Abstract
The vtc2 mutant of Arabidopsis thaliana is vitamin C deficient and is defective in the ascorbate biosynthesis enzyme GDP-l-Galactose phosphorylase/l-Galactose guanylyltransferase. The connection between the VTC2 gene and this enzyme has only recently been established, and little is known about the molecular characteristics of the VTC2 gene. In this paper, the expression of the VTC2 gene was characterized on both the RNA and the protein level. The VTC2 gene was expressed in all developmental stages, and the mRNA was expressed at a higher level in green tissues than in the root. VTC2 mRNA expression was induced strongly by exposing dark-grown seedlings to light, to levels higher than found in light-grown seedlings. A VTC2:GUS fusion protein was detected only in green tissues. Unexpectedly, a VTC2:YFP fusion protein was found not only in the cytosol, but also in the nucleus, which suggests that GDP-l-Galactose phosphorylase/l-Galactose guanylyltransferase might be a dual-function protein, which has both enzymatic and regulatory function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ascorbate is most known for its role as an antioxidant (Smirnoff 2000), but it is important for a multitude of functions in plants (Smirnoff 1996, 2000). For example, it is a precursor for the synthesis of oxalate and tartrate (Loewus 1988) and as a co-factor it is involved in the synthesis of hydroxy-proline (Arrigoni et al. 1977) and plant hormones (Arrigoni and deTullio 2002). It also plays a role in electron transport (Smirnoff 1996; Asada 1999) and in cell division (Horemans et al. 2000). The importance of ascorbate for the plant is underlined by the fact that, so far, no viable mutant has been found that is totally devoid of ascorbate (Smirnoff et al. 2001).
The biosynthesis of ascorbate in plants has only recently been elucidated and multiple pathways have been suggested (Hancock and Viola 2005). The best-described pathway (Smirnoff–Wheeler) consists of the formation of ascorbate from guanosine diphosphate-mannose (GDP-Man) with the intermediates GDP-l-galactose (GDP-l-Gal), l-Gal 1-phosphate, l-Gal, and l-galactone-1,4-lactone (Wheeler et al. 1998). All enzymes of this pathway have been identified, many of them by cloning vitamin C-deficient (vtc) mutants. The vtc1 mutant is defective in the GDP-d-Man pyrophosphorylase gene (Conklin et al. 1999; Lukowitz et al. 2001), and the vtc4 mutant in l-Gal-1-phosphate phosphatase (Laing et al. 2004; Conklin et al. 2006). Finally, the last missing enzyme of the Smirnoff–Wheeler pathway, GDP-l-Gal phosphorylase/l-Gal guanylyltransferase (Laing et al. 2007; Linster et al. 2007), was recently identified as the VTC2 gene (Jander et al. 2002). Three other pathways have been described (reviewed in Hancock and Viola 2005): the l-Gulose pathway (Wolucka and Van Montagu 2003), the d-Galacturonic acid pathway (Agius et al. 2003), and the Myo-Inositol pathway (Lorence et al. 2004), but these seem to be of minor importance, as a vtc2vtc5 double mutant that lacks two enzymes with GDP-l-Gal phosphorylase activity is unable to grow past the seedling stage if not supplemented with ascorbate (Dowdle et al. 2007).
The VTC2 gene has orthologs in invertebrates, vertebrates and plants and has a low similarity with members of the histidine triad (HIT) protein superfamily (Linster et al. 2007). Proteins of the HIT protein superfamily are characterized as nucleotide-binding proteins and hydrolases. The closest homologue in Arabidopsis thaliana is the At5g55120 gene, which was shown by Dowdle et al. (2007) to encode a second GDP-l-Gal phosphorylase with similar properties to VTC2. An existing insertion mutant, vtc5, had about 80% of the wild-type ascorbate level.
The VTC2 protein has two different enzymatic activities (Wolucka and Van Montagu 2007), both of which lead to the production of l-Gal-1-phosphate: It can function as GDP-l-Gal phosphorylase (Linster et al. 2007) and as l-Gal-hexose-1-phosphate guanylyltransferase (Laing et al. 2007). For that reason the VTC2 protein will be called by its two possible enzymatic activities, GDP-l-Gal phosphorylase/l-Gal guanylyltransferase, throughout this paper.
