Abstract
The calcineurin B-like proteins (CBLs) represent a unique family of calcium sensors in plants. Although extensive studies and remarkable progress have been made in Arabidopsis (Arabidopsis thaliana) CBLs, their functions in other plant species are still quite limited. Here, we report the cloning and functional characterization of ZmCBL4, a novel CBL gene from maize (Zea mays). ZmCBL4 encodes a putative homolog of the Arabidopsis CBL4/SOS3 protein, with novel properties. ZmCBL4 has one copy in maize genome and harbors seven introns in its coding region. ZmCBL4 expressed differentially in various organs of the maize plants at a low level under normal condition, and its expression was regulated by NaCl, LiCl, ABA and PEG treatments. Expression of 35S::ZmCBL4 not only complemented the salt hypersensitivity in Arabidopsis sos3 mutant, but also enhanced the salt tolerance in Arabidopsis wild type at the germination and seedling stages. Moreover, the LiCl tolerance in all of the ZmCBL4-expressing lines increased more significantly as compared with the NaCl tolerance, and in consistent with this, it was found that the expression of Arabidopsis AtNHX8, a putative plasma membrane Li+/H+ antiporter gene identified recently, was induced in these transgenic lines under LiCl stress. The ZmCBL4-expressing Arabidopsis lines accumulated less Na+ and Li+ as compared with the control plants. This study has identified a putative maize CBL gene which functions in the salt stress-elicited calcium signaling and thus in the tolerance to salinity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
High soil salinity results in both ionic and osmotic stresses for plants. The ionic stress disrupts ion homeostasis in plant cells, resulting in excess toxic ions, for example, Na+, in the cytosol and a deficiency of essential ions such as K+ (Zhu 2001a, b, 2002). Therefore, plants have to develop mechanisms to maintain the ion homeostasis in the cytoplasm. Previous studies have indicated that various ion transporters, pumps and channels play crucial roles in these processes, and their expression and activities are regulated by some signaling pathways (Xiong et al. 2002; Zhu 2003; Reddy and Reddy 2004). Salt stress is known to trigger a rapid and transient increase of free calcium concentration in plant cells (Lynch et al. 1989; Knight et al. 1997; Knight 2000; Pauly et al. 2000). As such, Ca2+ signaling processes are one of the earliest events in salt signaling and may play an essential role in the ion homeostasis and salt tolerance in plants (Lynch et al. 1989; Knight et al. 1997; Zhu 2003; Reddy and Reddy 2004). Little was known about the salt stress-elicited calcium signaling processes in plant cells until the SOS pathway was revealed in Arabidopsis (Zhu 2000). This study began with genetic screening for SOS (salt overly sensitive) Arabidopsis mutants from an EMS or fast-neutron mutagenized M2 population or T-DNA insertion lines, which resulted in the discovery of five genes namely, AtSOS1–5 (Wu et al. 1996; Liu and Zhu 1998; Liu et al. 2000; Shi et al. 2000, 2002b, 2003; Shi and Zhu 2002c). Subsequent studies indicated that SOS1, 2 and 3 function in the same signaling pathway and their functions are Ca2+ dependent (Halfter et al. 2000). Loss-of-function mutation in each of them causes the plants to be hypersensitive both to NaCl and LiCl stresses (Liu and Zhu 1997, 1998; Liu et al. 2000; Shi et al. 2000). The Arabidopsis AtSOS3, also known as AtCBL4, encodes a small N-myristoylated calcium-binding protein with four predicted EF-hands (Sánchez-Barrena et al. 2005). The SOS3 protein has sequence similarities with both the regulatory B-subunit of calcineurin (CNB) in yeast and the neuronal calcium sensor (NCS) in animals (Liu and Zhu 1998). Both N-myristoylation and Ca2+ binding are required for its function in perceiving and relaying the Ca2+ signal elicited by salt stress (Ishitani et al. 2000). The Arabidopsis AtSOS2 (AtCIPK24) encodes a serine/threonine protein kinase consisting of a conserved N-terminal catalytic kinase region and a unique regulatory C-terminal region that is required for interacting with SOS3 and functioning in plant (Albrecht et al. 2001; Guo et al. 2001, 2004). The SOS2 protein kinase is the target of SOS3 calcium sensor in vivo (Shi et al. 1999; Halfter et al. 2000). The expression of AtSOS2 was up-regulated by salt stress (Liu et al. 2000). The Arabidopsis AtSOS1 encodes a plasma membrane Na+/H+ antiporter, which is directly involved in removing the excess Na+ out of the cell. The expression of AtSOS1 in the wild-type roots was up-regulated by NaCl stress and partially controlled by AtSOS3 and AtSOS2 (Shi et al. 2000). The activation of SOS1 Na+/H+ antiporter also required SOS3 and SOS2 proteins (Qiu et al. 2002). In the SOS pathway, a short chain of calcium signal transduction was proposed in that the SOS3 senses cytosolic calcium changes elicited by salt stress and physically interacts with and activates SOS2. The SOS3–SOS2 complex may be recruited to the plasma membrane by the myristoylated SOS3, where the SOS1 Na+/H+ antiporter activity is activated through phosphorylation by the activated SOS2 and thereby regulates ion homeostasis (Halfter et al. 2000; Zhu 2002; Qiu et al. 2002). Reconstruction of the Arabidopsis SOS system in yeast provided additional evidences for the interaction between SOS3 and SOS2 and their regulation of SOS1. The SOS3–SOS2 complex phosphorylated and activated SOS1 and further reduced Na+ accumulation in yeast cells. Coexpression of AtSOS1, 2, and 3 dramatically increased the Na+ tolerance of a yeast mutant lacking all the endogenous Na+ transporters (Quintero et al. 2002).
Very recently, it has been reported that the SOS salt tolerance pathway was identified from rice (Oryza sativa). OsSOS1, the rice functional homolog of the Arabidopsis SOS1 displayed the ability for Na+ excluding from yeast cells and suppressed the salt sensitivity of a sos1-1 mutant of Arabidopsis. OsCIPK24 and OsCBL4, the putative rice counterparts of the Arabidopsis SOS2 and SOS3 proteins, also exhibited the functional similarity to AtSOS2 and AtSOS3 (Martínez-Atienza et al. 2007).
The Arabidopsis CBL1 and CBL9 calcium sensors also play roles in response to salt stress (Cheong et al. 2003; Pandey et al. 2004). These two CBL proteins share ∼90% sequence identity (Kolukisaoglu et al. 2004). Although having distinct response to exogenous ABA, the expressions of both AtCBL1 and AtCBL9 were highly inducible by salt, cold and drought stresses. Both cbl1 and cbl9 null mutants displayed reduced salt tolerance and CBL1-overexpressing transgenic plants showed enhanced salt tolerance (Cheong et al. 2003; Pandey et al. 2004). These findings suggest that either AtCBL1 or AtCBL9 functions as a positive regulator in salt stress response pathway. Xu et al. (2006) demonstrated that the Arabidopsis CBL1 and CBL9 proteins are also involved in the CIPK23 activated, AKT1-mediated K+ uptake pathway. Both CBL1 and CBL9 localize to the plasma membrane. Low-K+ stress signals may trigger the cytosolic Ca2+ signal and lead to the activation of CBL1 and CBL9, and then the CBL proteins interact with and activate CIPK23 and may recruit CIPK23 to the plasma membrane, where AKT1, a K+ transporter, is phosphorylated by CIPK23. As the result, AKT1 is activated for K+ uptake under low-K+ conditions.
