Abstract
Perakine reductase (PR) catalyzes an NADPH-dependent step in a side-branch of the 10-step biosynthetic pathway of the alkaloid ajmaline. The enzyme was cloned by a “reverse-genetic” approach from cell suspension cultures of the plant Rauvolfia serpentina (Apocynaceae) and functionally expressed in Escherichia coli as the N-terminal His6-tagged protein. PR displays a broad substrate acceptance, converting 16 out of 28 tested compounds with reducible carbonyl function which belong to three substrate groups: benzaldehyde, cinnamic aldehyde derivatives and monoterpenoid indole alkaloids. The enzyme has an extraordinary selectivity in the group of alkaloids. Sequence alignments define PR as a new member of the aldo-keto reductase (AKR) super family, exhibiting the conserved catalytic tetrad Asp52, Tyr57, Lys84, His126. Site-directed mutagenesis of each of these functional residues to an alanine residue results in >97.8% loss of enzyme activity, in compounds of each substrate group. PR represents the first example of the large AKR-family which is involved in the biosynthesis of plant monoterpenoid indole alkaloids. In addition to a new esterase, PR significantly extends the Rauvolfia alkaloid network to the novel group of peraksine alkaloids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the past two decades, systematic investigation of the enzymes of plant alkaloid biosynthesis has been carried out for a handful of alkaloid groups. For instance, extensive research has been conducted on single enzymes participating in the complex pathways leading to isoquinoline alkaloids such as morphine or berberine (see Kutchan 1998; Zenk 1995 for reviews; Unterlinner et al. 1999). In addition, a set of enzymes and genes involved in the biosynthesis of the diterpenoid taxus alkaloid taxol has been described in detail (see Kaspera and Croteau 2006; Croteau et al. 2006, and cited literature). In this context, the monoterpene-derived alkaloids, including the monoterpenoid isoquinolines and monoterpenoid indoles, are of specific interest. For the monoterpenoid isoquinolines, two enzymes of Ipecacuanha alkaloid biosynthesis have previously been detected and characterized (De-Eknamkul et al. 1997, 2000). However, almost all enzymes and several genes of a separate multi-step biogenetic pathway have recently been detected and cloned: those pertaining to the generation of the Rauvolfia alkaloid ajmaline and a number of its derivatives which possess the typical sarpagan and ajmalan skeleton types (for reviews see Stöckigt 1995; Ruppert et al. 2005a). This entailed comprehensive alkaloid profiling of in vitro systems, in particular, cell cultures of the Indian medicinal plant Rauvolfia serpentina Benth. ex Kurz (Apocynaceae), was employed in this experimental setting for the last 15 years. The resulting knowledge has allowed description of one of the largest and most complicated metabolic networks of plant natural product biosynthesis: with particular focus on alkaloid biosynthesis, it further represents the first, in depth functional characterization of individual Rauvolfia enzymes.
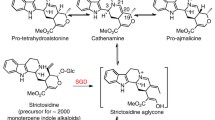
The broad alkaloidal metabolome of Rauvolfia is due to the existence of several side-routes branching from the ajmaline biosynthetic pathway, the latter encompassing ten enzyme-catalyzed steps (Fig. 1). In one of these branches, the indolenine alkaloid vomilenine serves as the precursor of the aldehydic perakine, which in turn is reduced at the expense of NADPH to raucaffrinoline, a typical Rauvolfia alkaloid (Sabino et al. 2006). This enzyme, detected in crude protein mixtures of Rauvolfia cell suspensions, has been designated as perakine reductase (PR): expression, preliminary characterization (Stöckigt 1995) and crystallization of a methylated PR triple mutant (Rosenthal et al. 2006) have been carried out for this protein. The current paper describes the purification, “reverse genetics”-mediated cloning, and functional expression of His6-tagged wild type PR in detail. Sequence analysis groups PR as a novel member of the aldo-keto reductase (AKR) enzyme family. Extensive investigation of the substrate specificity of PR not only points to multi-functionality of the enzyme, which has not been observed for any other Rauvolfia enzyme known to date, but also links and significantly extends the large Rauvolfia alkaloid metabolome to the recently detected novel group of peraksine alkaloids (Sheludko et al. 2002).
Part of the alkaloidal network of the Rauvolfia plant. The 10-step biosynthetic route to the monoterpenoid indole alkaloid ajmaline is connected at the stage of the intermediate vomilenine with perakine. Vomilenine can be isomerized into perakine under acidic conditions. The corresponding enzyme has not yet been detected. Perakine reductase (PR) and acetylesterase (AE) are the central enzymes for the metabolic scheme to the peraksines—a novel group of Rauvolfia alkaloids recently detected (framed). XR represents an unknown reductase. Other enzymes of this network are strictosidine synthase (STR1), strictosidine glucosidase (SG), sarpagan bridge enzyme (SBE), polyneuridine aldehyde esterase (PNAE), vinorine synthase (VS), vinorine hydroxylase (VH), cytochromeP450 reductase (CPR), vomilenine reductase (VR), 1,2-dihydrovomilenine reductase (DHVR), acetylajmalan esterase (AAE) and norajmalan methyltransferase (NAMT) (* means heterologously expressed enzymes). A further branch of the main pathway leads to raucaffricine alkaloids and derivatives (framed by dashed line, structures not illustrated)
Materials and methods
Chemicals
Benzaldehyde, phenylacetaldehyde, phenylpropionaldehyde, 2-nitrobenzaldehyde, 4-hydroxy-3-nitrobenzaldehyde, 4-nitrobenzaldehyde, 2-hydroxy-5-nitrobenz-aldehyde were bought from Acros (New Jersey, USA), ρ-anisaldehyde, coniferyl aldehyde from Sigma-Aldrich (Steinheim, Germany), cinnamic aldehyde from Roth (Karlsruhe, Germany), glucose and pyridoxal from AppliChem (Darmstadt, Germany). ρ-Coumaryl-, coniferyl-, sinapyl- and caffeyl aldehyde were gifts of Prof. Norman G. Lewis (Washington State University, USA). The synthetic cinnamic aldehyde derivatives, 3-(3,4,5-trimethoxy-phenyl)-propenal, 3-(3,4,5-tris-methoxy-methoxy-phenyl)-propenal and 3-[2-bromo-3,5-dimethoxy-4-(3,7,11-trimethyl- dodeca-2,6,10-trienyloxy)-phenyl]-propenal were synthesized by Jingxu Gong, Lihong Hu, Hongbin Zou (Zhejiang University, China). Gardneral was kindly provided by Profs. Mariko Kitajima and H. Takayama (Chiba University, Japan), 3-oxo-tabersonine by Prof. Bruno Danieli (University of Milan, Italy) and vallesine by Prof. Carl Djerassi (Stanford University, USA). The remaining alkaloids were taken from our alkaloid collection at the Department of Phamaceutical Biology.
