Abstract
To study the functions of nuclear genes involved in chloroplast development, we systematically analyzed albino and pale green Arabidopsis thaliana mutants by use of the Activator/Dissociation (Ac/Ds) transposon tagging system. In this study, we focused on one of these albino mutants, designated apg3-1 (for a lbino or p ale g reen mutant 3). A gene encoding a ribosome release factor 1 (RF1) homologue was disrupted by the insertion of a Ds transposon into the APG3 gene; a T-DNA insertion into the same gene caused a similar phenotype (apg3-2). The APG3 gene (At3g62910) has 15 exons and encodes a protein (422-aa) with a transit peptide that functions in targeting the protein to chloroplasts. The amino acid sequence of APG3 showed 40.6% homology with an RF1 of Escherichia coli, and complementation analysis using the E. coli rf1 mutant revealed that APG3 functions as an RF1 in E. coli, although complementation was not successful in the RF2-deficient (rf2) mutants of E. coli. These results indicate that the APG3 protein is an orthologue of E. coli RF1, and is essential for chloroplast translation machinery; it was accordingly named AtcpRF1. Since the chloroplasts of apg3-1 plants contained few internal thylakoid membranes, and chloroplast proteins related to photosynthesis were not detected by immunoblot analysis, AtcpRF1 is thought to be essential for chloroplast development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The translational system in plant chloroplasts is similar to that in prokaryotes (Sugiura 1992; Zerges 2000). Chloroplast mRNAs are not m7G-capped and can be polycistronic (Sugita and Sugiura 1996; Rochaix 1996). Several chloroplast protein synthesis factors have been characterized, e.g., the elongation factor G (EF-G) homologue (Hernandez-Torres et al. 1993; Akkaya et al. 1994; Albrecht et al. 2006), ribosome recycling factor (Rolland et al. 1999), initiation factor 1 (IF1) (Hirose et al. 1999), and initiation factor 2 (IF2) (Campos et al. 2001). Among the translation factors identified so far, only initiation factors from Chlorella and some plants have been shown to be encoded in the chloroplast genome (Hirose et al. 1999), while IF1 genes from soybean and Arabidopsis are found in their nuclear genomes and have a transit peptide that targets chloroplasts (Millen et al. 2001).
Protein synthesis on ribosomes stops in response to a sense stop codon in the ‘decoding’ site (A site) and, in both prokaryotes and eukaryotes, ribosome release factors are required for termination of translation (Scolnick et al. 1968; Nakamura et al. 1996). Two structurally similar class 1 release factors, RF1 and RF2, have been identified in bacteria (Klein and Capecchi 1971; Lee et al. 1988), and both have been shown to catalyze the release of the nascent polypeptide chain in a codon-specific manner by recognizing UAA/UAG and UAA/UGA stop signals, respectively. Mutations in these genes often cause misreading of stop signals, increased frameshifting, and temperature-sensitive growth of cells (Nakamura et al. 1995). The class 2 release factor RF3, a guanosine triphosphate-binding protein, is known to accelerate the dissociation of RF1 and RF2 after the release of polypeptides from the ribosomal A-site. Defects in RF3 cause misreading of three stop codons, whereas excessive RF3 stimulates the formation of ribosomal termination complexes and increases RF1 and RF2 activity (Matsumura et al. 1996). The Hemk protein, which has high amino acid similarity with DNA (adenine-N6)-methyltransferase (MTase), methylates RFs in E. coli, and the hemk mutant shows an enhanced rate of read-through of stop codons and induction of transfer-mRNA-mediated tagging of proteins within the cell. Thus, the hemk mutant induces defects in translational termination (Nakahigashi et al. 2002). Recently, the prmC gene was shown to encode an N5-glutamine S-adenosyl-L-methionine-dependent methyltransferase of RFs in the obligate intercellular pathogen Chlamydia trachomatis (Pannekoek et al. 2005).
Compared with bacterial ribosome release factors, the biological functions of chloroplast homologues of RF have not been completely elucidated. Recently, in Arabidopsis thaliana, 34 recessive photosynthetic mutants of the high-chlorophyll-fluorescence phenotype (hcf) were isolated by screening of 7700 M2 progenies of ethyl methane sulfonate-treated seeds (Meurer et al. 1996b). One of these mutants, hcf109, was shown to be seedling-lethal, with a pale green phenotype and defects in the stability of UGA-containing transcripts in Arabidopsis chloroplasts (Meurer et al. 1996b, 2002). The hcf109 mutation was later identified as a peptide chain release factor 2 (AtprfB), and shown to terminate transcripts with UGA/UAA stop codons and regulate both mRNA stability and protein synthesis (Meurer et al. 2002). On the other hand, chloroplast RF1 has not yet been identified in Arabidopsis, in which the detailed functions of the plastid translation system have yet to be revealed.
