Protoplasts suspended in enzyme solution (0.1% cellulase Onozuka R10 (Yakult; http://www.yakult.co.jp/)), 0.05% pectolyase Y-23 (Kyowa Chemical Products, Japan; http://www.kyowachemical.co.jp), 400 mM mannitol, 10 mM CaCl2, 20 mM KCl, 5 mM EGTA, 20 mM MES, pH 5.7) were gently shaken for 1 h at room temperature. After being washed with Wash buffer (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7), the isolated protoplasts were suspended with MaMg buffer (400 mM mannitol, 15 mM MgCl2, 5 mM MES, pH 5.7). The isolated protoplasts were transformed with 10 μg of each of the plasmid and 100 μl of PEG solution (0.4 M mannitol, 1 M Ca(NO3)2, and 40% (w/v) polyethylene glycol 4000), followed by incubation on ice for 20 min. After being washed with Wash buffer, the protoplasts were suspended in MS medium containing 0.4 M mannitol. The suspended protoplasts were incubated overnight at 23°C with gentle agitation. To visualize mitochondria, protoplasts were treated briefly with 0.5 μM MitoTracker Orange in MS medium containing 0.4 M mannitol. The protoplasts were examined using a fluorescent microscope (Olympus BX51 equipped with DP-50 digital camera) with a B-excitation filter set (for GFP) and G-excitation filter set (for chlorophyll autofluorescence and MitoTracker).
Isolation of mitochondria and yeast total protein extract
Arabidopsis mitochondria were isolated from leaves of 8-week plants according to the procedure described previously by Urantowka et al. (2005). Yeast mitochondria were isolated from 100 ml of overnight culture of BY4742 strain according to Daum et al. (1982). Yeast protein extract was generated from 5 ml of overnight culture by alkaline lysis (Herlan et al. 2003).
In vitro translation, processing and import assays
Precursor proteins were synthesized by in vitro transcription and translation using the TNT-coupled reticulocyte lysate system (Promega; http://www.promega.com/) in the presence of [35S]methionine, according to the manufacturer’s instruction. Import assay into yeast mitochondria according to Daum et al. (1982). In vitro processing reactions were performed according to Whelan et al. (1991). To generate mitochondrial extracts, we used n-Dodecyl β-D-maltoside (DDM; 2 g/g). Subfractionation of mitochondrial proteins was performed using carbonate extraction procedure (Herlan et al. 2003).
Electrophoresis, western blotting and autoradiography
Total protein extracted from yeast culture, as well as Arabidopsis mitochondrial extracts after in vitro processing or import were separated on SDS-PAGE (10% gel) according to Schagger and von Jagow (1987). For Western blotting, proteins were transferred on PVDF membrane (Bio-Rad, Hercules, CA; http://www.bio-rad.com), and probed with anti-Ccp1 and anti-Mgm1 sera. Immunodetection was performed using the Enhanced Chemiluminescence Plus Detection System (ECL, Amersham Pharmacia Biotech, Little Chalfont, UK; http://www1.gelifesciences.com). After in vitro processing and import, separated proteins were visualized by autoradiography.
Identification of plant rhomboid-like proteins and sequence analysis
The preliminary list of rhomboid-like proteins from Physcomitrella patens, Oryza sativa, Arabidopsis thaliana and Populus trichocarpa was made based on sequences reported by others (Kanaoka et al. 2005; Garcia-Lorenzo et al. 2006; Tripathy and Souwdhamini 2006) and our own search of the NCBI BLAST server (for A. thaliana and O. sativa; http://www.ncbi.nlm.nih.gov/BLAST/) and JGI database (for P. trichocarpa and P. patens; http://genome.jgi-psf.org) using the blastp algorithm (http://ncbi.nlm.nih.gov/blast/) with the sequences of non-plant rhomboids as queries. A sequence with an expectation value (E) of <0.05 was considered as a potential rhomboid-like protein. Subsequently, all selected plant sequences were evaluated on the basis of the presence of the rhomboid domain (Pfam accession number PF01694) using InterProScan (http://www.ebi.ac.uk/InterProScan/; Quevillon et al. 2005) and Pfam (http://www.sanger.ac.uk/Software/Pfam/; Finn et al. 2006) with the cutoff level set on 0.01. Transmembrane domains were predicted according to the TMHMM (http://www.cbs.dtu.dk/services/TMHMM/; Krogh et al. 2001) and TMPred (http://www.ch.embnet.org/software/TMPRED_form.html; Hofmann and Stoffel 1993) algorithms. Only proteins containing at least six transmembrane domains and amino acids essential for proteolytic activity, serine residue in the fourth TMD and histidine residue in the sixth TMD, were considered as active plant rhomboids-like proteins.
For sequence alignments Vector NTI® Advance 9.1 program (Invitrogen, San Diego, CA) and for construction of phylogenic trees ClustalX v.1.83 (Jeanmougin et al. 1998) and MEGA3.1 (Kumar et al. 2004) were used. Subcellular localization was predicted with TargetP (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al. 2000), PREDOTAR (http://urgi.infobiogen.fr/predotar/; Small et al. 2004) and iPSORT (http://hc.ims.utokyo.ac.jp/iPSORT/; Bannai et al. 2002) algorithms.