In spite of the extensive studies and remarkable progress in the Arabidopsis salt-elicited SOS/CBL-CIPK calcium signaling pathways, information about these signaling modules in other plant species is still quite limited. Here we present the cloning and characterization of ZmCBL4, a novel CBL gene from maize. ZmCBL4 encodes a putative homolog of the Arabidopsis CBL4/SOS3 protein, with novel properties. ZmCBL4 expressed differentially in various plant organs under normal condition. The expression of ZmCBL4 in young seedling roots was up-regulated by salt stress and ABA, but down-regulated by PEG treatment. Expression of 35S::ZmCBL4 not only complemented the salt hypersensitivity in Arabidopsis sos3 mutant, but also enhanced the salt tolerance in Arabidopsis wild type. Moreover, the LiCl tolerance in all of the ZmCBL4-expressing lines increased more significantly as compared with the NaCl tolerance, and in consistent with this, it was found that the expression of Arabidopsis AtNHX8, a putative plasma membrane Li+/H+ antiporter gene identified recently, was induced in these transgenic lines under LiCl stress. To the best of our knowledge, this is the first report of maize CBL genes.
Materials and methods
Maize material and stress treatments
Maize inbred line Han21 was used in this study. The grains were surface-sterilized in a solution of 0.5% Clorox plus 0.05% Triton X-100 for 20 min and rinsed five times with tap water and then sown in pots filled with sand. The pots were placed in a greenhouse and watered once every 3 days with tap water. The three-leaf-stage seedlings were carefully removed from the sand and washed clean with tap water. After the three-day adaptive culture in Hoagland solution as described by Zheng et al. (2004), the seedling roots were immersed in MS nutrient solution supplemented with 20% PEG(w/v), 100 μΜ(±)-cis,trans-ABA, 250 mM NaCl or 20 mM LiCl for 0, 2, 6, 12 and 24 h, respectively. The shoots and roots from all the treated seedlings were harvested separately at the indicated time points and stored at −76°C for RNA extraction.
Database searches and isolation of ZmCBL4 gene
To identify maize CBL genes, we performed database searches for the predicted CBL mRNA sequences, expressed sequence tags (ESTs), or genomic sequences by sequence comparison with previously identified Arabidopsis CBLs in different maize databases using TBLASTN. The sequences or ESTs obtained were further assembled and compared pairwise to identify nonredundant sequences. Among the identified putative maize CBL unique genes, one mRNA sequence (BT018770), which encodes a predicted protein with the highest similarity to Arabidopsis SOS3/CBL4, was chosen for further study and designated as ZmCBL4.
The complete coding region of the cDNA and the genomic DNA of ZmCBL4 were amplified by PCR approach using the specific primer pairs, 5′-GATCCATGGGCTGCGCGACGTCCAA-3′ (forward) and 5′-GTGGGTCACCATACGCAGATGTACGCAAAC-3′ (reverse). The condition for amplification was at 94°C for 3 min followed by 30 cycles at 94°C for 30 s, at 62°C for 30 s and at 72°C for 1 min and 30 s, plus a final extension at 72°C for 10 min. Three independent PCR products were purified and cloned into pGEM T-easy vector (Promega) and sequenced.
Real-time quantitative PCR
Total RNA was extracted from the stored (−76°C) maize samples using the hot-phenol method and was treated with RQI DNase (Promega) to remove the genomic DNA contamination. The first strand cDNA was synthesized from the total RNA (5 μg) with M-MLV reverse transcriptase (Promega) and used as the template for subsequent PCR amplification.
The real-time quantitative PCR (RT-qPCR) for examination of ZmCBL4 expression was carried out with an ABI Prism 7900 HT sequence detection system (Applied Biosystems). The maize α-tubulin gene was used as internal control for normalization of the template cDNA. The specific primers were designed according to the guidelines with help of the Primer Express 2.0 software (Applied Biosystems). The primers for ZmCBL4 were 5′-TCAGTGTGTTCCACCCTAAAGCA-3′ (forward) and 5′-ATCAAGCAGCGCCAAGACCAT-3′ (reverse). The primers for maize α-tubulin were 5′-GAGCATGGCATTCAGGCTGACG-3′ (forward) and 5′-TCAACAAAAACAGCACGGGGCA-3′ (reverse). The expected sizes of the amplified fragments were all 128 bp. Each PCR was repeated at least four times in a total volume of 25 μl containing 1 × SYBR Green I PCR Master Mix (ABI), 200 nM of each primer and 1 μl 1:10 diluted template cDNA, using 96-well optical-grade PCR plate and the matched optical-grade membrane. The PCR protocol was as follows: an initial denature step consisting of 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 30 s at 94°C, 30 s at 60°C and 18 s at 72°C, and an additional cycle of 15 s at 95°C, 15 s at 60°C and 15 s at 95°C for melting curve analysis. The specificity of the PCR products was verified by their migrating rate on a 2.0% (w/v) agarose gel at the expected size and by sequencing. The data obtained were analyzed with SDS 2.2 software (Applied Biosystems). The relative expression of ZmCBL4 under each stress condition or in different organs was calculated using the relative 2−ΔΔCt method (Livak and Schmittgen 2001) and the error bars indicate SD (n = 4).
Arabidopsis transformation and stress tolerance characterization
The ZmCBL4 cDNA in the sense orientation was cloned into the NcoI/BstEII restriction enzyme site of vector p3301 (pCAMBIA3301) under the control of 35S promoter from the cauliflower mosaic virus. The p3301-ZmCBL4 construct was introduced into Agrobacterium tumefaciens GV3101 strain cells. The Arabidopsis sos3 mutant and wild type (ecotype Columbia) were used in this study. The plants were grown at 22°C under long-day conditions (16-h-light/8-h-dark cycle) to the flowering stage for plant transformation. The transformation was carried out by the floral dip method (Clough and Bent 1998). The T1 transgenic plants were screened by spraying with 0.5‰ (v/v) phosphinothricin (ppt) solution and confirmed by PCR approach. The T2 seeds were plated on MS (Murashige and Skoog 1962) agar plates containing 7 mg/l ppt and the transgenic lines with a 3:1(resistant: sensitive) segregation ratio were selected to produce T3 seeds. The T3 lines displaying 100% ppt resistance were considered homozygous and used for further experiments. Northern blot analysis was performed to confirm the expression of ZmCBL4 in the transgenic lines. The sos3 mutant and the transformed wild type with empty vector p3301 were used as the controls.