Cell suspension cultures and purification of PR
Plant cell suspension cultures of R. serpentina were grown as described previously (Warzecha et al. 1999). Extraction and purification of PR was carried out by procedures previously applied to Rauvolfia enzymes (Ruppert et al. 2005b). In brief, 1.7 kg of R. serpentina cells were extracted with 2 l of Tris/HCl buffer (0.1 mM, pH 7.5). Following filtration and centrifugation of the resulting slurry, proteins in the supernatant were precipitated with (NH4)2SO4 in two consecutive fractions (0–30% and 30–70% (NH4)2SO4 saturation, respectively). The protein pellet obtained in the second fraction was re-dissolved in extraction buffer, dialyzed and subjected to three subsequent activity-guided separation steps via ion exchange by diethylaminoethyl (DEAE) sepharose, AcA54 size exclusion and ion exchange chromatography on a MonoQ column. After the final purification step, samples of the fraction exhibiting reductase activity were subjected to non-denaturing polyacrylamide gel electrophoresis (PAGE) (as described by Ornstein 1964). Sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE), showed several protein bands.
Determination of protein concentrations and partial amino acid sequence analysis
Protein concentrations were measured according to Bradford (1976) using bovine serum albumin as the reference protein.
From the Coomassie-brilliant blue-stained SDS-PAGE, major protein bands were digested in situ with proteinase LysC for sequence analysis and further processed as reported (Ruppert et al. 2005b). Partial peptide sequences obtained from the main protein band (∼38 kDa) of the putative PR, were (a) QIYFRIEALAQK and (b) FGISELDFSGLFAK, and were used for cloning.
Isolation of a full-length cDNA of PR
Total cDNA was obtained from R. serpentina cell suspensions by reverse-transcription PCR (RT-PCR) (Qiagen, Hilden, Germany). The oligo peptides mentioned above were used for primer design. The primer sequences were as follows: PR (b)-forward, TTY GGI ATH WSI GAR YTI GAY TTY WSI GG; PR (a)-reverse, TTY TGI GCI ARI GCY TCD ATI CWR AAR TA; R = A/G, S = C/G, W = A/T, Y = C/T, D = A/G/T, H = A/C/T, I = Inosine. PCR was performed using the following conditions: 1 × (94°C 5 min), 4 × (94°C 1 min, 41°C 1 min, 72°C 2 min), 30 × (94°C 1 min, 46°C 1 min, 72°C 2 min), 1 × (72°C 10 min). A 510 bp DNA fragment was obtained and cloned into the pGEM-Teasy vector (Promega, Mannheim, Germany).
The DNA fragment was sequenced and the following primers were designed to identify the sequence of the complete 3′- and 5′-ends by rapid amplification of cDNA ends (RACE-PCR). The following primers were used: 5′-RACE-PR CT CCA GAC TAT GTC CGC TCC TGC TGT GAG G, 3′-RACE-PR CAT CAC ATC CCA GAT TTG TTG GGG AGA ACC. cDNA of the R. serpentina cell suspension culture, produced with the GeneRacer Kit (Invitrogen, Karlsruhe, Germany), was used as template. The conditions were 1 × (94°C 5 min), 5 × (94°C 30 s, 71°C 2 min), 5 × (94°C 30 s, 69°C 30 s, 72°C 2 min), 20 × (94°C 30 s, 67°C 30 s, 72°C 2 min), 1 × (72°C 10 min).
The full-length cDNA was amplified by PCR using the following primers: PR for-pQE-2 GCA TGC GCA AAT GCC AAG AGT TAA GCT GGG, PR rev-pQE-2 CCG CGG TTA CTT CAA TGG TGG TGT ATT. The template mentioned above was used. These primers generated restriction sites (SphI and SacII) at the ends of the full-length clone for sub-cloning into the pQE-2 expression vector (Qiagen, Hilden, Germany). The following PCR conditions were used: 1 × (94°C 5 min), 33 × (94°C 1 min, 55°C 1 min, 72°C 3 min), 1 × (72°C 10 min).
Sequence alignments
Multiple sequence alignments were performed with CLUSTALW and BLAST for database searches.
Expression and purification of PR
Full-length PR cDNA (1014 bp; Gene Bank accession number AY766462) from R. serpentina was cloned into the pQE-2 vector and transformed in the M15 Escherichia coli strain (Qiagen, Hilden, Germany), resulting in a PR fusion protein harboring six histidine residues at its N-terminal. Bacteria were grown at 37°C in LB-medium in 500 ml batches (1000 ml Erlenmeyer flasks) with 50 μg/ml ampicillin and 30 μg/ml kanamycin up to a cell density of OD600 ∼ 0.8. Expression was induced by addition of 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG) to the medium. Following 24 h growth at 25°C, bacteria were harvested by centrifugation and the bacterial pellets stored at −20°C.