Physiological characterization of vtc2 has shown that it has 10–25% of the wild-type ascorbate level (Conklin et al. 2000; Müller-Moulé et al. 2002), is non-photochemical quenching (NPQ)-deficient (Noctor et al. 2000; Smirnoff 2000; Müller-Moulé et al. 2002), and that it bleaches in high light (HL) when transferred from low light (LL) to HL (Müller-Moulé et al. 2003). In response to transfer to HL, the thylakoid proteome showed a higher level of Cu,Zn-superoxide dismutase (SOD) and heat shock protein-70, but a lower level of Fe-SOD and PsbS than the wild type (WT) (Giacomelli et al. 2006). When the vtc2 mutant was grown in HL from seedling age on, however, no bleaching occurred in the mutant, but mutant plants had a lower maximum photosystem II efficiency and lower oxygen evolution rates (Müller-Moulé et al. 2004). Interestingly, mutant plants also had a higher glutathione content than the wild type, which shows that plants compensated for the deficiency in one water-soluble antioxidant by increasing the levels of another.
In this paper I characterize the molecular properties of the VTC2 gene and protein by analyzing the expression of the gene by quantitative real time (qRT)-PCR in various tissues and developmental stages, and determine the localization of the protein within the plant and the cell by using VTC2:β-glucuronidase (GUS) and VTC2:yellow fluorescent protein (YFP) protein fusions. I present evidence that the gene is expressed at a higher level in green tissues and is light induced. In addition, the protein appears to be present only in green tissues and is localized in both the cytosol and the nucleus, making it possible that it is a dual-function protein.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) and the vitamin C-deficient mutant, vtc2-1, also Col-0 ecotype, were grown in soil for 4 weeks (qRT-PCR analysis of alleles, Fig. 2) or 10 weeks (plants used for tissue and development studies, Fig. 3) in controlled growth conditions of 10 h light, 20°C/14 h dark, 18°C with a light intensity of 150 μmol photons m−2 s−1. Plants grown longer than 4 weeks were nutrient supplemented once a week. For the light-induction analysis, plants were grown for 5 days on minimal plant nutrient agar plates (Haughn and Somerville 1986) at a constant temperature of 23°C either in the light (14 h light/10 h dark; 30 to 40 μmol photons m−2 s−1 light intensity) or wrapped in foil in the same growth chamber. Plants used for confocal microscopy were grown for 4 to 7 days on agar plates. Finally, the GUS assays were performed on plant seedlings or older plants grown on soil in long day conditions (14 h light/10 h dark) with a temperature of 22°C, a light intensity of 60 to 70 μmol photons m−2 s−1 and a humidity of 70%.
RNA preparation
RNA was prepared from snap-frozen tissue using the RNEasy Plant Mini kit (QIAGEN).
Preparation of cDNA
cDNA was prepared from total RNA using oligo(dT)18 primer and RevertAidTM H Minus M-MuLV Reverse Transcriptase from Fermentas. If the cDNA was used for qRT-PCR the RNA was treated with Desoxyribonuclease I (Fermentas) prior to reverse transcription.
qRT-PCR analysis of gene expression
Transcript levels were assayed using the ABIPrism® 7700 Sequence Detection system (Applied Biosystems) and SYBR® GreenER™ qPCR SuperMix (Invitrogen). 1 μg total RNA was transcribed into cDNA as described above and 1 μl cDNA (45 ng) was used in a total volume of 20 μl for qRT-PCR. For qRT-PCR, following primers (5′–3′) were used:VTC2-F (final concentration 300 nM): TTCGCTATGATGTCACTGCCTG; VTC2-R (final concentration 900 nM): GCAACGAAACCATACTTCCCC. For control the At1g13320 gene was used. This control gene is described in Czechowski et al. (2005), and has a similar expression level as the VTC2 gene. Control-F (final concentration 200 nM): TAACGTGGCCAAAATGATGC; Control-R (final concentration 200 nM): GTTCTCCACAACCGCTTGGT. Each of three to five (see caption below figure) independent cDNA preparations was assayed twice for each transcript analyzed. Transcript abundance was calculated using the relative expression software tool (REST-MCS; Pfaffl et al. 2002), which takes into account the different efficiencies and calculates the statistical significance of the observed differences.
Ascorbate measurement
Total ascorbate was determined by a spectrophotometric method using UV absorption by reduced ascorbate at 265 nm (Conklin et al. 1996). In this assay, ascorbate oxidase is used to oxidize all ascorbate and the amount of ascorbate is determined by the difference in absorption at 265 nm before and after addition of the enzyme (only reduced ascorbate absorbs at 265 nm). To obtain values for total ascorbate, samples were first reduced by addition of dithiotreitol (final concentration of 4 mM).