Results
Prediction of catalytically active, plant mitochondrial rhomboid-like proteases
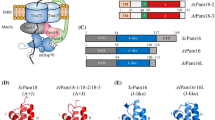
The main goal of this paper was to identify catalytically active rhomboid-like proteases in plant mitochondria. Our strategy described in “Materials and methods” was based on two criteria. First, we searched for Physcomitrella patens, Oryza sativa, Arabidopsis thaliana and Populus trichocarpa rhomboid like proteins containing at least six TMDs as well as residues believed to be essential for catalysis. Using this criterion, we identified 13 rhomboid-like proteins in each plant species (Table 1). Second, subcellular localization of the selected plant RBLs was predicted using three organelle-targeting programs. We considered RBL as potentially mitochondrial when it was predicted by at least one algorithm. We found that eight of moss (e_gw1.84.66.1, estExt_fgenesh1_pg.C_120050, estExt_Genewise1.C_5090005, PpRBL10, PpRBL12, PpRBL14.1, PpRBL14.2, PpRBL15), three of rice (Os11g47840, OsRBL12, OsRBL14), five of Arabidopsis (AtRBL6, AtRBL11, AtRBL12, AtRBL14 and AtRBL15) and four of poplar (PtRBL10, PtRBL12, PtRBL14, PtRBL15) putative catalytically active rhomboid-like proteins may reside in mitochondria (Table 1). To avoid any ambiguity in nomenclature, we have termed rhomboid-like genes from Arabidopsis thaliana and Populus trichocarpa according to proposed names in literature (Kanaoka et al. 2005; Garcia-Lorenzo et al. 2006). In the case of Physcomitrella patens and Oryza sativa, we proposed a nomenclature based on their closest homologues in Arabidopsis (Fig. 1). We have not named the rice and moss RBLs which do not exhibit a clear orthological relationship with the Arabidopsis rhomboids. Searching for rice RBLs, we found that the gene at the locus Os3g44830 encodes three proteins differing in N-termini (Table 1) and all of them are most homologous to AtRBL15. Therefore, we named them OsRBL15a–OsRBL15c.
Unrooted N-J tree computed from multiple sequence alignments of Physcomitrella patens (Pp), Oryza sativa (Os), Arabidopsis thaliana (At) and Populus trichocarpa (Pt) potentially active rhomboid-like proteases and non-plant mitochondrial members of PARL subfamily (DmRho7:Drosophila melanogaster; HsPARL:Homo sapiens; ScPcp1:Saccharomyces cerevisiae; TgROM6:Toxoplasma ghondii). Bootstrap values are based on 1000 replicatives. For a clear view the branches for non-plant mitochondrial rhomboids and names of AtRBLs predicted as mitochondrial are in bold. The name of yeast Pcp1 is underlined. We indicated proteins predicted to localize in mitochondria with (m); proteins localized experimentally in mitochondria and in Golgi apparatus with (M) and (G), respectively. For the other abbreviations see Table 1
The topology of the phylogenetic tree shown in Fig. 1 clearly indicates that the plant catalytically active rhomboid-like proteins segregate into two groups. One of them contains AtRBL1 and AtRBL2, rhomboids experimentally localized to the Golgi apparatus in Arabidopsis cell and have been designated as members of the RHO subfamily (Kanaoka et al. 2005). The second group includes plant rhomboid-like proteins along with all known mitochondrial members of the PARL subfamily: Pcp1 (yeast), PARL (human), Rhomboid-7 (D. melanogaster) and TgROM6, which was characterized as the closest Toxoplasma ghondii orthologue of the mitochondrial rhomboids (Dowse and Soldati 2005).
Sub-cellular location of RBL proteins examined by GFP transient assay
To verify the subcellular localization of the Arabidopsis rhomboid–like proteases predicted as mitochondrially targeted (AtRBL6, AtRBL11, AtRBL12, AtRBL14, and AtRBL15), we conducted the GFP transient expression assay. In this analysis we also included AtRBL13 because based on the phylogenetic analysis (Fig. 1) this protease is closely related to other plant RBLs predicted as mitochondrial. Two types of constructs were used in our experiments. In the first set of constructs, the GFP reporter was fused in frame to the C terminus of the full-length coding sequences (FL), while the second set of constructs contained GFP attached to sequences coding for N-terminal regions of proteins (N) (Table 2). These constructs were used for transfection of protoplasts prepared from Arabidopsis mesophyll cells.
Based on the clustering of the phylogenetic analysis, it became evident that the AtRBL11, AtRBL12 are most closely related to the non-plant mitochondrial rhomboids (Fig. 1). Therefore, we started our experiments by looking for sub-cellular localization of these two enzymes. First, we found that FL-AtRBL11-GFP was localized in chloroplasts, as the green fluorescence co-localized with chlorophyll autofluorescence (Fig. 2a). Sometimes dotted signals were seen as located on chloroplast envelope, but they were never found in the cytosol. The chloroplast-targeting was consistent when N-AtRBL11-GFP was used. These results suggest that AtRBL11 is a chloroplast protein.