For enzyme purification, in a typical experiment, 10 g of the harvested bacteria were resuspended in 50 ml extraction buffer (50 mM KPi, 300 mM NaCl, pH 8.0) containing 10 mM imidazole and 1 mg/ml lysozyme. Following bacterial sonication (4°C) and centrifugation (14,000g, 30 min, 4°C) the supernatant was added to a 5 ml column containing 2 ml Ni-nitrilo-tri-acetic acid (Ni-NTA) superflow (Qiagen, Hilden, Germany). The column was then washed with 70 ml of extraction buffer containing 50 mM imidazole. PR protein was eluted with 250 mM imidazole in the above mentioned buffer. Enzyme containing fractions (1 ml each) were combined and dialyzed for 12 h at 4°C against 4 l of 50 mM KPi buffer (pH 7.0) containing 10 mM of EDTA.
Enzyme assay and substrate specificity studies of PR
For measurement of PR activity during enzyme isolation and purification, 21-hydroxyraumacline and NADPH were used in excess as substrates. In brief, aliquots of fractions from chromatographic separations of the crude protein (after (NH4)2SO4 precipitation) were incubated in a total of 200 μl KPi buffer (40 mM, pH 8.5) in the presence of 0.5 mM 21-hydroxyraumacline and 0.6 mM NADPH at 37°C for 60 min (the alkaloid 21-hydroxyraumacline was used as a substrate instead of perakine, because the latter was only available in trace amounts). The reaction was terminated by adding 200 μl methanol. After centrifugation (14,000g, 5 min), 30 μl of the supernatant was subjected to HPLC analysis.
Purified His6-tagged PR enzyme (5 μg enzyme for benzaldehyde derivatives, 100 μg for cinnamic aldehyde derivatives and for alkaloids) was incubated in 200 μl KPi buffer (50 mM, pH 7.0) in the presence of 0.8 mM substrate and 4.0 mM NADPH at 30°C for 15 min. Following addition of 200 μl methanol and centrifugation (14,000g, 5 min), 50 μl of the supernatant was subjected to HPLC analysis. The reverse reaction was measured under the same conditions in the presence of NADP+ instead of NADPH. All assays were performed in triplicate with controls in the absence of NADPH or NADP+.
Kinetic data of PR
Enzyme kinetics were measured by monitoring product formation rates by HPLC and 6–8 different substrate concentrations were used under the same conditions as above, but in the presence of excess NADPH (25 mM). The enzyme amounts were between 1 and 25 μg.
The optimum temperature of PR was determined in the forward direction between 10 and 70°C in 10°C intervals, and the pH optimum between 5.5 and 9.0 in intervals of 0.5 units.
Analysis and identification of PR products
For quantitation of products by HPLC, peak areas were determined; wavelengths of 280 nm were used for cinnamic aldehyde derivatives, and 254 nm for alkaloids and benzaldehyde derivatives respectively. A Lichrospher 60 RP-select B column (250 × 4 mm, Merck, Darmstadt, Germany) was used and the HPLC conditions were: 20% acetonitrile and 80% water, pH 2.3 and 1 ml/min flow.
The identity of enzymatic reaction products was proven by mass spectrometry (Finnigan MAT 44S quadrupole instrument (Bremen, Germany)) in EI-MS (electron impact) mode at 70 eV (ion source 200°C, evaporation temperature was 30–320°C with a gradient of 100°C/min). A five-fold incubation mixture was extracted with CHCl3. The organic layer was evaporated, the residue purified by thin layer chromatography (TLC) and the appropriate product measured by EI-MS.
Analysis and identification of R. serpentina cells for occurrence of cinnamic aldehydes by TLC, HPLC, GC-MS
To check for the presence of cinnamic aldehydes, 200 g freeze dried (4 kg fresh weight) R. serpentina cells were suspended in 2 l of MeOH (HPLC grade, Roth, Karlsruhe, Germany) and divided into two equal parts. 20 mg of coniferyl aldehyde and 20 mg of cinnamic aldehyde was added to one part as a positive extraction control. The two parts were then separately processed under the same conditions. After extracting with MeOH (22°C) for 24 h, the two parts were centrifugated (10,000g, 30 min, 4°C), and the supernatant allowed to evaporate under vacuum at 40°C. The residue obtained was resuspended in 50 ml H2O (pH 2.0) and extracted four times with 50 ml CHCl3. The organic fractions were combined and evaporated under vacuum at 40°C. Using coniferyl-, and cinnamic aldehyde as control, the CHCl3 fraction was analysed by TLC, HPLC, and GC-MS.
TLC plates (20 × 20 cm, Silica gel 60 F254, Merck, Darmstadt, Germany) were developed in a solvent system consisting of CHCl3/MeOH/formic acid 150:5:0.2, and analysed by UV and spraying with aldehyde reagent (2,4-dinitrophenylhydrazine solution; ethanol and concentrated H2SO4).
HPLC conditions for detecting the cinnamic aldehyde derivates were: Lichrospher 60 RP-select B column (250 × 4 mm, Merck, Darmstadt, Germany); mobile phase (A = acetonitrile, B = water pH 2.3), 0–15.0 min 30% A and 70% B, 15.1–18.0 min 50% A and 50% B, 18.1–20.0 min 70% A and 30% B, 20.1–25.0 min 30% A and 70% B; wavelength 280 nm; 1 ml/min flow.