The salt tolerance analysis of the transgenic lines at the seedling stage was carried out using the root-bending assay described as Liu and Zhu (1998). The MS agar plates containing 1% (w/v) agar and 2% (w/v) sucrose, pH 5.7 were supplemented with different concentrations of NaCl (100, 125, 150 or 175 mM) and LiCl (12, 15, 17 or 20 mM). For germination test, the surface-sterilized seeds were sown on the MS agar plates supplemented with different concentrations of NaCl (125, 150, or 175 mM), LiCl (12, 15, 17, or 20 mM), KCl (90 or 120 mM), ABA (0.75, 1.0, 1.25, 1.5, or 1.75 μM) and mannitol (300, 350, or 400 mM). The plates were maintained at 4°C for 4–5 d and then incubated in a growth room at 22°C under long-day conditions (16-h-light/8-h-dark cycle) at ∼100μE m−2 s−1. At least 60 seeds were sown for each line and the percentage of germinated seeds that developed green cotyledons were calculated in all assays 5 d after germination. For seedling fresh weight assay, 30 ten-day-old seedlings growing on vertical MS agar plates (1.2%) supplemented with different concentrations of NaCl (125, 150, or 175 mM) and LiCl (12, 15, or 20 mM) were weighed. All experiments were performed in triplicate and the error bars indicated the standard deviation (SD).
Southern blot and Northern blot analyses
For Southern blot analysis, genomic DNA (30 μg) from maize inbred line Han21 seedlings was digested with EcoRI, EcoRV, HindIII BamHI, KpnI, or SacI, separated by electrophoresis on a 0.8% agarose gel, transferred to nylon membrane (Amersham), and hybridized for 16 h at 65°C with α-32p-dCTP-labeled complete coding region of the ZmCBL4 cDNA as a probe. The membrane was washed with 2 × SSC, 0.1% SDS; 1 × SSC, 0.1% SDS and 0.5 × SSC, 0.1% SDS for 15 min at 65°C, respectively.
For Northern blot analyses, the transgenic and control Arabidopsis seedlings growing on vertical MS agar plates for 2 weeks were treated by immersing the roots in MS nutrient solution supplemented with 250 mM NaCl or 20 mM LiCl for 5 h. The seedlings were collected and washed two times with double distilled water (ddH2O), and then stored at −76°C for RNA extraction. Total RNA was extracted using hot-phenol method, fractionated in denaturing formaldehyde agarose gel (1.2%), blotted onto nylon membranes (Amersham), and hybridized for 20 h at 65°C with α-32p-dCTP-labeled specific probes. For ZmCBL4 expression, the complete coding region of ZmCBL4 cDNA was used as a probe. For AtSOS1 expression, a 500 bp cDNA fragment in the 5′ region of the open reading frame was used as the probe and amplified by PCR using the primers, 5′-TTGTGAAGGTCACGTTTCCGTAT-3′ (forward) and 5′-TGGTAACTTTCGCTTGGTAGGC-3′ (reverse); for AtNHX8 expression, a 321 bp cDNA fragment was used as the probe and amplified by PCR using the primers, 5′-TCTGACTTGAAGAAGCTCCTGAG-3′ (forward) and 5′-TCAAAGCCAAAAAGGATTGATTGAA-3′ (reverse). The membranes were washed with 2 × SSC, 0.5% SDS; 1 × SSC, 0.5% SDS; 0.5 × SSC, 0.5% SDS and 0.1 × SSC, 0.1% SDS for 15 min at 65°C, respectively.
The hybridization signals for both Southern- and Northern-blotting were imaged using a phosphorimager. The ethidium bromide-stained rRNA bands in the agarose gel were shown as a RNA loading control.
Measurement of ion contents
Twelve-day-old Arabidopsis seedlings growing on vertical MS agar plates (1.0%) were transferred onto MS, MS supplemented with 125 mM NaCl or 12 mM LiCl agar plates and placed vertically. After growing for 6 d, the seedlings were collected and washed five times with ddH2O, and then dried at 80°C for 3 days. Approximately 4 mg (dry weight) of the samples were dry-ashed for 6 h in quartz dishes at 570°C in a muffle furnace. The samples were digested in 2 ml 1:1 diluted concentrated HCl with ddH2O and then diluted to a total volume of 20 ml with ddH2O prior to ion measurement. The K+, Na+, and Li+ contents in the solutions were determined with an inductively coupled plasma atomic emission spectrometer (ICP-AES, Perkin-Elmer, Boston, MA, USA). All the experiments were repeated three times and the SD was showed as the error bar.
Results
Isolation of ZmCBL4 gene and comparison of its amino acid sequence
Database searches together with bioinformatics analysis led to the identification of nine putative maize CBL unique mRNA sequences, and one of them (BT018770), which encodes a predicted protein with the highest sequence similarity (57% of identities) to Arabidopsis CBL4/SOS3, was designated as ZmCBL4 and chosen for further study.
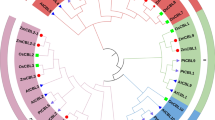
ZmCBL4 has an open reading frame (ORF) of 633 bp and encodes a predicted polypeptide of 211 amino acids. The cDNA and genomic DNA fragments with the complete coding region of ZmCBL4 were cloned by PCR approach and sequenced. The cDNA ORF sequences from three independent positive clones were identical. This sequence was compared with the original sequence in the public databases (BT018770) and four nucleotide differences were found between them, which resulted in the changes of three amino acid residues (data not shown). Alignment with several representatives of Arabidopsis and rice CBLs exhibited that the predicted ZmCBL4 contains all of the four elongation factor (EF) hand motifs found in other CBL proteins (Fig. 1A). ZmCBL4 protein also carries the consensus myristoylation sequence MGXXXS/T at its amino terminus. The clustering pattern of ZmCBL4 with 10 AtCBLs showed that it is more similar to AtCBL4 than to others (Fig. 1B).
Sequence analyses of ZmCBL4. (A) Alignment of the deduced amino acid sequences of maize CBL4, Arabidopsis CBL1, CBL2, CBL4/SOS3, CBL9, and rice OsCBL2. Hyphens indicate gaps introduced to maximize the sequence alignment. Identical residues are highlighted in black, and similar residues are highlighted in gray (60% similarity), respectively. The location of the myristoylation sequence at N- terminus was lined above. The four predicted EF hands are showed as I–IV. (B) Phylogenetic relationship of ZmCBL4 with 10 Arabidopsis CBLs. Multiple sequence alignment was performed using the program ClustalX1.83, and the phylogenetic tree was constructed using the Neighbor-Joining method. The accession numbers of Arabidopsis and rice CBLs are as follows: AtCBL1, AAC26008; AtCBL2, AAC26009; AtCBL3, AAC26010; AtCBL4/SOS3, AAG28402; AtCBL5, AAG28401; AtCBL6, AAG28400; AtCBL7, AAG10059; AtCBL8, AAL10300; AtCBL9, AAL10301; AtCBL10, AAO72364; the rice OsCBL2, AL713904
Genomic organization of ZmCBL4
Sequencing results demonstrated that the genomic DNA of ZmCBL4 is 1994 bp (accession number: EF405963). Comparison of the genomic sequence with its corresponding cDNA sequence revealed that the genomic copy of ZmCBL4 harbors seven introns in its coding region (Fig. 2A), which is similar to the case in most AtCBLs and OsCBLs. Southern blot analysis showed that ZmCBL4 has only one copy in maize genome (Fig. 2B).