AtRBL11 locates in chloroplast, and AtRBL12 in mitochondria of Arabidopsis mesophyll protoplasts. (a) Protoplasts expressing GFP N-terminally fused with FL-AtRBL11 (208 aa) and N-AtRBL11 (70 aa). As a control of chloroplast targeting, a protoplast expressing FtsH1-GFP (Sakamoto et al. 2003) is also shown. Close-up pictures of a chloroplast observed by GFP, chlorophyll fluorescence and the merged image are shown at the bottom. (b) Protoplasts expressing GFP N-terminally fused with N-AtRBL12 (95 aa) and N-ATPδ, as a control of mitochondrial targeting. A high-magnification image of protoplasts expressing N-AtRBL12 is displayed on the bottom of (b) to show the co-localization of GFP with MitoTracker. (c) A protoplast expressing GFP without the targeting signal is shown as a negative control. In each panel, fluorescent signals corresponding to GFP (left) and chlorophyll autofluorescence (middle) are shown together with images of protoplasts taken by light microscopy (right)
Next, localization of AtRBL12 was examined. In our assay, however, we were unable to obtain transformants expressing FL-AtRBL12-GFP. Two possible reasons could be considered: one, inefficient expression of the GFP fused to FL-AtRBL12 or alternatively, that overexpression of the fusion protein was toxic, so that very little or no transformants were visible. In contrast, we obtained transformants using N-AtRBL12-GFP. These transformants contained GFP signals that were globular or rod-shaped, similar to those obtained by the control GFP fused to a presequence from ATPδ (δ subunit of mitochondrial ATPase) (Fig. 2b). A high-magnification image of protoplasts expressing N-AtRBL12 shows indeed the co-localization of the GFP with MitoTracker signals. However, we could not completely rule out possibility that RBL12 might be also targeted into other cellular compartments since we only used its N-terminal region.
We also detected GFP signals when GFP fused to FL-AtRBL6 and FL-AtRBL14 was expressed in mesophyll protoplasts. However, the detected signals appear to correspond neither to chloroplast nor to mitochondria, and were not co-localized with chlorophyll autofluorescence or MitoTracker-orange signals (see Supplemental Fig. 2). Although they co-localized with some membrane structures such as the Golgi apparatus, the actual location of these GFP fusion proteins were undetermined in this study. Therefore, we considered that AtRBL6 and AtRBL14 were neither chloroplastic nor mitochondrial proteins. For AtRBL15 and AtRBL13, we were unable to obtain transformants regardless of using FL- or N-GFP fusions (data not shown). In summary, our transient assay allowed us to identify two Arabidopsis RBL proteins, AtRBL11 and AtRBL12, to be localized in chloroplasts and mitochondria, respectively.
Ability of AtRBL12 to cleave Pcp1 substrates
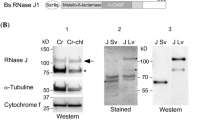
Given that AtRBL12 is the closest Arabidopsis homologue of Pcp1 (Fig. 1, protein sequence identity of 29%) and is localized in mitochondria, we wondered whether AtRBL12 might also share substrate specificity with the yeast mitochondrial rhomboid. To address this issue, we tested the ability of AtRBL12 to cleave the Pcp1 substrates, Mgm1 and Ccp1, in yeast cells lacking a functional copy of the yeast mitochondrial rhomboid. In the first approach, Δpcp1 cells were transformed with plasmids containing the full open reading frame of AtRBL12 (construct A in Fig. 3) or PCP1 sequence as a positive control. Then, mitochondrial fractions were isolated and subjected to western blot analysis using anti-Ccp1 serum. As shown in Fig. 6a, the band corresponding to the mature cytochrome c peroxidase (mCcp1) is visible in mitochondrial fractions isolated from the wild type and Δpcp1 cells transformed with the Pcp1-coding construct, but not from the Δpcp1 cells transformed with the construct containing the full open reading frame of AtRBL12 (Fig. 6a, construct A).