GC-MS analyses were performed on a Viking 573 instrument (Bruker Daltonics, Billerica, MA, USA) in EI mode, using a Restec XTI-5 column (30 m × 0.25 mm I.D., 0.25 μm film thickness). The injection port and the transfer line were maintained at 270°C, the MS transfer line at 285°C, and the GC-oven temperature from 80 to 270°C (at 10°C/min). EI (70 eV) ionization was used and mass spectra were collected full scan over the range 40–400 m/z with ion trace filtering on molecular ions M+ of the phenylpropanoids cinnamic aldehyde (M+ 132 m/z) and coniferyl aldehyde (M+ 178 m/z).
Site directed mutagenesis for generation of PR mutants
Mutagenesis was performed with the QuickChange™ Site-Directed Mutagenesis kit (Stratagene, La Jolla, USA). The PR-pQE-2 plasmid was used as a template for PCR with Phusion-Polymerase (NEB, Frankfurt, Germany). For the generation of the four single mutants: Asp52Ala, Tyr57Ala, Lys84Ala, His126Ala the following primers were used (exchanged triplet in bold):
Asp52Ala_for 5′-A ACA TTC TTT GCT ACA TCA G-3′; Asp52Ala_rev 5′-ATC TGA TGT AGC AAA GAA TG-3′; Tyr57Ala_for 5′-TCA GAT ATT GCT GGC GAG AAC-3′.
Tyr57Ala_rev 5′-C GTT CTC GCC AGC AAT ATC TG-3′; Lys84Ala_for 5′-TG GGA ACG GCG TTT GGT ATA C-3′; Lys84Ala_rev 5′-T ACC AAA CGC CGT TCC CAC C-3′; His126Ala_for 5′-TAC ATA GCT CGC ATA GAT AC-3′; His126Ala_rev 5′-TC TAT GCG AGC TAT GTA GAA G-3′.
Results
Enrichment of PR from Rauvolfia cells and isolation of a full length PR cDNA
Perakine reductase could be purified according to the protocol developed for another Rauvolfia enzyme, the raucaffricine glucosidase (Warzecha et al. 1999). However, as judged by SDS-PAGE, after the third chromatographic step (MonoQ), the fractions showing reductase activity still contained several proteins. To assign the reductase activity to one of the protein bands visible on SDS-PAGE, the enzyme solution was subjected to native PAGE in duplicate: proteins from sections of one gel lane were eluted and tested for enzyme activity. In parallel, proteins in the second lane were subjected to SDS-PAGE. PR activity was found to correspond to the presence of a ∼38 kDa protein in SDS-PAGE. Peptides obtained from in situ digestion of the respective protein band were sequenced and yielded two partial amino acid sequences. Degenerate primer pairs based on these fragments were designed and generated a ∼500 bp PR cDNA fragment by PCR. Following ligation into the pGEM-T vector, cloning in E. coli and sequencing, the PCR product was found to contain the coding region for both peptide fragments indicating that the correct fragment corresponding to PR cDNA had been isolated. RACE-PCR resulted in isolation of the full-length cDNA of 1014 bp (accession number AY766462). cDNA translation yielded a polypeptide of 337 amino acids with a calculated molecular mass of 37159.8 Da and a theoretical isoelectric point of 5.9.
PR sequence alignment
The highest amino acid sequence identities for PR were found in the group of hypothetical plant oxidoreductase proteins e.g. from Oryza sativa (acc. number gb EAY78505.1 and gb EAZ16093.1, each 71% identity), an auxin-induced protein from O. sativa (dbj BAD61512.1, 66% identity), for the F8A5.21, At1g60710 and F8A5.24 protein from Arabidopsis thaliana (gb AAB71981.1, gb AAM20506.1, both 56% identity, gb AAB71982.1, 55%) and an AKR from Manihot esculenta (gb AAX84672.1, 53%) (see also Fig. 2).
Alignment of the perakine reductase amino acid sequence with the sequences of proposed aldo-keto reductases from various plant species and with plant AKR’s with known function. The predicted catalytic residues are marked red (Asp52, Tyr57, Lys84, His126). Identical amino acids are marked with * and homolog amino acid exchanges are marked with : or : PR: perakine reductase from Rauvolfia serpentina (gbAAX11684.1) (100%); AIOS: putative auxin-induced protein, from Oryza sativa (dbjBAD61512.1) (66%); AIAT: auxin-induced protein, putative, from Arabidopsis thaliana (gbAAM20506.1) (56%); AKRME: aldo-keto reductase AKR, from Manihot esculenta (gbAAX84672.1) (53%); 2A1: NADP+-dependent sorbitol-6-phosphate reductase from Malus domestica (sp P28475) (14%); 4A2: chalcone polyketide reductase from Medicago sativa (pir S48851) (15%); 4B3: codeinone reductase from Papaver somniferum (gb AAF13736) (14%); 4C5: aldose reductase from Digitalis purpurea (emb CAC32834) (15%). Identities of the amino acid sequences towards PR is given in brackets
Alignment with sequences of all members of the plant AKRs (http://www.med.upenn.edu/akr/; Hyndman et al. 2003) revealed that PR is related to aldose reductase from Digitalis (∼15% identity), followed by chalcone polyketide reductase of various plant origins with 14% identity, deoxy-mugineic acid synthase1 of Zea, Oryza and Hordeum (12–14% identity), and codeinone reductase from Papaver somniferum (∼14% identity) (Fig. 2). Comparison of PR amino acid sequence with those of cinnamic aldehyde reductases involved in lignin biosynthesis revealed identities inferior to 13%.