Gene structure and genomic organization of ZmCBL4. (A) Gene structure of ZmCBL4. Filled boxes indicate exons, and lines between boxes indicate introns. (B) Southern blot analysis of ZmCBL4 to determine the copy number in the maize genome. Genomic DNA (30 μg) was completely digested with the enzyme indicated
Expression of ZmCBL4 is regulated by abiotic stresses and ABA
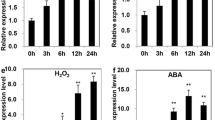
To gain insight into the possible function of ZmCBL4, we initially examined the expression patterns of this gene. However, because the transcript level of ZmCBL4 was too low to be detected by Northern blot analysis (data not shown), the real-time PCR approach was used to detect its expression. Under NaCl stress, the ZmCBL4 expression was down-regulated in shoots (Fig. 3A). At 2 h after treatment, the transcript level of ZmCBL4 was decreased to 0.33 ± 0.20 times that at the initial time point (time 0 h), and it reached the lowest level at 12 h after treatment. However, the ZmCBL4 expression was up-regulated in roots under NaCl stress (Fig. 3B). At 12 h after treatment, the ZmCBL4 mRNA level increased to 4.01 ± 0.56 times that at the initial time point, and it reached more than 11 times at 24 h after treatment. Under LiCl stress, the expression pattern of ZmCBL4 in roots was similar to that of NaCl stress, whereas the expression in shoots increased during the first 2 h and then decreased to the basal level (Fig. 3A, B). ABA treatment (100 μM) caused a more rapid and more obvious up-regulation of ZmCBL4 expression as compared with the induction by NaCl and LiCl. This change was stronger in roots than that in shoots and showed a wavelike pattern during the whole treatment period (Fig. 3A, B). In addition, a slight kinetic difference was observed between roots and shoots in response to ABA. By contrast, PEG treatment (20%) resulted in a rapid and strong down-regulation of ZmCBL4 both in shoots and roots (Fig. 3A, B).
Expression of ZmCBL4 in maize. The maize α-tubulin gene was used as the internal control for normalization of the template cDNA. Each PCR was repeated at least four times and the error bars represent the SD. (A) and (B) Expression of ZmCBL4 in shoots and roots, respectively, under different treatments. The transcript level at time 0 h (untreated control) was used as the calibrator whose ZmCBL4 mRNA level was given as 1. (C) Expression of ZmCBL4 in various organs. 1, young roots; 2, shoots; 3, mature roots; 4, stalks; 5, mature leaves; 6, ears; 7, tassels; 8, silks. The transcript level in mature leaves was used as the calibrator and its ZmCBL4 mRNA level was given as 1
We also examined the expression of ZmCBL4 in shoots and roots of young seedlings and in various organs of adult maize plants. As shown in Fig. 3C, the expression level was most abundant in ears but was very low in mature leaves, stalks and tassels. When the transcript level in mature leaves was used as the calibrator (its ZmCBL4 mRNA level was given as 1), the relative fold changes in other organs were between 1.05 ± 0.13 and 10.98 ± 0.92. Interestingly, the transcript level in shoots was noticeably higher than that in roots at the seedling stage while the transcript level in adult roots was almost three times that in adult leaves. We performed three independent assays with RNA isolated from different samples of shoots/leaves and roots and obtained similar results each time.
Taken together, the expression patterns of ZmCBL4 suggest its specific or differential regulatory roles in stress responses and developmental signaling processes in maize.
Constitutive expression of ZmCBL4 complements the salt hypersensitivity of Arabidopsis sos3 mutant
As mentioned above, among the nine putative maize CBL proteins, CBL4 had the highest sequence similarity to Arabidopsis SOS3 protein. The salt-inducible expression also suggested a possible role of ZmCBL4 in salt stress signaling pathway. These findings led us to hypothesize that ZmCBL4 may be a homolog of AtSOS3 and may function underlying the mechanism similar to SOS3 in the salt responsiveness. To test this hypothesis, we first constructed transgenic lines of 35S::ZmCBL4 in the Arabidopsis sos3 mutant background. Thirty independent transgenic homozygous T3 lines were obtained and six of them were selected to examine the expression of ZmCBL4 by Northern blot analysis. As shown in Fig. 4A, ZmCBL4 was strongly expressed in all six transgenic lines and all of them were selected for phenotypic characterization.
Constitutive expression of ZmCBL4 complemented the NaCl and LiCl hypersensitive phenotypes of the Arabidopsis sos3 mutant. sos3, Arabidopsis sos3 mutant; WT, wild-type; SS1, 13, 23, 24, 28 and 29, six independent transgenic lines, respectively. (A) Expression of ZmCBL4 in Arabidopsis sos3 mutant background. Twenty micrograms of total RNA from each material was loaded. The ethidium biomide-stained rRNA bands in the agarose gel were shown as a RNA loading control. (B), (C) and (D) The results of root-bending assays: 5-day-old seedlings were transferred from MS medium to the MS agar plates (B), MS agar plates supplemented with 100 mM NaCl (C) or 12 mM LiCl (D), and then the plates were placed vertically on a rack with roots upside down. The photographs were taken at 12 d after transferring to 22°C. (E), (F) and (G) Seed germination and subsequent growth on MS agar plates (E), MS agar plates supplemented with 125 mM NaCl (F) or 15 mM LiCl (G), and the photographs were taken at 8, 12 and 15 d, respectively, after transferring to 22°C. (H) and (I) Seed germination percentages on MS agar plates supplemented with different concentrations of NaCl and LiCl, respectively. The seeds (each with at least 60 seeds) that developed green cotyledons were scored at 5 d after transferring to 22°C. (J) and (K) Fresh weight (mg/30 seedlings) of seedlings on vertical MS agar plates supplemented with different concentrations of NaCl and LiCl, respectively. All the values are means±SD (n = 3), and SD was shown as error bar. * and ** represent significantly different from the control WT at P < 0.05 and 0.01 level, respectively, by student’s t-test
A preliminary assay on the salt tolerance of the six transgenic lines showed that these transgenic lines had similar salt tolerance. Therefore, the detailed analyses were later performed using three independent transgenic lines, SS1, 13 and 23. On MS agar plates supplemented with various concentrations of NaCl (100–175 mM), the root-bending phenotypes of the transgenic lines were similar to those of wild type seedlings, whereas the sos3 seedlings exhibited no significant root growth. Under different concentrations of LiCl (10–20 mM), the transgenic seedlings displayed a better growth both in roots and shoots than the wild type seedlings, whereas the root growth of the sos3 seedlings was inhibited completely (Fig. 4C, D).