Structure of constructs coding for Pcp1, AtRBL12 and various hybrids of these two proteins. All inserts were under control of the Pcp1 promoter (PCP1 (Prom); red). Except the construct A coding for AtRBL12, all other contained Pcp1 signal sequence (PCP1 (SS); yellow). The remaining colors represent fragments derived from AtRBL12 (green), PCP1 (brown), GFP (blue). The top construct: the full-length of PCP1; A: the full-length of AtRBL12; B: AtRBL12 in which native signal sequence was replaced with PCP1 (SS); C: PCP1 with three N-terminal TMDs replaced by respective TMDs from AtRBL12; D: PCP1 with three C-terminal TMDs replaced by respective TMDs from AtRBL12; E: PCP1 in which the fourth, catalylic TMD was replaced by respective TMD from AtRBL12; F: the hybrid B fused with GFP
At that time we assumed that lack of Ccp1 processing by AtRBL12 might be because of an inefficient transport of plant protease into yeast mitochondria or an improper sub-mitochondrial targeting. Therefore, we decided to replace the native signal peptide of AtRBL12 with putative mitochondrial targeting sequence of PCP1 (Fig. 3, construct B). Due to the lack of an antibody directed against the plant rhomboid, we tested mitochondrial targeting of a hybrid protein encoded by the construct B using two methods. Firstly, we performed an import into isolated yeast mitochondria of a radioactive-labeled hybrid protein B (Fig. 4a). A protein band smaller than the precursor and resistant to proteinase K digestion was detected after the import, suggesting that the hybrid protein B was targeted into mitochondria and that the import was accompanied by processing. No import was observed in the presence of valinomycin, supporting an expectation that the import of AtRBL12 involves the protein translocation system of the inner membrane. To further verify this finding, yeast mitochondria were isolated and separated into the soluble and membrane fractions using sodium carbonate procedure which discriminates between peripheral and integral membrane proteins. The purity of both fractions was monitored using antibodies directed against marker proteins (Fig. 4b). This experiment confirmed that the mature product of the hybrid B processing is an integral membrane protein (Fig. 4c). In the second approach, we fused the sequence coding for the hybrid protein B at its 3′-end to the GFP gene and the resulting construct F (Fig. 3) was used for yeast transformation. The fusion protein of expected size, ∼55 kD, was detected exclusively with the mitochondrial membrane fraction by the anti-GFP antibody (Fig. 4c). Taken together, these results demonstrate that the hybrid protein B is efficiently imported into mitochondria and it is totally located in the membrane fraction.
Mitochondrial targeting of the hybrid protein B. (a) Import of radio-labeled hybrid protein B into isolated yeast mitochondria. Upon import the hybrid is processed, which results in a mass drop of about 8.5 kDa. The import did not occur after addition of valinomycin (0.5 μg/ml). TP, translation product; IMP, import; IMP/PK20, import + Proteinase K 20 μg/ml; IMP/PK100, import + PK 100 μg/ml; IMP/VAL, import reaction carried out with addition of valinomycin; IMP/PK20/VAL, import + valinomycin + Proteinase K 20 μg/ml. (b) Carbonate fractionation of mitochondria. Fraction containing soluble (SOL) and membrane (MEM) proteins were probed with antibodies directed against marker proteins: porin (29 kDa, OM), Mcr1 (34/32 kDa; OM/IMS); Tim23 (23 kDa, IM). (c) Detection of the Pcp1-AtRBL12 fusions in membrane and soluble fractions. Sub-fractionation was carried out for mitochondria after the import reaction (left panel; autoradiography), but also for mitochondria isolated from Δpcp1 cells transformed with construct E (middle panel; immunodetection using anti-Pcp1 antibodies) and F (right panel; immunodetection using anti-GFP antibodies)
Subsequently, Δpcp1 cells (both the haploid and diploid strain) were transformed with the construct B. Total protein extracts from the transformed cells and controls were subjected to western blot analysis using anti-Ccp1 serum as well as antibodies against Mgm1. It is known that the Pcp1 protease generates the short isoform of Mgm1 and the mature form of Ccp1, respectively. As expected, the large (L) and the short (S) isoforms of Mgm1 and the mature form of Ccp1 were observed in wild type cells and in the Δpcp1 cells transformed with the yeast mitochondrial rhomboid. However, plasmid-borne expression of the hybrid protein B did not rescue cleavage of either substrate and the observed patterns were the same as those in Δpcp1 (Fig. 5). Both, Ccp1 and Mgm1, were processed only to the intermediate forms. In the case of both substrates, we also observed additional bands, which were distinct in size from the isoforms described above (asterisks). The occurrence of additional bands in the Δpcp1 cells as products of nonspecific degradation of Mgm1 or Ccp1 has already been reported (Herlan et al. 2003; Michaelis et al. 2005; Tatsuta et al. 2007).
Expression of the hybrid construct B did not restore processing of either Ccp1 or Mgm1 in Δpcp1 cells. Δpcp1 cells of both haploid and diploid strains were transformed with pRS316 plasmids carrying the hybrid construct B or PCP1 as a positive control. Untransformed Δpcp1 cells are marked by “–”. Protein extracts from the yeast cells were probed using Mgm1- (the upper panel) and Ccp1-directed (the bottom panel) sera. ‘L’ and ‘S’ correspond to the long and short form of Mgm1 respectively, while M – to a mature form of Ccp1 protein. Additional, unspecific forms for both substrates are marked by asterisks
The experiments described above imply that the plant mitochondrial rhomboid is not able to generate the mature form of either of the Pcp1 substrates in yeast cells. To identify regions responsible for apparently divergent functional properties of plant AtRBL12 and yeast Pcp1, several hybrids of Pcp1 and AtRBL12 were generated and expressed in Δpcp1 cells (Fig. 3). Replacement of three N-terminal TMDs of Pcp1 (construct C) or three C-terminal TMDs of Pcp1 (construct D) by the corresponding domains of AtRBL12 resulted in loss of the Pcp1 processing activity towards its native substrate, cytochrome c peroxidase (Fig. 6a). Moreover, the PCP1 gene in which only the fourth TMD containing the catalytic serine was replaced by the respective AtRBL12 sequence (construct E) did not display catalytic activity against Ccp1 (Fig. 6a). Using an anti-Pcp1 antibody, we confirmed that the hybrid protein E is integrated into the mitochondrial membrane fraction (Fig 4c). This hybrid protein migrates slower than native Pcp1 (Fig. 6b). We think this may be due to the difference in hydrophobicity between these two proteins, caused by the presence of four addition polar residues in the exchanged fourth domain. Below, the band corresponding to Pcp1 or the hybrid protein E we detected additional bands (asterisks), which as we assume, may result from degradation of these proteins.