We previously used the Activator/Dissociation (Ac/Ds) two-component transposon system (Ds2 389-13 and Ds4 391-20; Smith et al. 1996) in Arabidopsis to prepare a large collection of tagged lines carrying a single Ds insertion (Ito et al. 1999, 2002, 2005; Seki et al. 1999; Kuromori et al. 2004). To analyze the functions of nuclear genes involved in chloroplast development, we screened albino and pale green Arabidopsis mutants from Ds-tagged lines. Among 2,739 Ds-tagged lines, we identified 11 lines with albino or pale green phenotypes closely linked to insertion sites of the Ds element, and termed them apg (for a lbino or p ale g reen mutant). In this study, we focus on one of these albino mutants, named apg3-1, which has a disrupted gene for an RF1 orthologue. Physiological analysis of the mutant revealed that APG3 has an important function in chloroplast development and plays essential roles in the termination of translation in plastids.
Results
Isolation of an apg3-1 mutant
One of the apg mutants, apg3-1, isolated from our Ds-tagged lines, showed an albino phenotype on agar medium containing 1% sucrose (Fig. 1A). In addition, when germinated on soil the mutant was seedling-lethal (data not shown). Two hundred and sixty-one progenies obtained from self-pollination of a green plant of the Ds-inserted line apg3-1 were segregated at a green to albino ratio of 199:62 [χ2 (3:1) = 0.10, p > 0.05], which is not significantly different from that expected for a recessive Mendelian trait. On an agar plate without sucrose, the apg3-1 plants stopped growing at the germination stage, showing a white cotyledon phenotype (Fig. 1B). The apg3-1 plants grown on agar plates supplemented with sucrose grew better than apg3-1 grown on the plates without sucrose, suggesting that the albino apg3-1 cannot grow photoautotrophically.
Albino phenotype of the Ds-tagged mutant, apg3-1, and T-DNA-tagged mutant, apg3-2. (A) Albino phenotype of the Ds-tagged mutant, apg3-1. apg3-1 mutants (black arrows) germinated on agar medium showed an albino phenotype, whereas the heterozygous plants (red arrow) did not show any unusual phenotypes. The apg3-1 plants were much smaller than heterozygous plants. (B) Growth of apg3-1 mutant plants cultured on various concentrations of sucrose for 3 weeks. (C) Growth of apg3-1 mutant plants cultured under various light conditions for 3 weeks. (D) Revertant green sectors in an apg3-1 mutant plant carrying the Ac transposase gene. Albino leaves with green sectors are indicated by arrows. The reversion was due to the excision of Ds from the APG3-1 gene locus. (E) Albino phenotype of the T-DNA-tagged mutant, apg3-2. The apg3-2 mutants (black arrows) showed an albino phenotype like apg3-1
To analyze PSII activity, we measured the minimum (Fo) and maximum (Fm) chlorophyll-a fluorescence of dark-adapted leaves of apg3-1 plants, and calculated the maximum PSII activity and the steady-state PSII yield during photosynthesis, Fv/Fm [=(Fm Fo)/Fm] (Krause et al. 1988). The Fv/Fm value in 3-week-old leaves of Ds donor line plants was 0.72 ± 0.03; in contrast, the value was 0.08 ± 0.10 in apg3-1 plants grown on agar plates with 2% sucrose. This result indicates that apg3-1 plants do not have PSII activity.
We also examined various pigments in apg3-1 and control plants using high-performance liquid chromatography. Because the white color of the cotyledons differs from that of true leaves in apg3-1 plants, the pigments in the two organs were analyzed separately. The total amount of pigment in the cotyledons was less than 20% of that in true leaves. The amounts of chlorophyll-a, chlorophyll-b, cis-neoxanthin, trans-violoxanthin, lutein, and β-carotene in apg3-1 were 19, 26, 36, 58, 40, and 10% of those in wild-type plants, respectively. The decrease in these plastidial pigments suggests an alteration in the photosensitivity of apg3-1 plants. Therefore, we cultured apg3-1 plants under various light conditions for 3 weeks on agar plates with sucrose (Fig. 1C). However, the growth of apg3-1 plants under various light conditions were essentially similar, although the pale-green true leaves grown under dim light (7 μmol quanta m−2 s−1) showed slightly better development than those under stronger light, indicating that the growth phenotypes of mutant seedlings are simply based on the fact that wild-type plants perform photosynthesis while mutant plants do not.