Heterologous expression and properties of His6-PR
In order to simplify enzyme purification, full-length PR cDNA was ligated into SphI and SacII restriction sites of the expression vector pQE-2, resulting in synthesis of a fusion protein harboring an N-terminal 6 histidine tag. The vector harboring the putative PR sequence was expressed in E. coli M15 cells yielding approximately 10 mg homogeneous protein per liter of bacterial culture, after Ni2+-NTA chromatography (Fig. 1, supplementary material). In an NADPH-dependent reaction, the native His6-PR converted the aldehyde perakine into raucaffrinoline. This compound was identified by TLC and by mass spectrometry on the basis of its molecular ion M+ 352 m/z and fragmentation pattern, which was identical to a reference sample of raucaffrinoline (Fig. 2, supplementary material). The enzyme was found to exhibit a pH optimum centered at pH 7.0 with half maximal activity at pH 6.5 and 8.5, and the temperature optimum was at 50°C. As shown for a range of substrates, NADPH could not be replaced by NADH, and in presence of the oxidized cofactor NADP+ the reverse reaction of the reductase was low, with <0.5% relative activity of the forward reaction only.
Substrate acceptance of PR
Since the PR sequence showed homology to other enzymes belonging to the AKR superfamily, we investigated its substrate specificity. In order to clearly define the specificity of this novel recombinant plant enzyme, a comprehensive substrate study was carried out. Three main groups of compounds were investigated: six benzaldehyde derivatives (A1–A6), phenylacetaldehyde (B), phenylpropionaldehyde (C), including pyridoxal, glucose and progesterone (Table 1); nine cinnamic aldehyde derivatives (D1–D9) (Table 2), and nine indole alkaloids with reducible carbonyl functions (Tables 3a, 3b). With the exception of benzaldehyde and 2-nitrobenzaldehyde, the enzyme accepted all the other benzaldehyde derivatives (Table 1). 4-Nitrobenzaldehyde and phenyl-propionaldehyde were found generate the highest relative enzyme activity (100% and 38.9% respectively), whereas 4-hydroxy-3-nitrobenzaldehyde was reduced at a very low rate (0.5%). In the second group (Table 2), the naturally occurring cinnamic aldehyde derivatives were generally well accepted. Sinapyl-, 5-hydroxy-coniferyl- and p-coumaryl aldehyde were converted at rates of 100%, 76.5% and 36.8%, respectively. Synthetic derivatives of this group with large substituents were not reduced, e.g. D9 (see the legend of Table 2). Of the nine alkaloids comprising the third group (Tables 3a, 3b), the natural substrate of PR, perakine and 19(S),20(R)-di-hydroperaksine-17,21-al were reduced, whereas all remaining alkaloids did not act as enzyme substrates. Typical substrates of enzymes of the AKR superfamily, such as pyridoxal, progesterone and glucose, were also not accepted as substrates by the PR.
Analysis of Rauvolfia cells for presence of cinnamic aldehydes
As cinnamic aldehydes and their derivatives are converted in favor of the indole alkaloids by recombinant PR (see Tables 2, 3a), it cannot be excluded that PR also acts in the metabolism of phenylpropanoids at their aldehydic stage. Whether this PR exercises this function in vivo during Rauvolfia cell growth is uncertain. Although alkaloid formation in these cell cultures has been investigated in the past in significant detail, the presence of cinnamic aldehydes was not previously proven. Rauvolfia cells were therefore analyzed for the presence of coniferyl- and cinnamic aldehyde by three analytical techniques (TLC, HPLC and GC-MS) all of which resulted in good separation of both aldehydes: coniferyl- and cinnamic aldehyde (TLC R f 0.4 and 0.6; HPLC R t 7.34 and 16.4 min; GC-MS R t 12.12 and 6.20 min). Based on the detection limit under the HPLC and GC-MS conditions described (∼10−5% and ∼10−6% cell dry weight, respectively) neither of the two aldehydes could be identified as constituents of Rauvolfia cell cultures (data not shown).
Kinetic data of the recombinant enzyme and site-directed mutagenesis
K m and V max were measured in order to characterize the enzyme by its kinetic properties (Table 4). K cat values were calculated from K m , V max and enzyme’s molecular mass. In addition to the alkaloid perakine, one representative substrate from each group was selected, i.e. 4-nitrobenzaldehyde and coniferylaldehyde. Both were reduced by PR with high relative activity. Table 4 shows that apparent K m values were in the same range for each substrate tested. The lowest K m was determined for 4-nitrobenzaldehyde, whereas for perakine and coniferyl aldehyde the values were 3 and 4.6 times higher, respectively. The catalytic efficiencies (K cat /K m ) strongly differed, resulting in ∼1750 and ∼900-fold efficacy for 4-nitrobenzaldehyde compared to perakine and coniferyl aldehyde, respectively.
In order to determine catalytically active amino acids of PR, the residues belonging to the putative tetrad, as deduced from sequence alignment of plant AKR enzymes (Fig. 2), were substituted by a non-reactive alanine residue. The mutants generated by directed mutagenesis were Asp52Ala, Tyr57Ala, Lys84Ala, His126Ala. These were well expressed in E. coli at comparable levels to the wild-type (data not shown), purified, and their activity for direct comparison determined under the same conditions as those used for recombinant, wild type PR. All four mutants showed a loss of activity in the range between ∼98 and ∼100% (Table 5), which clearly indicates that the amino acids selected represent the functionally important PR residues.