Generation of transgenic plants
To generate a VTC2:GUS fusion protein, the VTC2 gene was amplified by PCR using the primers VTC2gDNA-attB1-F: AAAAAGCAGGCTGAAATTGATGTTTACTGACGCACGA and VTC2attB2-R: AGAAAGCTGGGTACTGAAGGACAAGGCACTCG, as well as the adapter primer attB1 and attB2 (Invitrogen). The PCR product was cloned into the Gateway donor vector pDONR™ 201 (Invitrogen) using BP clonase™ Mix (Invitrogen). The VTC2 entry vector was recombined using LRClonase™ mix (Invitrogen) with the destination vector pGWB3 (kindly provided by T. Nakagawa, Shimane University, Izumo, Japan) to yield the final vector VTC2:GUS, which consists of a C-terminal fusion between the native gene including its promoter and the GUS gene. To generate a C-terminal fusion with YFP, the VTC2 cDNA was amplified using the primers VTC2cDNA-attB1-F: AAAAAGCAGGCTGAATGTTGAAAATCAAAAGA, VTC2attB2-R, attB1 and attB2. The PCR product was cloned into the Gateway donor vector pDONR™ 201 (Invitrogen) using BP clonase™ Mix (Invitrogen). The VTC2 entry vector was recombined using LRClonase™ mix (Invitrogen) with the destination vector pEarleyGate101 (Earley et al. 2006) to yield the final vector 35S:VTC2:YFP. The vectors were transformed into Agrobacterium GV3101 (VTC2:GUS) or AGL-1 (35S:VTC2:YFP) using electroporation. Plants (WT or the vtc2 mutant) were transformed with Agrobacterium using the floral-dip method (Clough and Bent 1998). Transgenic plants were selected on PN-plates with either Kanamycin (GUS) or Basta (YFP) and then transferred onto soil.
GUS assay
4 different lines (M2 generation) were used for the GUS assay. Plant tissue was pre-stained in pre-staining solution (100 mM NaPO4, pH 7.0, 2% formaldehyde, 1 mM EDTA) for 5 min in the vacuum and 5 min without vacuum. Then tissue was stained in staining solution (100 mM NaPO4, pH 7.0, 0.1% (v/v) Trion X-100, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 1 mM EDTA, and 1 mM X-gluc) for 10 min in the vacuum and then overnight at 37°C. After overnight staining the dye was fixed with 100 mM NaPO4, pH 7.0 and 3.7% (v/v) formaldehyde, after which it was chlorophyll-destained for several hours using 70% ethanol with frequent changes of the ethanol solution.
Confocal Laser-Scanning Microscopy
Seedlings were analyzed with an upright Zeiss LSM 510 metaconfocal laser-scanning microscope, Plan-Apochromat 63×/1.4 mm Oil differential interference contrast objective, using a 405-nm diode laser and an Argon/2 458, 477, 488, 514 nm laser with a LP650 filter, a BP420-480 filter, and a 514 nm filter. Data were acquired in 8-bit depth. Images were processed with Volocity 4.2.0 (Improvision Ltd) and Photoshop 7.0 (Adobe Systems) software. Plants were stained with (0.001% (w/v) 4,6-diamidino-2-phenylindole (DAPI; Sigma) in 100 mM sodium phosphate, pH 7.0 for staining of nuclei.
Immunodetection
Eight day-old plants grown on PN plates were ground using mortar and pestle and ground tissue was mixed with 0.5× extraction buffer (4% SDS, 25 mM Tris pH 8.8, 2.5% glycerol). 2X extraction buffer (16% SDS, 100 mM Tris pH 8.8, 10% glycerol) was added and the sample was boiled for 4 min, after which 10% (v/v) β-mercaptoethanol was added. After 30 min at room temperature the sample was centrifuged in a micro centrifuge for 5 min at maximum speed. The supernatant was transferred to a new tube and the protein concentration was determined using the Bio-Rad Protein Assay solution. Proteins (12 μg) were separated on a 10% Lämmli Tris-glycin gel. Proteins were blotted to a PVDF membrane and decorated with anti-GFP antibody (Roche Applied Science).