We also examined the salt tolerance of these materials at seed germination and post-germination stages. On the MS agar plates supplemented with different concentrations of NaCl (125–175 mM), the germination rates and subsequent growth of the transgenic lines were similar to the wild type (Fig. 4F, H, J). Under LiCl stress (12–20 mM), however, all of the transgenic lines showed much higher germination rates and significantly better subsequent growth than the wild type (Fig. 4 G, I, K). For example, at 15 mM LiCl, the average germination rate of the transgenic lines was 69.81 ± 4.36% after 5 d of incubation at 22°C, whereas the germination rate of the wild type was only 13.66 ± 2.51% on the same conditions. After 10 days under the same LiCl stress, the average fresh weight of the transgenic lines was 45.12 ± 2.58 mg per 30 seedlings, which was significantly higher than that of the wild type seedlings (14.63 ± 1.76 mg per 30 seedlings). The sos3 mutant displayed hypersensitivity to both NaCl and LiCl stresses, just a few of the seeds developed green cotyledons when the concentration of NaCl or LiCl was increased to 150 mM or 15 mM, respectively (Fig. 4F–K).
The evidences presented thus far clearly indicate that the expression of ZmCBL4 completely complemented the salt hypersensitive phenotype of the sos3 mutant and that ZmCBL4 is a functional homolog of AtSOS3. No significant difference was observed between the transgenic lines and the sos3 mutant or wild type on MS medium (Fig. 4B, E, H–K) or MS media containing different concentrations of KCl, mannitol or ABA (data not shown).
Constitutive expression of ZmCBL4 enhances the salt tolerance of wild type Arabidopsis
To further investigate the function of ZmCBL4, we also constructed transgenic lines of 35S::ZmCBL4 in the Arabidopsis wild-type background and obtained 20 independent homozygous T3 lines. To confirm the expression of ZmCBL4, Northern blotting was performed on six of them. As shown in Fig. 5A, all of the transgenic lines tested produced the ZmCBL4 transcript and the four lines showing higher expression levels, namely SW16, 17, 18 and 19, were selected for further study.
Constitutive expression of ZmCBL4 in Arabidopsis wild type enhanced the salt tolerance. WT, wild type; SW8, 9, 16, 17, 18, and 19, six independent transgenic lines, respectively. (A) The expression of ZmCBL4 in Arabidopsis wild type background. Twenty micrograms of the total RNA from each material was loaded. The ethidium biomide-stained rRNA bands in agarose gel were shown as a RNA loading control. (B) and (C) Seed germination and subsequent growth of each material on MS agar plates supplemented with 150 mM NaCl or 17 mM LiCl, respectively. The photographs were taken at 10 d after transferring to 22°C. (D) and (E) Seed germination percentage of each material on MS agar plates supplemented with different concentrations of NaCl and LiCl, respectively. The seeds that developed green cotyledons were scored at 5 d after transferring to 22°C. (F) and (G) Seedling fresh weight (mg/30 seedlings) growing on vertical MS agar plates supplemented with different concentrations of NaCl and LiCl, respectively. All the values are means ± SD (n = 3), and SD was shown as error bar. * and ** are significantly different from the control WT at P < 0.05, 0.01, respectively, by student’s t-test
The seed germination and subsequent growth of the transgenic lines and the control wild type were compared under different conditions. No significant differences were observed in all the materials examined on MS medium or MS media supplemented with different concentrations of KCl, mannitol, or glucose (data not shown). However, under various concentrations of NaCl or LiCl, the transgenic lines showed significantly higher germination rates and better seedling growth than the wild type (Fig. 5B–G). Figure 5B and C illustrated the germination and subsequent seedling growth of the transgenic lines and wild type on MS medium supplemented with 150 mM NaCl or 17 mM LiCl, respectively. Figure 5D and E presented the results of more detailed analyses of germination rates on MS media supplemented with different concentrations of NaCl and LiCl, separately, after 5 days of incubation at 22°C, and the significant differences between the transgenic lines and wild type existed. Figure 5F and G further showed the fresh weights of the transgenic and control wild-type seedlings on these stress media after incubating for 10 d at 22°C, and the differences between them were also significant. All of the results indicate that constitutive expression of ZmCBL4 in Arabidopsis enhanced their NaCl and LiCl tolerance during the early developmental stages. Moreover, it is necessary to mention that the salt-tolerant properties of the transgenic lines remained for at least 1 month, when the transgenic plants were still green and quite strong but the control wild-type plants became weak and yellow (under 150–175 mM NaCl stress) or died (under 15–20 mM LiCl stress) (data not shown). Later, the media became dried.
Constitutive expression of ZmCBL4 induces the transcription of Arabidopsis AtSOS1 and AtNHX8
Previous study has shown that expression of AtSOS1 is inducible in the wild-type seedling roots by NaCl stress, but in sos3 mutant, no up-regulation of SOS1 expression was found in either shoots or roots under NaCl stress (Shi et al. 2000). To investigate whether the ZmCBL4 expression really rescued the AtSOS1 expression in the transgenic sos3 lines, we examined the AtSOS1 mRNA level in the ZmCBL4-expressing sos3 seedlings by Northern blot analysis. As shown in Fig. 6A, the transcripts of AtSOS1 were detected in the transgenic lines SS1 and SS23, but no signal was seen in the control sos3 mutant, wild type and the transgenic line SS13 under NaCl stress. In normal growth condition, the AtSOS1 mRNA was not detectable in all the materials tested (data not shown). This result indicates that ZmCBL4 at least partially rescued the transcription of AtSOS1 in the transgenic sos3 lines.
Constitutive expression of ZmCBL4 in Arabidopsis induced the transcription of AtSOS1 and AtNHX8. (A) AtSOS1 expression in the ZmCBL4-expressing transgenic sos3 mutant seedlings under NaCl stress (250 mM NaCl for 5 h). (B) and (C) AtNHX8 expression in the ZmCBL4-expressing sos3 mutant and wild type seedlings, respectively, under LiCl stress (20 mM LiCl for 5 hours). Forty (for AtSOS1) or twenty (for AtNHX8) micrograms of total RNA from each material was loaded. The ethidium biomide-stained rRNA band in the agarose gel was shown as a RNA loading control
During phenotype characterization of the transgenic lines, we noticed that the LiCl tolerance in all of the transgenic lines including SS1-23 and SW16-19 increased more significantly as compared with the NaCl tolerance. Moreover, Northern blot analysis showed that the expression level of AtSOS1 in the transgenic seedlings was very low. This contradiction might imply that new monovalent cation:proton antiporter(s) other than SOS1 be activated by ZmCBL4 and responsible for LiCl tolerance. Recent study has shown that Arabidopsis AtNHX8, a highly homologous gene of AtSOS1, encodes a putative plasma membrane Li+/H+ antiporter and functions specifically in lithium extrusion and ion homeostasis in Arabidopsis (An et al. 2007). To the best of our knowledge, this is the only specific Li+/H+ antiporter identified so far. Therefore, the AtNHX8 was selected as the candidate and Northern blotting was performed for its expression. Under normal growth condition, the AtNHX8 transcript was not detectable in any of the seedlings examined including wild type, the sos3 mutant, and the transgenic lines (data not shown). Under LiCl stress, however, all the transgenic lines produced moderate or low levels of AtNHX8 mRNA, whereas no signal was detected in the sos3 and wild type seedlings (Fig. 6B, C). This finding indicates that constitutive expression of ZmCBL4 induced the transcription of AtNHX8, which might account for the enhanced LiCl tolerance of ZmCBL4-transgenic lines.