Discussion
Alkaloid biosynthesis in Rauvolfia cell suspension cultures is well understood at the enzymatic level for the major pathway leading to ajmaline. Side reactions of this pathway have, to date, only been subject of cursory investigations. Knowledge of these pathways would allow additional exploration and likely extension of the metabolic and genetic network of this medicinal plant. The formation of raucaffrinoline by reduction of perakine, the former alkaloid having been isolated several times from Rauvolfia plants (Kahn and Siddiqui 1972; Subhadhirasakul et al. 1994; Sabino et al. 2006), is an important side reaction which, until now, remained unexplored at the enzymatic level. During NADPH-dependent reduction-based substrate-consuming chromatography of PR, the use of unnatural 21-hydroxyraumacline was favored over the natural substrate perakine for two reasons. Firstly, the raumacline derivative can easily be obtained from ajmaline by biotransformation, whereas perakine is a very rare alkaloid. Secondly, 21-hydroxyraumacline mimics the reducible sub-structure of perakine and can be used as an unnatural substrate for PR (for details see Fig. 3). A “reverse genetic” approach, applying sequence-based, degenerate primers for PCR combined with RACE-PCR, allowed isolation of a novel full-length cDNA from a Rauvolfia cell suspension derived cDNA library. Since the isolated clone showed significant similarities to enzymes of the AKR superfamily, it may constitute the first example of this enzyme family identified in Rauvolfia species. If so, it would also be a novel member of the large AKR superfamily (Bashir et al. 2006; Gavidia et al. 2002, 2007) which, at present, consists of >5000 members (InterPro database family IPR001395, November 2007).
Expression of the N-terminal His6-tagged protein allowed the protein to be functionally analyzed. Protein-dependent conversion of perakine to raucaffrinoline was ascertained, and the result provided clear evidence that the correct clone was functionally expressed. In addition to a recently heterologously expressed cytochrome P450 reductase (our unpublished data), the PR described here constitutes the second example of an over-expressed reductase from Rauvolfia cells and seems to be involved in alkaloid metabolism.
An amino acid alignment of PR performed with UniProt confirmed its relationship to plant AKR enzymes, with the three closest AKR relatives (identities of up to 71%) being of plant origin (Fig. 2), from Oryza, Arabidopsis and Manihot. However, in contrast to the PR described here, the biochemical functions of these proteins are yet to be determined.
Enzymes in plant natural product biosynthesis usually have rather limited, or even exclusive, substrate specificity. In Rauvolfia for instance, acetylajmalan esterase accepts only two out of six closely structurally related alkaloids as substrates (Ruppert et al. 2005b) and vinorine hydroxylase converts only two of ten structurally highly related alkaloids (Falkenhagen et al. 1995). Similarly, polyneuridine aldehyde esterase of the ajmaline pathway (Fig. 1) exclusively hydrolyzes its natural substrate (polyneuridine aldehyde), even though a range of 13 esters including eight naturally occurring alkaloid esters, have been tested (Dogru et al. 2000). Most other functionally investigated enzymes of the Rauvolfia system likewise exhibit remarkable substrate selectivity (Ruppert et al. 2005a).
Close homology of PR to the AKR family prompted the current study, which represents one of the most comprehensive substrate studies carried out to date in this enzyme family. The AKR proteins are ubiquitous enzymes which are known for their low substrate specificity (Jez et al. 1997; Jez and Penning 2001). From a total of 28 compounds, 16 were found to be accepted by PR, indicating a remarkably low substrate-specific behavior. The putative substrates included some compounds which are usually regarded as typical AKR substrates, in particular nitrobenzaldehydes and other benzaldehyde derivatives. Moreover, a variety of cinnamic aldehydes and a group of indole alkaloids structurally related to perakine were tested. Though to varying degrees, nearly all nitrobenzaldehydes served as substrates (Table 1). These are highly reactive compounds that, due to “activation” of the aldehyde group by the electron-withdrawing nitrogroup in positions 2- and 4-, can be easily reduced chemically. Surprisingly however, even though it represents a typical AKR substrate and has been shown to be an excellent substrate of the AKR YakC protein from Schizosaccharomyces (Morita et al. 2002), 2-nitrobenzaldehyde was not converted. The selectivity observed for the nitrobenzaldehydes may exclude electronic effects. Other typical AKR substrates such as pyridoxal, glucose and progesterone, were also not accepted by PR, demonstrating a relatively high degree of AKR-related substrate intolerance. The selectivity of PR may in large part depend on a combination of steric effects and characteristics of its binding pocket.
The cinnamic aldehyde group and structurally related derivatives were tested for two reasons. Firstly, they are significantly larger in size than the benzaldehydes, clearly needing a bigger binding pocket. Secondly, they serve as biosynthetic precursors in the metabolism of phenylpropanoids in higher plants. Without exception, all naturally occurring cinnamic aldehydes assayed are accepted as substrates by PR. However, substrate selectivity is different, in that sinapyl aldehyde is effectively reduced and cinnamic aldehyde is the weakest substrate. The results indicate a surprisingly low overall selectivity of the enzyme in this particular group of aldehydes. It is obvious that the three-carbon chain makes all these phenylpropanoid aldehydes acceptable to the site of reduction, which is likely not to be located on the enzyme surface, but deep in the PR structure. Increasing enzyme activity with extension of the side chain as observed for benzaldehyde, phenylacetaldehyde, phenylpropionaldehyde (Table 1) support such an assumption.
The most pronounced substrate selectivity was, however, observed in the alkaloid group, because only two out of nine alkaloids belonging to the same family (monoterpenoid indoles) exhibiting reducible carbonyl functions could be converted, including the natural substrate perakine. Interestingly, of the structurally related 19(S),20(R)-dihydroperaksine alkaloids, the dialdehyde 19(S),20(R)-dihydroperaksine-17,21-al alone is reduced, giving rise to the recently isolated 19(S),20(R)-dihydroperaksine-17-al from Rauvolfia hairy roots (Sheludko et al. 2002). The second aldehyde group, located on the opposite side of the molecule at C17, was not reduced by the enzyme. Identification of 19(S),20(R)-dihydroperaksine alkaloids with both reduced aldehyde functions points to the putative existence of additional reductases in the Rauvolfia metabolome, involved in 19(S),20(R)-dihydroperaksine alkaloid conversion (XR, see Fig. 1).