Results
The vtc2-1 allele shows the strongest loss-of-function
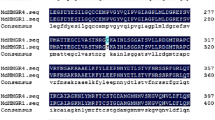
When the vtc2 mutant was isolated, four different alleles of the VTC2 gene were found (Conklin et al. 2000), of which three encompassed different mutations. The vtc2-1 and the vtc2-4 allele have the same G to A change, the vtc2-2 allele also has a G to A change and the vtc2-3 allele an A to C change (Fig. 1a; Jander et al. 2002). The base changes of the vtc2-2 and the vtc2-3 alleles cause a change in the amino acid sequence (Fig. 1c), whereas the vtc2-1 allele is predicted to be an intron-splice mutant. As 3′ splicing occurs behind AG at the end of the intron in almost all cases (Brown et al. 1996), it was conceivable that the vtc2-1 mRNA was spliced just one base pair later (see DNA sequence, Fig. 1a). In order to test this hypothesis, mRNA was reverse transcribed into cDNA and sequenced. The mutant cDNA was indeed shorter than the wild-type cDNA by only a single base pair (Fig. 1b). This change in reading frame caused a pre-mature stop codon just after the HIT motif, shortening the protein to less than 2/3 of wild-type length (Fig. 1c). In addition, mRNA quantification of vtc2-1 showed that the mRNA level was significantly reduced to only about 20% of the wild type (Fig. 2). This reduction in mRNA level is likely due to nonsense-mediated mRNA decay, coming from the accumulation of stop codons in the mRNA sequence (Isshiki et al. 2001; Conti and Izaurralde 2005). The mRNA level of the vtc2-2 allele was reduced to about 55% of the wild type, whereas the mRNA level of the vtc2-3 allele was not significantly different from wild type (Fig. 2). Taken together, the vtc2-1 allele appeared to be the strongest loss-of-function allele of the three different alleles and therefore all further studies were performed using the vtc2-1 mutant, hereafter referred to simply as vtc2. However, it has recently been shown by Dowdle and co-authors that vtc2-2 is also a null mutant because it produces an inactive enzyme (Dowdle et al. 2007).
VTC2 DNA, cDNA, and VTC2 protein sequence of the wild type and the vtc2 mutant. (a) VTC2 genomic DNA sequence. The changes in sequence of the vtc2 alleles (Jander et al. 2002) are noted. Exons are shown in capital letters, introns in small letters. (b) VTC2 cDNA sequence of the region showing the difference between wild type and the vtc2-1 mutant. (c) Protein sequence alignment between the wild type and the vtc2-1 mutant allele. The amino acids changed in the vtc2-2 and the vtc2-3 mutants are also shown. HIT—histidine triad motif, NLS—nuclear localization sequence
VTC2 mRNA levels of VTC2 alleles. Three mutant alleles (vtc2-1, vtc2-2 and vtc2-3) were compared to the wild-type allele. Data shown are the average of five measurements (vtc2-3 four measurements) with standard errors. Results were calculated using the REST program. Asterisks denote results that are significantly different from the WT (**P-value < 0.005; *P-value < 0.05)
The VTC2 mRNA is expressed at a higher level in all green tissues
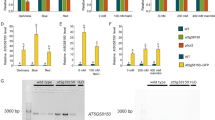
To learn more about the function of the VTC2 gene, its mRNA expression in different tissues and at different developmental stages was determined by qRT-PCR. As shown in Fig. 3, VTC2 mRNA was found in all tissues (seedling, leaf, root, stem, flower, and silique). The difference in expression between the various green tissues was not significant, however, the VTC2 mRNA level in roots was significantly reduced (0.066 ± 0.031; P-value < 0.05) compared to seedling levels (Fig. 3a). Leaves of different ages showed a trend of increasing VTC2 mRNA levels with age, however, this difference was statistically not significant (Fig. 3b). As an extension to the mRNA analysis, I also measured ascorbate levels in the same tissues used for the mRNA quantification. Roots had the lowest VTC2 mRNA level and lowest measured ascorbate content, however, no correlation was found between the high ascorbate content and the VTC2 mRNA expression in siliques (Fig. 3c, d). There was also no correlation of VTC2 mRNA levels with the amount of ascorbate found in mutant tissues compared to wild-type tissues. These apparent discrepancies might not really be such, as protein levels and enzyme activities could be quite different from the mRNA levels. The high amount of ascorbate in 10-week-old leaf samples might be due to the onset of flowering at this time (Gander 1982; Horemans et al. 2000; Attolico and De Tullio 2006).
VTC2 mRNA and ascorbate levels in wild-type and vtc2 mutant plants. (a) VTC2 mRNA levels in different wild-type tissues normalized to seedling. All tissues except seedling were taken from 10 week-old plants. Data shown are the average of three to four measurements with standard errors. Results were calculated using the REST program. The asterisk denotes result that is significantly different from the seedling (P-value < 0.05). (b) mRNA levels of wild-type leaves during development normalized to seedling. Data shown are the average of three to four measurements with standard errors. Results were calculated using the REST program. (c) Ascorbate content in different tissues of wild type and the vtc2 mutant. All tissues except seedling were taken from 10 week-old plants. Data shown are the average of three measurements with standard errors. (d) Ascorbate content of wild-type and vtc2 mutant leaves during aging. Data shown are the average of three measurements with standard errors
VTC2 mRNA expression is induced by light
As the VTC2 gene is expressed mainly in green tissues, I tested whether VTC2 gene expression is light induced. The expression of the VTC2 gene was compared in 5-day-old seedlings that had been grown either in the light or the dark, or that had been grown for 5 days in the dark and were then transferred to light for 1 h. As can be seen in Fig. 4, VTC2 mRNA levels were higher in light-grown seedlings than in dark-grown seedlings. In addition, the mRNA levels in the dark-grown seedlings that had been exposed to light for 1 h were even higher (by a factor of 4) than the mRNA levels in light-grown seedlings.