The transgenic seedlings accumulate less Na+ or Li+ under NaCl or LiCl stress
To investigate whether constitutive expression of ZmCBL4 altered the accumulation of Na+, Li+, and K+, we measured the contents of these cations in control and transgenic seedlings under normal growth condition, NaCl and LiCl stresses. As shown in Fig. 7A, when the seedlings of the controls and ZmCBL4-expressing sos3 lines were grown on MS medium supplemented with 125 mM NaCl or 12 mM LiCl, the K+ contents in all of the seedlings were reduced. Further, the reductions were less strong in the transgenic and wild type seedlings as compared with the sos3 mutant seedlings. The Na+ contents were similar in all of the seedlings grown on MS medium or MS medium containing 12 mM LiCl. Under NaCl stress, however, the transgenic and wild type seedlings accumulated significantly less Na+ than did the sos3 mutant seedlings (Fig. 7B). No Li+ was detected in the seedlings grown on MS medium or MS medium supplemented with 125 mM NaCl. After 6 days subjected to 12 mM LiCl stress, however, both the sos3 mutant and wild type seedlings accumulated significantly more Li+ than did the transgenic lines (Fig. 7C).
ZmCBL4-expressing Arabidopsis seedlings accumulated less Na+ or Li+ than did the control Arabidopsis seedlings under salt stress. (A), (B) and (C) K+, Na+ and Li+ contents of the ZmCBL4-expressing sos3 mutant seedlings on MS agar plates, MS agar plates supplemented with 125 mM NaCl or 12 mM LiCl, respectively. (D), (E) and (F) K+, Na+ and Li+ contents of the ZmCBL4-expressing wild type seedlings on MS agar plates, MS agar plates supplemented with 125 mM NaCl or 12 mM LiCl, respectively. Values are means ± SD (n = 3), and SD was shown as error bar. * and ** are significantly different from the control WT at P < 0.05, 0.01, respectively, by student’s t-test
The changes of K+, Na+ and Li+ contents in the transgenic wild type and control wild type seedlings were shown in Fig. 7D–F, respectively. The transgenic lines also accumulated significantly less Na+ or Li+ but relatively more K+ as compared with the control wild type seedlings under NaCl or LiCl stress.
Together, these results indicate that ZmCBL4 is involved in the Na+, Li+ and K+ homeostasis in the transgenic Arabidopsis lines under the corresponding salt stresses.
Discussion
ZmCBL4 may be involved in different calcium signaling pathways
Calcium plays a crucial role as a second messenger in various environmental and developmental signaling pathways in plant cells. The calcium sensors are immediate downstream components after calcium changes and function as a threshold factor in linking the calcium signature to the downstream components in the pathways (Cheong et al. 2003; Reddy and Reddy 2004). CBLs are newly identified calcium sensors unique to plants. Recent studies have demonstrated that most CBL genes are differentially involved in various stress and/or developmental signaling processes (Kudla et al. 1999; Albrecht et al. 2003; Cheong et al. 2003; Pandey et al. 2004; Hwang et al. 2005; Mahajan et al. 2006). We show here that ZmCBL4 expressed differentially in various maize organs tested under normal growth conditions, and that the expression level of ZmCBL4 changed when the seedlings were subjected to different abiotic stresses and ABA treatment. Unlike many other salt tolerance genes that are preferentially expressed in roots but not in aerial tissues, the ZmCBL4 transcript levels in maize shoots and ears were obviously higher than those in roots under normal growth condition (Fig. 3C). This tissue-specific expression pattern suggests its other function in addition to that for response to salt stress. Salt stress is known to elicit a rapid increase in the free calcium concentration in the cytosol (Lynch et al. 1989; Knight et al. 1997; Knight 2000; Pauly et al. 2000). The maize CBL4, a putative homolog of Arabidopsis SOS3 calcium sensor, is proposed to sense this salt-triggered calcium signal and relay it downstream. The up-regulation of ZmCBL4 expression in roots under NaCl or LiCl stress seems consistent with its function in salt response (Fig. 3B). However, the significant induction appeared 12 h after exposure to NaCl or LiCl stress if a 2-fold induction criterion was used. This is actually too slow to account for any significant contribution toward the rapid transduction of the stress signal. There exists a low level of ZmCBL4 expression in maize roots under normal growth condition (Fig. 3C), and perhaps this basal mRNA and its corresponding protein function during the early exposure to salt stress, while for a sustained response, the induced transcript and its protein may be required and thus play a major role.
Unlike many other stress-regulated genes that are induced not only by various stresses but also by ABA, the expression of AtSOS1, an ultimate component in the SOS pathway, was not inducible by exogenous ABA (Shi et al. 2000). In addition, the Arabidopsis sos mutants, including the sos1, 2 and 3 mutants, did not show altered response to ABA treatment (Liu and Zhu 1998; Shi et al. 2000; Liu et al. 2000). This phenotype again suggests that the Arabidopsis SOS pathway is not regulated by ABA, or not involved in ABA-mediated signaling processes. However, our data indicate that ZmCBL4 expression is up-regulated rapidly and obviously by ABA treatment (Fig. 3A, B), which suggests its involvement in ABA-mediated signaling pathway. Some other CBL genes, for example, the Arabidopsis AtCBL9, are also implicated in the plant response to ABA (Pandey et al. 2004).
By contrast to ABA, PEG treatment negatively regulated the expression of ZmCBL4. Although the meaning of this down-regulation is unknown, it is clear that ZmCBL4 is responsive to osmotic stress and may further influence the related signaling processes in maize.
Taken together, the expression patterns of ZmCBL4 in various organs or under stresses suggest its involvement in the developmental and stress signaling pathways in maize. Like AtSOS3 (Liu et al. 2000), the expression level of ZmCBL4 is also low, which is consistent with its regulatory role.