These substrate studies make the classification of PR as a typical AKR protein difficult, but the AKR characteristic tetrad Asp52, Tyr57, Lys84 and His126 are also found in the PR sequence (Fig. 2), which were finally identified as the functional amino acids by way of site-directed mutagenesis experiments (Table 5). PR must therefore be unambiguously regarded as a novel member of the AKR enzyme superfamily.
Based on the cofactor requirements, the AKR family may be divided into three sub-groups: the major group which is predominantly NADPH-dependent, a small subgroup with dual NADPH/NADH specificity and very few examples which show exclusive specificity for NADH (Di Luccio et al. 2006). Given its cofactor dependence, PR clearly belongs to the first of these three subgroups.
It must however be regarded as an enzyme with an inherently low substrate specificity (Tables 1–3), with pronounced preference for the small nitrobenzaldehydes over medium-sized cinnamic aldehydes, and the structurally bulky alkaloids. Though a large binding site is required, additional structural requirements necessary for PR to convert such structurally diverse substrates remain undefined.
Moreover, PR represents the first example of this enzyme family which possesses a functional relationship to the indole alkaloid biosynthetic network. However, multi-functionality such as the detoxification of reactive aldehydes seen in human AKRs (Jin and Penning 2007) or in the metabolism of phenylpropane aldehydes, may not be excluded for the Rauvolfia cells. Since sequence analysis of PR shows no significant identity (<13%) or homology to well known cinnamic aldehyde reductases linked to lignin biosynthesis (Kim et al. 2004; Zhang et al. 2006), the enzyme may however exemplify a novel component of phenylpropanoid metabolism. However, extensive analysis of Rauvolfia cells by TLC, HPLC and GC-MS clearly indicates the absence of cinnamic and coniferyl aldehyde, in contrast to the known presence of the indole alkaloid perakine and its reduction product, a finding which favors a significant role of PR in alkaloid biosynthesis. Further alkaloid profiling of Rauvolfia resulted in the detection of the novel peraksines (Sheludko et al. 2002) and an additional, new Rauvolfia enzyme, acetyl esterase (AE), was identified (see Fig. 1) (Gerasymenko et al. unpublished). In conclusion, it is very likely that the both enzymes, PR and AE, significantly expand the alkaloidal network of Rauvolfia, now resulting in one of the biggest and best understood metabolomes in plant natural product biosynthesis.
Abbreviations
- AKR:
-
Aldo-keto reductase
- DEAE:
-
Diethylaminoethyl
- KPi:
-
Potassium phosphate
- NTA:
-
Nitrilo-tri-acetic acid
- PAGE:
-
Polyacrylamide gel electrophoresis
- PR:
-
Perakine reductase
- RACE:
-
Rapid amplification cDNA ends
- Rt :
-
Retention time
- RT-PCR:
-
Reverse-transcription PCR
- SDS:
-
Sodium dodecylsulphate
- TLC:
-
Thin layer chromatography
References
Bashir K, Inoue H, Nagasaka S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2006) Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J Biol Chem 281:32395–32402
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Croteau R, Ketchum REB, Long RM, Kaspera R, Wildung MR (2006) Taxol biosynthesis and molecular genetics. Phytochem Rev 5:75–97
De-Eknamkul W, Ounaroon A, Tanahashi T, Kutchan TM, Zenk MH (1997) Enzymatic condensation of dopamine and secologanin by cell-free extracts of Alangium lamarckii. Phytochemistry 45:477–484
De-Eknamkul W, Suttipanta N, Kutchan TM (2000) Purification and characterization of deacetylipecoside synthase from Alangium lamarckii Thw. Phytochemistry 55:177–181
Di Luccio E, Elling RA, Wilson DK (2006) Identification of a novel NADH-specific aldo-keto reductase using sequence and structural homologies. Biochem J 400:105–114
Dogru E, Warzecha H, Seibel F, Haebel S, Lottspeich F, Stöckigt J (2000) The gene encoding polyneuridine aldehyde esterase of monoterpenoid indole alkaloid biosynthesis in plants is an ortholog of the α/β hydrolase super family. Eur J Biochem 267:1397–1406
Falkenhagen H, Polz L, Takayama H, Kitajima M, Sakai S-I, Aimi N, Stöckigt J (1995) Substrate specificity of vinorine hydroxylase, a novel membrane-bound key enzyme of Rauvolfia indole alkaloid biosynthesis. Heterocycles 41:2683–2690
Gavidia I, Perez-Bermudez P, Seitz HU (2002) Cloning and expression of two novel aldo-keto reductases from Digitalis purpurea leaves. Eur J Biochem 269:2842–2850
Gavidia I, Tarrio R, Rodriguez-Trelles F, Perez-Bermudez P, Seitz HU (2007) Plant progesterone 5β-reductase is not homologous to the animal enzyme. Molecular evolutionary characterization of P5βR from Digitalis purpurea. Phytochemistry 68:853–864
Hyndman D, Bauman DR, Heredia VV, Penning TM (2003) The aldo-keto superfamily homepage. Chem Biol Int 143–144:621–631
InterPro database (2007) (http://www.ebi.ac.uk/interpro/DisplayIproEntry?ac=IPR001395)
Jez JM, Penning TM (2001) The aldo-keto reductase (AKR) superfamily: an update. Chem Biol Int 130–132:499–525