Light induction of VTC2 mRNA normalized to light-grown seedlings. Tissue samples were taken from 5 day-old seedlings grown in the dark, in the light, or in the dark and then transferred to light for 1 h. Data shown are the average of five measurements with standard errors. Results were calculated using the REST program. Asterisks denote results that are significantly different from light-grown seedlings (**P-value < 0.005; *P-value < 0.05)
The VTC2:GUS fusion protein is expressed in all green tissues
To learn more about the expression of the protein in different tissues, vtc2 mutant plants were transformed with a construct expressing a VTC2:GUS fusion under the control of the native VTC2 promoter. Several independent transgenic lines were isolated and checked for expression of the VTC2:GUS protein. In young seedlings the protein was found in leaves (Fig. 5a, b), but not in roots. Because VTC2 mRNA was found in the roots, it is possible that the protein level was below the detection level. Older plants that already flowered and had siliques also had the protein in leaves, but not in stems, flowers, or siliques (Fig. 5c). Again, roots did not stain for the GUS protein. It is possible that stems, flowers, and siliques did not show staining because it is more difficult for the substrate to penetrate the tissue. However, even when only staining the stems, the flowers, or the siliques themselves, no staining was visible. In a leaf cross section it was apparent that the whole leaf except the epidermal layers stained (Fig. 5d), indicating that the VTC2 protein is only found in green cells.
The VTC2:YFP fusion protein is found in the cytoplasm and the nucleus
To learn more about the intracellular distribution of the VTC2 protein, transgenic plants carrying a Pro35S:VTC2:YFP construct were generated. Several independent transgenic lines were isolated and imaged under a confocal microscope. Figure 6 shows false-color images of root and cotyledon of two different transgenic lines, and the VTC2:YFP fusion protein was clearly visible in both the cytoplasm and the nucleus. It should be noted that the VTC2-YFP fusion protein was only present in roots because the 35S promoter and not the native promoter was used, as roots usually have a very low VTC2 mRNA level and no detectable VTC2 protein (see previous section). Panels A to C show the root tip stained with 4,6-diamidino-2-phenylindole (DAPI), which stains nuclei. The co-localization of the red (DAPI) with the green (VTC2:YFP) is clearly visible as yellow color. Panels D to I show microscopic images of a cotyledon. In red, DAPI staining (nuclei) can be seen, in blue the autofluorescence of the chloroplasts and in green the VTC2:YFP protein. The YFP protein is clearly visible in the cytoplasm (notice the cytoplasmic strands surrounding the chloroplasts) and in the nucleus, where it again co-localizes with the DAPI stain. It is also very clear that the VTC2:YFP protein is excluded from the chloroplasts. However, because the fusion protein is driven by the 35S promoter, it is not possible to determine the quantitative partitioning of the VTC2 protein between cytoplasm and nucleus.
False-color image of the intracellular localization of the VTC2:YFP fusion protein in root and cotyledon of two independent transgenic Arabidopsis lines. Root tip line 3 (a–c) and cotyledon line 9 (d–i). The third column is the composite image of the first two columns. a and d—DAPI staining in red (nuclei); b and e—YFP in green; g—chlorophyll autofluorescence in blue; h—composite of e and g; i—composite of f and g. Bars denote 10 μm. Seedlings were 4 to 7 days old at time of measurement
To make sure that the localization of the VTC2:YFP fusion protein in the nuclei was not due to unspecific diffusion into the nuclei, Western Blots were performed to determine the molecular size of the fusion protein. Figure 7 confirms that the YFP protein is fused to the VTC2 protein in vivo. In addition, PSORT (Nakai and Horton 1999) found a putative nuclear localization signal (NLS) at position 115 (Fig. 1c).