Expression of various hybrids of Pcp1 and AtRBL12 did not restore processing of Ccp1 in Δpcp1 cells. (a) Δpcp1 cells were transformed with full-length AtRBL12 (construct A) and various Pcp1-AtRBL12 hybrids (constructs B–E) and mitochondrial extracts from transformed cells were probed with anti-Ccp-1 antibodies. The structures of used constructs are presented in Fig. 3. As positive controls, mitochondrial extracts from the wild type yeast (WT), and Δpcp1 cells transformed with PCP1 (Δpcp1 PCP1) were used. As negative controls yeast lacking functional Pcp1(“-”) or Ccp1 (ccp1 Δ) were utilized. The intermediate (iCcp1) and the mature form (mCcp) of Ccp1 are marked by arrows on the right. (b) Equal amounts of mitochondrial proteins of the wild type (WT), Δpcp1 cells (“-”) and Δpcp1 cells transformed with PCP1 or the hybrid construct E were loaded onto non-adjacent lanes of the same gel, fractionated and then subjected to immunoblotting using anti-Pcp1 serum. The Pcp1 protein of the expected size was detected in WT and Δpcp1 cells transformed with PCP1. The fusion protein Pcp1-AtRBL12 encoded by the hybrid E was also detected in mitochondria. We also detected additional bands (asterisks), which we suspect as degradation products
The above results suggest that the functional differences between AtRBL12 and Pcp1 appear to be at least caused by differences in their TM domains. In addition, it is possible, that other proteins not present in yeast cells are needed for activity of AtRBL12. To challenge this assumption, we decided to carry out in vitro processing of radio-labeled precursors of Mgm1 and Ccp1 using Arabidopsis mitochondrial extract (Fig. 7). To prove that the conditions used in the experiment do not prevent efficient Mgm1 and Ccp1 maturation, we used yeast mitochondrial extract as a positive control. As a negative control, mitochondrial extract generated from the Δpcp1 cells was also tested (Fig. 7a and b, right panel). Both Mgm1 and Ccp1 were processed by the yeast mitochondrial extract. In contrast, after in vitro processing, we did not receive the mature form of Mgm1 using Arabidopsis extract (Fig. 7a). Incubation of radio-labeled precursor of Ccp1 with plant extract resulted in a protein bands pattern different from the pattern observed with yeast extract, but a form in size of the mature Ccp1 was visible (Fig. 7b). Rather, different cleavage pattern suggests that the pathway connected with the appearance of a protein of mature Ccp1 size is not the same in plants and yeast. Moreover, a form similar in size to the mature Ccp1 was also present after processing with mitochondrial extract generated from the Δpcp1 cells. This result indicates that in the absence of the rhomboid protease, Ccp1 precursor is cleaved by unknown peptidase/s to the form with electrophoretic mobility of the mature Ccp1. In this context, interpretation of the proteolytic pattern obtained with Arabidopsis extract is difficult. Taken together, results obtained in vitro using plant mitochondrial extract, suggest that AtRBL12 does not recognize at least one yeast rhomboid substrate.
Processing of Mgm1 and Ccp1 precursors. Left panel: Autoradiography of in vitro processing products of radio-labeled Mgm1 (a) and Ccp1 (b) precursors using mitochondrial extracts isolated from Arabidopsis leaves (A.th.) and yeast wild-type cells (S.c./WT). Middle panel: Immunodetection of the mature and intermediate form of Mgm1 (a) and Ccp1 (b) in total cell extracts isolated from S.c./WT cells and yeast Δpcp1 cells (S.c./Δpcp1). Right panel: Autoradiography of in vitro processing products of radio-labeled Mgm1 (a) and Ccp1 (b) precursors using Arabidopsis, S.c./WT and S.c./Δpcp1 mitochondrial extracts. TP – translation product; L and S correspond to long and short form of Mgm1, respectively, while iCcp1 and mCcp1, to the precursor and mature forms of Ccp1, respectively
Discussion
This paper provides the first experimental evidence for the existence of plant mitochondrial (AtRBL12, At1g18600) and chloroplastic (AtRBL11, At5g25752) rhomboid proteases. The mitochondrial enzyme is also the first RIP protease identified in plant mitochondria. Quite recently, another type of RIP protease which belongs to the S2P family was identified in plastids (Bolter et al. 2006). Here, we showed that AtRBL12 does not cleave two known substrates of Pcp1, the yeast mitochondrial rhomboid. This result was unexpected since previous studies indicated that yeast substrates were efficiently processed by PARL, the mammalian orthologue of Pcp1 (McQuibban et al. 2003). Thus, our data imply that the mechanism of substrate recognition/processing by plant mitochondrial rhomboids has evolved in a specific manner, different from that of yeast and mammals.