Jez JM, Bennett MJ, Schlegel BP, Lewis M, Penning TM (1997) Comparative anatomy of the aldo-keto reductase superfamily. Biochem J 326:625–636
Jin Y, Penning TM (2007) Aldo-keto reductases and Bioactivation/detoxication. Ann Rev Phamacol Toxicol 47:263–292
Kahn MA, Siddiqui S (1972) Isolation and Structure of Raucaffrinoline, A New Alkaloid from Rauwolfia caffra Sonder. Experientia 28:127–128
Kaspera R, Croteau R (2006) Cytochrome P450 oxygenases of taxol biosynthesis. Phytochem Rev 5:433–444
Kim S-J, Kim M-R, Bedgar DL, Moinuddin SGA, Cardenas CL, Davin LB, Kang CH, Lewis NG (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. PNAS 101:1455–1460
Kutchan TM (1998) Molecular genetics of plant alkaloid biosynthesis. In: Cordell GA (ed) The alkaloids, vol 50. Academic Press, San Diego, pp 257–316
Morita T, Huruta T, Ashiuchi M, Yagi T (2002) Characterization of recombinant YakC of Schizosaccharomyces pombe showing YakC defines a new family of aldo-keto reductases. J Biochem 132:635–641
Ornstein I (1964) Disc electrophoresis I-background and theory. Ann NY Acad Sci 121:321–349
Rosenthal C, Mueller U, Panjikar S, Sun L, Ruppert M, Zhao Y, Stöckigt J (2006) Expression, purification, crystallization and preliminary X-ray analysis of perakine reductase, a new member of the AKR enzyme superfamily from higher plants. Acta Cryst F62:1286–1289
Ruppert M, Ma X, Stöckigt J (2005a) Alkaloid biosynthesis in Rauvolfia—cDNA cloning of major enzymes of the ajmaline pathway. Curr Org Chem 9:1431–1444
Ruppert M, Woll J, Giritch A, Genady E, Ma X, Stöckigt J (2005b) Functional expression of an ajmaline pathway-specific esterase from Rauvolfia in a novel plant-virus expression system. Planta 222:888–898
Sabino JR, Kato L, Braga RM, Vencato I (2006) Raucaffrinoline. Acta Cryst E62:o3181–o3183
Sheludko Y, Gerasimenko I, Kolshorn H, Stöckigt J (2002) New alkaloids of the sarpagine group from Rauvolfia serpentina hairy root cultures. J Nat Prod 65:1006–1010
Stöckigt J (1995) Biosynthesis in Rauvolfia serpentina, modern aspects of an old medicinal plant. In: Cordell GA (ed) The alkaloids, vol 47. Academic Press, San Diego, pp 115–172
Subhadhirasakul S, Takayama H, Aimi N, Ponglux D, Sakai SI (1994) Novel indole alkaloids from the leaves of Rauwolfia sumatrana JACK in Thailand. Chem Pharm Bull 42:1427–1431
Unterlinner B, Lenz R, Kutchan TM (1999) Molecular cloning and functional expression of codeinone reductase: the penultimate enzyme in morphine biosynthesis in the opium poppy Papaver somniferum. Plant J 18:465–475
Warzecha H, Obitz P, Stöckigt J (1999) Purification, partial amino acid sequence and structure of the product of raucaffrine-O-ß-d-glucosidase from plant cell cultures of Rauwolfia serpentina. Phytochemistry 50:1099–1109
Zenk MH (1995) Chasing the enzymes of alkaloid biosynthesis. In: Golding BT, Griffin RJ, Maskill H (eds) Organic reactivity: physical and biological aspects. The Royal Society of Chemistry, Newcastle upon Thyne, pp 89–109
Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L, Zeng D, Gu M, Chu C, Cheng Z (2006) Gold hull and internode2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140:972–983
Acknowledgements
We gratefully acknowledge Prof. Friedrich Lottspeich, Mrs. Isabella Mathes and Mr. Reinhard Mentele (Max-Planck-Institut für Biochemie, Martinsried, Germany) for amino acid sequence determination, the Deutsche Forschungsgemeinschaft (Bad-Godesberg, Germany) and Fonds der Chemischen Industrie (Frankfurt/Main, Germany) for financial support. Deutscher Akademischer Austauschdienst [Bonn, Germany, DAAD/D05/06969] and the China Scholarship Council [Beijing, People’s Republic of China CSC (2004) 3067] are acknowledged for support. We appreciate very much the help of Dr. Joachim Arend (Mainz, Germany) in measuring EI-mass spectra. GC-MS analyses were performed by SpectroData, company, Biebelsheim, Germany. We thank Dipl. Ing. Wilfried Löbel for measurements. We also thank Prof. Nikolaus Amrhein (ETH Zürich, Switzerland) for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequences reported in this article have been submitted to the Gene Bank under Accession No: AY766462.
Electronic supplementary material
11103_2008_9331_MOESM1_ESM.tif
SDS-PAGE after a typical purification of heterologously expressed His-tagged PR (arrow). Proteins were stained with Coomassie-blue. Lane 1: crude protein extract; 2: flow through; 3: washing fraction; 4: protein pellet; 5: protein marker; 6: fraction of highly purified PR eluted from Ni-NTA column (TIF 1217 kb)
11103_2008_9331_MOESM2_ESM.tif
(a) Chemical structure and EI-MS spectrum of standard raucaffrinoline. (b) EI-MS spectrum of the enzyme product raucaffrinoline of recombinant PR from Rauvolfia (TIF 45079 kb)
Rights and permissions
About this article
Cite this article
Sun, L., Ruppert, M., Sheludko, Y. et al. Purification, cloning, functional expression and characterization of perakine reductase: the first example from the AKR enzyme family, extending the alkaloidal network of the plant Rauvolfia . Plant Mol Biol 67, 455–467 (2008). https://doi.org/10.1007/s11103-008-9331-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-008-9331-7