Discussion
The VTC2 mRNA level is light-regulated
Light is an important factor in regulating ascorbate biosynthesis. The amount of ascorbate increases in HL (Grace and Logan 1996; Eskling and Åkerlund 1998; Müller-Moulé et al. 2004), and the ascorbate content also shows diurnal changes (Tamaoki et al. 2003). As can be seen in Figs. 3 and 4, similar to other ascorbate-related genes (Tabata et al. 2002), the VTC2 gene is clearly regulated by light. VTC2 mRNA levels are higher in all green tissues and very low in roots (Fig. 3). In addition, dark-grown seedlings had only half the amount of VTC2 mRNA compared with light-grown seedlings (Fig. 4). When transferred to light, dark-grown seedlings accumulated mRNA to 4-times higher levels than light-grown seedlings within 1 h of exposure to light. In a recent publication, VTC2 expression was measured during the light cycle. The authors showed that the VTC2 mRNA level peaked in the first few hours and was quite low in the dark, with the enzyme activity and the ascorbate level lagging behind (Dowdle et al. 2007). In constant light, the expression pattern stayed the same, suggesting a circadian regulation of VTC2 expression (Dowdle et al. 2007).
The VTC2 protein was only found in green tissues, although ascorbate biosynthesis occurs in green and non-green tissues. However, the VTC2:GUS protein could have simply been below the detection level, since VTC2 mRNA was found in roots (Fig. 3). Even then, the level of the VTC2 protein in roots, a tissue that is not light-exposed, was clearly lower. The fact that the expression level of the VTC5 gene was compared to VTC2 relatively higher in roots evokes the possibility that VTC2 is more important for ascorbate synthesis in leaves, whereas VTC5 is more important in non-photosynthetic tissues (Dowdle et al. 2007).
Interestingly, the VTC2 gene is not co-expressed with the other ascorbate-biosynthesis genes (co-expressed gene (CEG) analysis; Berardini et al. 2004; data not shown). GO terms that are over-represented among the 86 genes co-expressed with VTC2 are ubiquinone biosynthesis and photosynthesis light harvesting. Moreover, the lack of VTC2 gene expression in the vtc2 mutant does not influence the expression of other ascorbate biosynthesis genes. DNA chip analysis identified 41 differentially expressed genes between the wild type and the vtc2 mutant (Supplemental Table 1), of which only the VTC2 gene itself coded for an ascorbate biosynthesis enzyme; none of the other genes related to ascorbate metabolism. Instead, cell wall-related genes were over-represented among the differentially expressed genes, suggesting that VTC2 might be a link to cell wall biosynthesis as suggested by Wolucka and Van Montagu (2007).
The VTC2 protein might be a dual-function protein
The VTC2:YFP fusion protein was found in both the cytoplasm and the nucleus. Ascorbate biosynthesis occurs in the cytoplasm except for the last step, which occurs in the mitochondria. The presence of the VTC2 protein in the cytoplasm is therefore expected and makes sense, since many enzymes are found in functional enzyme complexes. However, the presence of the VTC2:YFP protein in the nucleus was unexpected. Western Blots clearly showed that the YFP protein is still fused to the VTC2 protein in vivo and that the presence of the VTC2:YFP protein in the nucleus is unlikely to result from simple diffusion. YFP alone (27 kDa) can diffuse into the nucleus because it is smaller than the size exclusion limit (Nigg 1997; Görlich 1998; Talcott and Moore 1999). If, however, it is bound to the VTC2 protein (VTC2—49 kDa), the fusion protein would be too large (VTC2:YFP—76 kDa) to diffuse into the nucleus. The fusion protein must instead be transported into the nucleus by means of a specific NLS in the VTC2 protein, and indeed one was predicted by PSORT (Fig. 1). It is therefore conceivable that VTC2 is a dual-function protein that has an additional, possibly regulatory, role in the nucleus. In order to confirm this hypothesis, more experiments will be necessary. First, and foremost, subcellular fractions will have to be isolated and tested for the presence of the VTC2:YFP fusion protein. Secondly, immunolocalization studies using an antibody against the VTC2 protein should be done to confirm its nuclear localization in wild-type plants. Analysis of wild-type plants will also make it possible to determine under which conditions the VTC2 protein is actually found in the nucleus; for example, its location could change in the light and the dark.
In the literature, other examples of dual-function proteins can be found that have both enzymatic and regulatory function in various eukaryotes, i.e. yeast, mammals and plants. For most of these, the function of the enzyme in the nucleus has not been established in spite of their clear presence in the nucleus (Wilson 2003; Hall et al. 2004). In plants, hexokinase1 (HXK1) is probably the best-studied example, for which a regulatory function has been established (Cho et al. 2006). HXK1 is a glucose sensor, which governs gene expression in response to environmental clues. Cho and colleagues were able to detect a small fraction of HXK1 in nuclear extracts and identify two direct interacting proteins using proteomic and yeast-two-hybrid screens, as well as a promoter to which the HXK1 complex binds to (Cho et al. 2006).