We searched for putative active and mitochondrial rhomboid-like proteases in the Arabidopsis thaliana, Oryza sativa, Populus trichocarpa genomes as well as in recently sequenced genome of moss, Physcomitrella patens. In this study, we excluded rhomboids that do not possess the proteolytic diad, and therefore our phylogenetic analysis differed in some details from previous reports predicting rhomboid-like sequences (Tripathi and Sowdhamini 2006; Garcia-Lorenzo et al. 2006). We found that several proteins match the searching criteria (presence of proteolytic diad and putative mitochondrial localization) in each of the analyzed species (Fig. 1). Then, we verified mitochondrial targeting of selected Arabidopsis proteins using GFP assay and found that one from the five possible candidates is targeted to mitochondria (AtRBL12). The other candidate was found to be located in chloroplasts (AtRBL11). The tree constructed in this work (Fig. 1) indicates that mitochondrial AtRBL12 or chloroplastic AtRBL11 are members of two independent, well-separated clades and have only one clear orthologue in other plant species. Moreover, all rhomboids from the AtRBL12 clade are predicted to be localized in mitochondria. It is worth noting that the AtRBL12 clade has none of the non-plant mitochondrial rhomboids, nevertheless it is very closely related to the clade encompassing all known the non-plant mitochondrial rhomboids (Fig. 1). In all organisms analyzed up to now there is only one mitochondrial rhomboid. It seems that this is also true for plants. Even if plants possess additional mitochondrial rhomboids, these enzymes are not closely related to any reported mitochondrial rhomboids.
Our results shed new light on the issue of conservation of substrate specificity between mitochondrial members of the rhomboid family. We showed that ectopic expression of the mitochondrial rhomboid from Arabidopsis restored neither Mgm1 nor Ccp1 processing in Δpcp1 cells, even though the plant rhomboid was efficiently imported to yeast mitochondria and located in the membrane fraction (Fig. 3b). Morover, we did not observe the mature form of Mgm1 when radio-labeled precursor of this protein was incubated with Arabidopsis mitochondrial extract. The result obtained for Ccp1 was uncertain because a protein corresponding in size to mature Ccp1 was also observed in the absence of yeast mitochondrial rhomboid. It has been reported for yeast that Ccp1 is degraded to protease-resistant fragment corresponding to its peroxidase domain in the absence of the rhomboid protease (Tatsuta et al. 2007). Therefore, we incline to the explanation that the plant rhomboid is not effective against Ccp1 during in vitro processing and the final proteolytic product is generated by other protease.
From both experiments, in vivo (complementation in yeast) and in vitro (processing with mitochondrial extract), we inferred that the plant mitochondrial rhomboid (AtRBL12) does not recognize yeast substrates. In contrast, the human mitochondrial rhomboid PARL was shown to restore proteolytic processing of Mgm1 and Ccp1 in Δpcp1 cells (McQuibban et al. 2003). The substrate conservation across species was also reported for rhomboids from the RHO subfamily. Members of this subfamily from diverse organisms, including bacteria, invertebrates, vertebrates and plants, have the ability to cleave Drosophila substrates such as Spitz and Keren (Urban et al. 2002). Thus, many rhomboid proteins maintain strong substrate specificity throughout evolution; however, no substrate inter-changeability between the rhomboids from different species was also described. The best known example concerns thrombomodulin, a specific substrate of vertebrate RHBDL2-like rhomboids (Lohi et al. 2004). Bacterial and Drosophila rhomboids are not able to cleave thrombomodulin, whereas the vertebrate RHBDL2-like rhomboids cleave Drosophila Spitz efficiently. A different example represents AtRBL1, an enzyme with highest homology to Drosophila Rho-1 among Arabidopsis rhomboids (Kanaoka et al. 2005). Specific substrate for this enzyme has not been identified, but it was found that AtRBL1 does not cleave the Drosophila substrates.
In this study, we showed that the mitochondrial plant rhomboid, AtRBL12, is not able to mediate processing of substrates recognized by other closely related rhomboids. Neither Pcp1 substrates such as Ccp1 nor Mgm1, can be found by homology in the genome of Arabidopsis (Praefcke and McMahon 2004; Logan 2006; Passardi et al. 2007). Probably, the newly identified Arabidopsis mitochondrial rhomboid evolved to fulfill some distinct functions specific for plants.