A calcium sensor for ion homeostasis and salt tolerance
The Arabidopsis SOS pathway is the first established calcium-dependent salt stress signaling pathway in plant cells. In the SOS pathway, the SOS3/CBL4 protein is known to perceive and relay the calcium signal elicited by salt stress to its target SOS2, which then activates SOS1 (Halfter et al. 2000; Quintero et al. 2002). SOS1 is a plasma membrane Na+/H+ antiporter and directly functions in removing excess Na+ from the cell (Qiu et al. 2002). In line with this module, the SOS1 gene expression and SOS1 transporting activity are partially controlled by SOS3 and SOS2, and in sos3 mutant plants, both the SOS1 transcription and the SOS1 activity cannot be induced or activated by salt stress (Shi et al. 2000; Qiu et al. 2002). Very recently, the SOS pathway was identified from rice and was demonstrated a high degree of functional similarity to its Arabidopsis counterpart (Martínez-Atienza et al. 2007). In the present study, the sequence similarity and the salt-induced expression pattern made ZmCBL4 a logical homolog of AtSOS3 and may function in salt stress. To test this hypothesis, we performed transgenic assays both in Arabidopsis sos3 mutant and wild type backgrounds. The root-bending assay, the seed germination and seedling growth experiments showed that constitutive expression of ZmCBL4 not only suppressed completely the NaCl hypersensitivity but also enhanced the LiCl tolerance in the transgenic sos3 lines as compared with the wild type Arabidopsis. We further provide evidence for ZmCBL4 rescuing the AtSOS1 transcription in the sos3 transgenic lines by Northern blot analysis, although the signals were weak (Fig. 6A). Shi et al (2000) reported that the AtSOS1 gene expression was salt-induced in the roots of wild-type Arabidopsis seedlings but not in the shoots. Unexpectedly, no AtSOS1 mRNA was detected in the NaCl treated-wild type seedlings in our Northern blotting assay. The failure, together with the weak Northern blotting signals of AtSOS1 detected in the transgenic lines (Fig. 6A), may result from the NaCl treatment and subsequent RNA loading. Perhaps the treatment duration (5 h) with 250 mM NaCl is not long enough and the total RNA loading of 40 μg is still not sufficient for successful detection of SOS1 transcripts by Northern blotting analysis. The whole-seedling RNA but not the root RNA used in our assay may be another possible reason. Constitutive expression of ZmCBL4 in the wild type background also enhanced the NaCl and especially LiCl tolerance during the seed germination and seedling growth. These data clearly demonstrate that ZmCBL4 is a functional homolog of AtSOS3 and it can increase the salt tolerance of the transgenic wild-type lines.
Maintaining low level of Na+ and a high K+/Na+ ratio in the cytosol is critical for cellular metabolism and salt tolerance in glycophytes (Zhu et al. 1998; Qiu et al. 2002; Zhu 2003). The AtSOS3 protein is required for maintaining sodium and potassium ion homeostasis and salt tolerance in Arabidopsis (Liu and Zhu 1998; Halfter et al. 2000; Qiu et al. 2002). Our data also indicate that constitutive expression of ZmCBL4 enable the transgenic Arabidopsis seedlings to maintain low levels of Na+ and Li+ but relatively high levels of K+ under salt stress, which provides additional evidence to certify that the ZmCBL4 is a functional homolog of AtSOS3. A yeast two-hybrid assay indicated that some rice CBLs could interact with CIPKs from Arabidopsis (Hwang et al. 2005). Perhaps the maize CBL4 protein heterogenously expressed in Arabidopsis can sense the calcium signal triggered by salt stress and interact with Arabidopsis SOS2, and then the ZmCBL1-AtSOS2 complex activates or enhances the activity of SOS1 in the sos3 mutant or wild type seedlings.
Overexpression of ZmCBL4 induces the transcription of Arabidopsis AtNHX8
In plants, the Na+/H+ antiporters play an important role in transporting monovalent cations including Na+, K+, and Li+. There are more than 40 genes that encode putative Na+/H+ antiporters in Arabidopsis genome (Maser et al. 2001; Brett et al. 2005). These antiporters fall into three families, and one of them is the monovalent cation:proton antiporter-1 (CPA1) family, which includes eight members from AtNHX1 to AtNHX8 (Maser et al. 2001). AtNHX1 and AtNHX7 (identical to SOS1) have been well characterized. AtNHX1 is localized to and contributes a major portion of the Na+/H+- exchange activity in tonoplast, where it mainly sequesters Na+ and K+ and to a much lesser extent Li+ (Apse et al. 2003; Qiu et al. 2004). Recent evidence indicates that the Arabidopsis tonoplast Na+/H+ exchanger is also a target for the SOS pathway. However, in this tonoplast SOS pathway, the Na+/H+ exchanger activity is regulated by SOS2 while not by SOS3. An unknown component regulates SOS2 (Qiu et al. 2003). AtNHX7/SOS1 is the first identified plasma membrane target for the Arabidopsis SOS pathway. SOS1 enables Na+ to efflux across the plasma membrane and controls long-distance Na+ transport from root to shoot (Shi et al. 2002a). Loss-of-function mutation in SOS1 renders the plants hypersensitive both to NaCl and LiCl stresses, although the transport experiment showed that SOS1 was specific for Na+ and could not transport Li+ or K+ (Zhu 2003 and the references therein). AtNHX8 is a highly similar homolog of AtNHX7/SOS1 and it is predicted to be a plasma membrane Na+/H+ antiporter with 756 amino acid residues, which exhibits ∼72% identity with the SOS1 sequence over a stretch of 760 amino acids (Maser et al. 2001; Ward 2001; Brett et al. 2005). Recent functional characterization demonstrated that AtNHX8 specifically functions in lithium but not sodium extrusion and in ion homeostasis in Arabidopsis (An et al. 2007). To the best of our knowledge, this is the only specific Li+/H+ antiporter identified so far. In the present study, overexpression of ZmCBL4 strongly increased the LiCl tolerance both in the Arabidopsis sos3 mutant (Fig. 4G, I, K) and the wild type backgrounds (Fig. 5C, E, G). In line with this phenotype, Northern blot analysis demonstrated that the AtNHX8 transcript obviously accumulated in all of the transgenic lines under LiCl stress, but was not detectable in the control sos3 mutant and wild type seedlings in the same condition (Fig. 6B, C). Although the AtNHX8 gene expression does not necessarily mean its corresponding product to be responsible for the enhanced LiCl tolerance, it is clear that the AtNHX8 expression is induced by ZmCBL4 at the transcription level in a LiCl stress-induced manner. Moreover, from the seed germination and seedling growth (Fig. 4F–K) and ion contents (Fig. 7B, C), it is not difficult to find that the difference in NaCl tolerance is more obvious than that in LiCl tolerance between the Arabidopsis sos3 mutant and wild type under our experimental conditions. These phenomena probably indicate that the SOS pathway or the SOS1 Na+/H+ exchanger regulated by SOS3 may function mainly in extruding Na+ but not Li+ in Arabidopsis. Our findings, together with other evidence mentioned above, make it conceivable that the AtNHX8 Li+/H+ antiporter may play a major role in removing Li+ out of the cell in the ZmCBL4-expressing Arabidopsis lines. Perhaps in these transgenic lines, the maize CBL4 protein cannot only induce the AtNHX8 gene expression, but also may activate the corresponding AtNHX8 transporting activity through interacting with and then activating some Arabidopsis CIPKs including SOS2 in a LiCl stress-dependent manner. Of course, this suggestion will need to be proved with more experimental data.