Conclusions
The experiments described in this paper have answered some of the questions regarding the expression of the VTC2 gene and protein. They have also raised one interesting new question: Is it possible that the VTC2 protein is a dual-function protein that has both a biosynthetic and a regulatory function? Confocal microscopy revealed the presence of a VTC2-YFP fusion protein in the cytosol and the nucleus. More experiments will be required that test the localization of the VTC2 protein using different methods, for example by subcellular fractionation and analysis of nuclear extracts. In addition, it would be helpful to isolate VTC2 alleles that separate enzymatic function from the hypothesized regulatory function.
Abbreviations
- CEG:
-
Co-expressed genes
- DAPI:
-
4,6-Diamidino-2-phenylindole
- Gal:
-
Galactose
- GDP:
-
Guanosine diphosphate
- GO:
-
Gene ontology
- GUS:
-
β-Glucuronidase
- HIT:
-
Histidine triad
- HL:
-
High light
- HXK1:
-
Hexokinase1
- LL:
-
Low light
- Man:
-
Mannose
- NPQ:
-
Non-photochemical quenching
- NLS:
-
Nuclear localization signal
- QRT-PCR:
-
Quantitative real-time polymerase chain reaction
- SOD:
-
Superoxide dismutase
- vtc:
-
Vitamin C-deficient
- WT:
-
Wild type
- YFP:
-
Yellow fluorescent protein
References
Agius F, Gonzalez-Lamothe R, Caballero JL, Munoz-Blanco J, Botella MA, Valpuesta V (2003) Engineering increased vitamin C levels in plants by overexpression of a d-galacturonic acid reductase. Nat Biotechnol 21:177–181
Arrigoni O, deTullio MC (2002) Ascorbic acid: much more than just an antioxidant. Biochim Biophys Acta 1569:1–9
Arrigoni O, Arrigoni-Liso R, Calabrese G (1977) Ascorbic acid requirement for biosynthesis of hydroxyproline-containing proteins in plants. FEBS Lett 82:135–138
Asada K (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50:601–639
Attolico AD, De Tullio MC (2006) Increased ascorbate content delays flowering in long-day grown Arabidopsis thaliana (L.) Heynh. Plant Physiol Biochem 44:462–466
Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, Moseyko N, Yoo D, Xu I, Zoeckler B, Montoya M, Miller N, Weems D, Rhee SY (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135:745–755
Brown JW, Smith P, Simpson CG (1996) Arabidopsis consensus intron sequences. Plant Mol Biol 32:531–535
Cho YH, Yoo SD, Sheen J (2006) Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127:579–589
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Conklin PL, Williams EH, Last RL (1996) Environmental stress sensitivity of an ascorbic acid-deficient Arabidopsis mutant. PNAS 93:9970–9974
Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL (1999) Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. PNAS 96:4198–4203
Conklin PL, Saracco SA, Norris SR, Last RL (2000) Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 154:847–856
Conklin PL, Gatzek S, Wheeler GL, Dowdle J, Raymond MJ, Rolinski S, Isupov M, Littlechild JA, Smirnoff N (2006) Arabidopsis thaliana VTC4 encodes l-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J Biol Chem 281:15662–15670
Conti E, Izaurralde E (2005) Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol 17:316–325
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N (2007) Two genes in Arabidopsis encoding GDP-l-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J 52:673–689
Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45:616–629
Eskling M, Åkerlund H-E (1998) Changes in the quantities of violaxanthin de-epoxidase, xanthophylls and ascorbate in spinach upon shift from low to high light. Photosynth Res 57:41–50
Gander JE (1982) Polyhydroxy acids: relation to hexose phosphate metabolism. In: Loewus FA, Tanner W (eds) Plant carbohydrates I: intracellular carbohydrates, chap 2 Springer-Verlag, Berlin, pp 77–102
Giacomelli L, Rudella A, van Wijk KJ (2006) High light response of the thylakoid proteome in Arabidopsis wild type and the ascorbate-deficient mutant vtc2-2. A comparative proteomics study. Plant Physiol 141:685–701
Görlich D (1998) Transport into and out of the cell nucleus. EMBO J 17:2721–2727
Grace SC, Logan BA (1996) Acclimation of foliar antioxidant systems to growth irradiance in three broad-leaved evergreen species. Plant Physiol 112:1631–1640
Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M (2004) Regulation of gene expression by a metabolic enzyme. Science 306:482–484
Hancock R, Viola R (2005) Biosynthesis and catabolism of l-ascorbic acid in plants. Crit Rev Plant Sci 24:167–188
Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204:430–434
Horemans N, Foyer CH, Potters G, Asard H (2000) Ascorbate function and associated transport systems in plants. Plant Physiol Biochem 38:531–540
Isshiki M, Yamamoto K, Satoh H, Shimamoto K (2001) Nonsense-mediated decay of mutant waxy mRNA in rice. Plant Physiol 125:1388–1395
Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129:440–450
Laing WA, Bulley S, Wright M, Cooney J, Jensen D, Barraclough D, MacRae E (2004) A highly specific l-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. PNAS 101:16976–16981
Laing WA, Wright MA, Cooney J, Bulley SM (2007) The missing step of the l-galactose pathway of ascorbate biosynthesis in plants, an l-galactose guanyltransferase, increases leaf ascorbate content. PNAS 104:9534–9539
Linster CL, Gomez TA, Christensen KC, Adler LN, Young BD, Brenner C, Clarke SG (2007) Arabidopsis VTC2 encodes GDP-l-Galactose phosphorylase, the last unknown enzyme in the Smirnoff-Wheeler pathway to ascorbic acid in plants. J Biol Chem 282:18879–18885
Loewus F (1988) Ascorbic acid and its metabolic products. In: Preiss J (ed) Carbohydrates, vol 14, chap 3. Academic Press, New York, pp 85–107
Lorence A, Chevone BI, Mendes P, Nessler CL (2004) myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134:1200–1205
Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR (2001) Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. PNAS 98:2262–2267
Müller-Moulé P, Conklin PL, Niyogi KK (2002) Ascorbate deficiency can limit violaxanthin de-epoxidase activity in vivo. Plant Physiol 128:970–977
Müller-Moulé P, Havaux M, Niyogi KK (2003) Zeaxanthin deficiency enhances the high light sensitivity of an ascorbate-deficient mutant of Arabidopsis. Plant Physiol 133:748–760
Müller-Moulé P, Golan T, Niyogi KK (2004) Ascorbate-deficient mutants of Arabidopsis grow in high light despite chronic photooxidative stress. Plant Physiol 134:1163–1172
Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24:34–36
Nigg EA (1997) Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779–787
Noctor G, Veljovic-Jovanovic S, Foyer CH (2000) Peroxide processing in photosynthesis: antioxidant coupling and redox signalling. Philos Trans R Soc Lond B 355:1465–1475
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36
Smirnoff N (1996) The function and metabolism of ascorbic acid in plants. Ann Bot 78:661–669
Smirnoff N (2000) Ascorbic acid: metabolism and functions of a multi-facetted molecule. Curr Opin Plant Biol 3:229–235
Smirnoff N, Conklin PL, Loewus FA (2001) Biosynthesis of ascorbic acid in plants: a renaissance. Annu Rev Plant Physiol Plant Mol Biol 52:437–467
Tabata K, Takaoka T, Esaka M (2002) Gene expression of ascorbic acid-related enzymes in tobacco. Phytochemistry 61:631–635
Talcott B, Moore MS (1999) Getting across the nuclear pore complex. Trends Cell Biol 9:312–318
Tamaoki M, Mukai F, Asai N, Nakajima N, Kubo A, Aono M, Saji H (2003) Light-controlled expression of a gene encoding l-galactono-γ-lactone dehydrogenase which affects ascorbate pool size in Arabidopsis thaliana. Plant Sci 164:1111–1117
Wheeler GL, Jones MA, Smirnoff N (1998) The biosynthetic pathway of vitamin C in higher plants. Nature 393:365–369
Wilson JE (2003) Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol 206:2049–2057
Wolucka BA, Van Montagu M (2003) GDP-mannose 3′,5′-epimerase forms GDP-l-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Biol Chem 278:47483–47490
Wolucka BA, Van Montagu M (2007) The VTC2 cycle and de novo biosynthesis pathways for vitamin C in plants: an opinion. Phytochemistry 68:2602–2613
Acknowledgements
First, I thank Professor Peter Westhoff, who offered me the possibility to work in his institute on my own project and who always was very supportive. I thank Yvonne Stahl and Marian Bureau for their help with the confocal microscope. For advice and/or critical reading of the manuscript I thank Peter Westhoff, Ute Höcker, Kris Niyogi, Talila Golan, David Huang, and two anonymous reviewers. Lastly, I thank the Deutsche Forschungsgemeinschaft for supporting me with an Emmy-Noether fellowship phase I (MU 1841/1-1 and 1-2).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Müller-Moulé, P. An expression analysis of the ascorbate biosynthesis enzyme VTC2. Plant Mol Biol 68, 31–41 (2008). https://doi.org/10.1007/s11103-008-9350-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-008-9350-4