Abbreviations
- Ccp1:
-
Cytochrome c peroxidase
- GFP:
-
Green fluorescent protein
- RBL:
-
Rhomboid-like protein
- RIP:
-
Regulated intramembrane proteolysis
- TMD:
-
Transmembrane domain
References
Baker RP, Young K, Feng L, Shi Y, Urban S (2007) Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc Natl Acad Sci USA 104:8257–8262. doi:10.1073/pnas.0700814104
Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18:298–305. doi:10.1093/bioinformatics/18.2.298
Ben-Shem A, Fass D, Bibi E (2007) Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci USA 104:462–466. doi:10.1073/pnas.0609773104
Bolter B, Nada A, Fulgosi H, Soll J (2006) A chloroplastic inner envelope membrane protease is essential for plant development. FEBS Lett 580:789–794. doi:10.1016/j.febslet.2005.12.098
Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K et al (2006) Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126:163–175. doi:10.1016/j.cell.2006.06.021
Daum G, Gasser SM, Schatz G (1982) Import of proteins into mitochondria. Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J Biol Chem 257:13075–13080
Dowse TJ, Soldati D (2005) Rhomboid-like proteins in Apicomplexa: phylogeny and nomenclature. Trends Parasitol 21:254–258. doi:10.1016/j.pt.2005.04.009
Duvezin-Caubet S, Koppen M, Wagener J, Zick M, Israel L, Bernacchia A et al (2007) OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA rotease in mitochondria. Mol Biol Cell 18:3582–3590. doi:10.1091/mbc.E07-02-0164
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016. doi:10.1006/jmbi.2000.3903
Esser K, Tursun B, Ingenhoven M, Michaelis G, Pratje E (2002) A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J Mol Biol 323:835–843. doi:10.1016/S0022-2836(02)01000-8
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T et al (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251. doi:10.1093/nar/gkj149
Garcia-Lorenzo M, Sjodin A, Jansson S, Funk C (2006) Protease gene families in Populus and Arabidopsis. BMC Plant Biol 6:30. doi:10.1186/1471-2229-6-30
Herlan M, Vogel F, Bornhovd C, Neupert W, Reichert AS (2003) Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem 278:27781–27788. doi:10.1074/jbc.M211311200
Herlan M, Bornhövd C, Hell K, Neupert W, Reichert AS (2004) Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J Cell Biol 165:167–173. doi:10.1083/jcb.200403022
Hofmann K, Stoffel W (1993) TMbase – a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 374:166
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405. doi:10.1016/S0968-0004(98)01285-7
Kanaoka MM, Urban S, Freeman M, Okada K (2005) An Arabidopsis Rhomboid homolog is an intramembrane protease in plants. FEBS Lett 579:5723–5728
Karakasis K, Taylor D, Ko K (2007) Uncovering a link between a plastid translocon component and rhomboid proteases using yeast mitochondria-based assays. Plant Cell Physiol 48:655–661. doi:10.1093/pcp/pcm031
Kaiser C, Michaelis S, Mitchel A (1994) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L (2003) The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol 4:R19. doi:10.1186/gb-2003-4-3-r19
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi:10.1006/jmbi.2000.4315
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163. doi:10.1093/bib/5.2.150
Lemberg MK, Freeman M (2007) Functional and evolutionary implications of enhanced genomic analysis of rhomboid intermembrane proteases. Genome Res 17:1634–1646. doi:10.1101/gr.6425307
Logan DC (2006) Plant mitochondrial dynamics. Biochim Biophys Acta 1763:430–441. doi:10.1016/j.bbamcr.2006.01.003
Lohi O, Urban S, Freeman M (2004) Diverse substrate recognition mechanisms for rhomboids; thrombomodulin is cleaved by Mammalian rhomboids. Curr Biol 14:236–241
McQuibban GA, Saurya S, Freeman M (2003) Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423:537–541. doi:10.1038/nature01633
McQuibban GA, Lee JR, Zheng L, Juusola M, Freeman M (2006) Normal mitochondrial dynamics requires rhomboid-7 and affects Drosophila lifespan and neuronal function. Curr Biol 16:982–989. doi:10.1016/j.cub.2006.03.062
Michaelis G, Esser K, Tursun B, Stohn JP, Hanson S, Pratje E (2005) Mitochondrial signal peptidases of yeast: the rhomboid peptidase Pcp1 and its substrate cytochrome c peroxidase. Gene 354:58–63. doi:10.1016/j.gene.2005.04.020
Passardi F, Bakalovic N, Teixeira FK, Margis-Pinheiro M, Penel C, Dunand C (2007) Prokaryotic origins of the non-animal peroxidase superfamily and organelle-mediated transmission to eukaryotes. Genomics 89:567–579. doi:10.1016/j.ygeno.2007.01.006
Praefcke GJ, McMahon HT (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5:133–147. doi:10.1038/nrm1313
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R et al (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–120. doi:10.1093/nar/gki442
Rose MD, Winston F, Hieter P (1990) Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Sakamoto W, Tamura T, Hanba-Tomita Y, Murata M, Sodmergen (2002) The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7:769–780. doi:10.1046/j.1365-2443.2002.00558.x