Taken together, our results demonstrate that ZmCBL4 is a functional homolog of AtSOS3, and that its function for improving the tolerance of transgenic plants to NaCl and LiCl is related to the positive regulation of AtSOS1 and AtNHX8 expression. The facts that overexpression of ZmCBL4 enhances NaCl and LiCl tolerance in wild type Arabidopsis, which is important for the improvement of crop salt tolerance by genetic manipulation, and that AtNHX8 is involved in the response of the ZmCBL4-expressing lines to LiCl stress represent the novel properties of ZmCBL4 because the similar functions have not been reported for either AtSOS3 or OsCBL4 (Guo et al. 2004; Martínez-Atienza et al. 2007).
Whether ZmCBL4 functions on salt tolerance and/or other physiological aspects in maize itself awaits further experimentation. At present, the transformation of ZmCBL4 into maize is on the way in our laboratory.
Abbreviations
- ABA:
-
Abscisic acid
- PEG:
-
Polyethylene glycol
References
Albrecht V, Ritz O, Linder S, Harter K, Kudla J (2001) The NAF domain defines a novel protein–protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20:1051–1063
Albrecht V, Weinl S, Blazevic D, D’Angelo C, Batistic O, Kolukisaoglu U, Bock R, Schulz B, Harter K, Kudla J (2003) The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J 36:457–470
An R, Chen QJ, Chai MF, Lu PL, Su Z, Qin ZX, Chen J, Wang XC (2007) AtNHX8, a member of the monovalent cation:proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li+/H+ antiporter. Plant J 49:718–728
Apse MP, Sottosanto JB, Blumwald E (2003) Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J 36:229–239
Brett CL, Donowitz M, Rao R (2005) Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol 288:C223–C239
Cheong YH, Kim KN, Pandey GK, Gupta R, Grant JJ, Luan S (2003) CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15:1833–1845
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Guo Y, Halfter U, Ishitani M, Zhu JK (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13:1383–1399
Guo Y, Qiu QS, Quintero FJ, Pardo JM, Ohta M, Zhang C, Schumaker KS, Zhu JK (2004) Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16:435–449
Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97:3735–3740
Hwang YH, Bethke PC, Cheong YH, Chang HS, Zhu T, Jones RL (2005) A gibberellin-regulated calcineurin B in rice localizes to the tonoplast and is implicated in vacuole function. Plant Physiol 138:1347–1358
Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12:1667–1677
Knight H, Trewavas AJ, Knight MR (1997) Calcium signaling in Arabidopsis thaliana responding to drought and salinity. Plant J 12:1067–1078
Knight H (2000) Calcium signaling during abiotic stress in plants. Int Rev Cytol 195:269–324
Kolukisaoglu U, Wein S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134:43–58
Kudla J, Xu Q, Harter K, Gruissem W, Luan S (1999) Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci USA 96:4718–4723
Liu J, Zhu JK (1997) An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA 94:14960–14964
Liu J, Zhu JK (1998) A calcium sensor homolog required for plant salt tolerance. Science 280:1943–1945
Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97:3730–3734
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lynch J, Polito VW, Läuchli A (1989) Salinity stress increases cytoplasmic Ca2+ activity in maize root protoplasts. Plant Physiol 90:1271–1274
Mahajan S, Sopory SK, Tuteja N (2006) Cloning and characterization of CBL-CIPK signaling components from a legume (Pisum sativum). FEBS J 273:907–925
Martínez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143:1001–1012
Maser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, Harper JF, Tchieu J, Gribskov M, Persans MW, Salt DE, Kim SA, Guerinot ML (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126:1646–1667
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15:473–497
Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D’Angelo C, Weinl S, Kudla J, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16:1912–1924
Pauly N, Knight MR, Thuleau P, van Der Luit AH, Moreau M, Trewavas AJ, Ranjeva R, Mazars C (2000) Control of free calcium in plant cell nuclei. Nature 405:754–755
Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99:8436–8441
Qiu QS, Barkla BJ, Vera-Estrella R, Zhu JK, Schumaker KS (2003) Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol 132:1041–1052
Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J Biol Chem 279:207–215
Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99:9061–9066
Reddy VS, Reddy ASN (2004) Proteomics of calcium-signaling components in plants. Phytochemistry 65:1745–1776
Sánchez-Barrena KJ, Martínez-Ripoll M, Zhu JK, Albert A (2005) The structure of the Arabidopsis thaliana SOS3:molecular mechanism of sensing calcium for salt stress response. J Mol Biol 345:1253–1264
Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11:2393–2405
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+antiporter. Proc Natl Acad Sci USA 97:6896–6901
Shi H, Quintero FJ, Pardo JM, Zhu JK (2002a) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14:465–477
Shi H, Xiong L, Stevenson B, Lu T, Zhu JK (2002b) The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 14:575–588
Shi H, Zhu JK (2002c) SOS4, a pyridoxal kinase gene, is required for root hair development in Arabidopsis. Plant Physiol 129:585–593
Shi H, Kim YS, Guo Y, Stevenson B, Zhu JK (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15:19–32
Ward JM (2001) Identification of novel families of membrane proteins from the model plant Arabidopsis thaliana. Bioinformatics 17:560–563
Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisitíon. Plant Cell 8:617–627
Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14:S165–S183
Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125:1347–1360
Zheng J, Zhao JF, Tao YZ, Wang JH, Liu YJ, Fu JJ, Jin Y, Gao P, Zhang JP, Bai YF, Wang GY (2004) Isolation and analysis of water stress induced genes in maize seedlings by subtractive PCR and cDNA macroarray. Plant Mol Biol 55:807–823
Zhu JK, Liu J, Xiong L (1998) Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell 10:1181–1191
Zhu JK (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124:941–948
Zhu JK (2001a) Plant salt tolerance. Trents Plant Sci 6:66–71
Zhu JK (2001b) Cell signaling under salt, water and cold stresses. Curr Opin Plant Biol 4:401–406
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Acknowledgements
We are grateful to Dr. Rui An (China Agricultural University) for kindly providing the cloning vector and primer pairs for Arabidopsis AtNHX8 cDNA. We also thank Dr. Zhizhong Chen (China Agricultural University) for kindly providing Arabidopsis sos3 mutants. This work was supported by the National High-tech Program of China (2006AA10A106 and 2006AA10Z103).
Author information
Authors and Affiliations
Corresponding author
Additional information
Database accession number: EF405963.
Rights and permissions
About this article
Cite this article
Wang, M., Gu, D., Liu, T. et al. Overexpression of a putative maize calcineurin B-like protein in Arabidopsis confers salt tolerance. Plant Mol Biol 65, 733–746 (2007). https://doi.org/10.1007/s11103-007-9238-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9238-8