Sakamoto W, Zaltsman A, Adam Z, Takahashi Y (2003) Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15:2843–2855. doi:10.1105/tpc.017319
Schagger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379. doi:10.1016/0003-2697(87)90587-2
Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4:1581–1590. doi:10.1002/pmic.200300776
Tatsuta T, Augustin S, Nolden M, Friedrichs B, Langer T (2007) m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J 26:325–335. doi:10.1038/sj.emboj.7601514
Tripathi LP, Sowdhamini R (2006) Cross genome comparisons of serine proteases in Arabidopsis and rice. BMC Genomics 7:200. doi:10.1186/1471-2164-7-200
Urantowka A, Knorpp C, Olczak T, Kolodziejczak M, Janska H (2005) Plant mitochondria contain at least two i-AAA-like complexes. Plant Mol Biol 59:239–252. doi:10.1007/s11103-005-8766-3
Urban S (2006) Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev 20:3054–3068. doi:10.1101/gad.1488606
Urban S, Lee JR, Freeman M (2001) Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107:173–182. doi:10.1016/S0092-8674(01)00525-6
Urban S, Schlieper D, Freeman M (2002) Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr Biol 12:1507–1512. doi:10.1016/S0960-9822(02)01092-8
Wang Y, Ha Y (2007) Open-cap conformation of intramembrane protease GlpG. Proc Natl Acad Sci USA 104:2098–2102. doi:10.1073/pnas.0611080104
Wang Y, Zhang Y, Ha Y (2006) Crystal structure of a rhomboid family intramembrane protease. Nature 444:179–180. doi:10.1038/nature05255
Whelan J, Knorpp C, Harmey MA, Glaser E (1991) Specificity of leaf mitochondrial and chloroplast processing systems for nuclear-encoded precursor proteins. Plant Mol Biol 16:283–292. doi:10.1007/BF00020559
Winzeler EA et al (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906. doi:10.1126/science.285.5429.901
Wolfe MS, Kopan R (2004) Intramembrane proteolysis: theme and variations. Science 305:1119–1123. doi:10.1126/science.1096187
Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, et al (2006) Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol 13:1084–1091. doi:10.1038/nsmb1179
Acknowledgements
We would like to thank Drs. Thomas Langer and Andreas Reichert for antisera. We also thank Chieko Hattori for her assistance in the GFP assay and Robert Wysocki for his help with yeast manipulations. This work was supported by a grant from the State Committee for Scientific Research (KBN), Poland (no. N303 3900/33 to HJ).
Author information
Authors and Affiliations
Corresponding author
Additional information
Beata Kmiec-Wisniewska, Katrin Krumpe, and Adam Urantowka contributed equally to this work and are listed in an alphabetical order.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2008_9359_MOESM1_ESM.pdf
Supplemental Figure 1: Multiple sequence alignment of all rhomboid-like proteins from Physcomitrella patens, Oryza sativa, Arabidopsis thaliana and Populus trichocarpa. Mode of identification of RBLs was described in Materials and methods in details. The alignment was performed using Vector NTI® Advance 9.1. In the alignment we used only fragments encompassing the ‘rhomboid core’, i.e. six-transmembrane domain (TMD) segment containing the active diad residues in the fourth and the sixth TMD (TM4 and TM6, respectively). To each TMD, predicted using TMHMM 2.0 and TMPred, we added a two-amino acid margin. Fragments of loops connecting the TMDs were omitted, since, due to their high sequence variability, they might have introduced noise in the alignment. A similar approach has already been used by Koonin et al. (2003). Numbers on the left side of the TM1 and the right side of the TM6 correspond to the position in the whole RBL sequence, whereas numbers between the TMDs correspond to the numbers of amino acids in the predicted loops. To designate RBLs we used protein IDs (according to JGI database) for moss (Pp) and poplar (Pt), gene loci (according to TIGR) for rice (Os) and protein IDs (according to NCBI) for Arabidopsis (At). Alignment of RBLs revealed that some of the sequences miss one or both catalytic diad residues (S in the fourth and H in the sixth TMD). Position of catalytic diad residues are marked under the sequences. RBLs without complete rhomboid domain (indicated with an asterisk), lacking complete six-TMD core (underlined) and inactive ones were excluded from further analysis. (PDF 90 kb)
11103_2008_9359_MOESM2_ESM.tif
Supplemental Figure 2: Cellular localization of the GFP fusion protein in Arabidopsis mesophyll protoplasts. Protoplasts expressing GFP N-terminally fused with FL-AtRBL6, FL-AtRBL14 are shown. Fluorescent signals corresponding to GFP (left) and chlorophyll autofluorescence (middle) were shown together with images of protoplasts taken by light microscopy (right). (TIF 263 kb)
Rights and permissions
About this article
Cite this article
Kmiec-Wisniewska, B., Krumpe, K., Urantowka, A. et al. Plant mitochondrial rhomboid, AtRBL12, has different substrate specificity from its yeast counterpart. Plant Mol Biol 68, 159–171 (2008). https://doi.org/10.1007/s11103-008-9359-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-008-9359-8