apg3-1 leaves contain abnormal plastids under light
Since apg3-1 plants showed an albino phenotype, we analyzed morphological changes in plastids in their leaves by electron microscopy (Fig. 2). The size and membrane structure of etioplasts in apg3-1 plants were similar to those in wild-type plants kept in dark conditions (Fig. 2A and B). In mature chloroplasts, the internal membrane was located in either single stroma or stacked grana thylakoids (Fig. 2C and E); however, abnormal plastids were observed in mature leaves of apg3-1 plants grown under light (Fig. 2D and F). The mutant plastids were more spherical than those in wild-type plants. Moreover, the mutant plastids contained internal membranes were considered to be abnormal thylakoids, which could not be distinguished into grana and stroma thylakoid, and many internal membranes that were unusual in size (Fig. 2D and F). The plastids observed in apg3-1 plants did not contain starch grains, and contained fewer densely stained globular structures than those in wild-type plants. These results suggest that accumulation of starch may be low in mature leaves of apg3-1 plants.
Electron micrographs of plastids of the apg3-1 mutant. Electron micrographs of chloroplasts from wild-type cells (A, C, E) and apg3-1 albino cells (B, D, F) of dark-grown plants (A, B) and light-grown plants (C, D). Close-up views of C and D (E, F). Lines are scale bars: 500 nm and 1 μm represent the size of the scale bars in each panel. ×10K, ×8,000 and ×15K represent the magnifications in each panel
Identification and molecular characterization of the APG3 gene
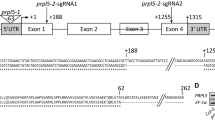
To identify the locus in which the Ds transposon was inserted, DNA fragments adjacent to the 5′ and 3′ ends of the inserted Ds were amplified from apg3-1 seedlings using the thermal asymmetric interlaced (TAIL) polymerase chain reaction (PCR) technique (Liu and Whittier 1995). Analysis of both flanking sequences revealed the same gene locus of At3g62910, which was annotated by the MIPS Arabidopsis thaliana database group (Schoof et al. 2004; http://mips.gsf.de/proj/plant/jsf/athal/index.jsp). The linkage between the albino phenotype of apg3-1 plants and the Ds insertion was confirmed by DNA gel blot analysis using HindIII-digested genomic DNA samples isolated from wild-type Columbia, wild-type Nossen, Ds2 392-13 donor lines, Ds-inserted green plants, and albino mutants (data not shown). The insertion duplicated an 8-bp target site of transposase as expected for a Ds insertion (Fig. 3A). In order to isolate revertants, we plated unstable apg3-1 mutant plants containing a fusion gene consisting of the cauliflower mosaic virus 35S RNA promoter (35S promoter) and a coding region for an Activator transposase protein (35S::Tpase). Accordingly, we identified green sectors in the albino leaves that correlated with restoration of the functional gene (Fig. 1D). Excision of Ds from the mutant allele occurred at various stages of plant development, and revertant sectors varying in size were observed (data not shown), as reported previously for other albino mutants (Motohashi et al. 2001, 2003). Moreover, we also isolated one mutant line (SALK 026458) possessing a T-DNA insertion within the second intron of APG3 from the SALK collection of T-DNA tag lines (http://signal.salk.edu/cgi-bin/tdnaexpress) (Fig. 3A). We named the T-DNA insertion mutant apg3-2. The apg3-2 plants showed an albino phenotype similar to apg3-1 plants (Fig. 1E). Taken together, these results indicate that disruption of the APG3 gene causes an albino phenotype.
The Ds insertion sites in the apg3 mutant. (A) The Ds and T-DNA insertion sites and sequences of flanking regions of Ds insertion sites in the apg3 mutants. Boxes show exons of APG3. The nucleotide sequences at the Ds insertion sites are shown for wild-type and apg3-1 plants. The underlined 8-bp sequence is the target site for duplication. (B) Alignment of the deduced amino acid sequences from Arabidopsis APG3 and RF1-like proteins of various species. The arrow is the putative cleavage site of the transit peptide in the APG3 amino acid sequence. The open triangle shows the Ds insertion site. The underline shows the tRNA-mimicry region. Conserved motifs, PXT and GGQ, are also indicated. Synec, RF1 in Synechocystis sp. (strain PCC 6803); Clostridium, Clostridium perfringens; E. coli, Escherichia coli; Arabidopsis, a putative RF1 (ACC34223) in A. thaliana; YeastMt, RF1 in the mitochondria of S. cerevisiae
Next, we isolated a full-length APG3 cDNA clone from the Arabidopsis cDNA library. The APG3 cDNA (DDBJ/EMBL/GenBank Accession No. AB109893) contains 15 exons and an open reading flame that encodes a protein of 422 amino acids (Fig. 3B). In apg3-1 plants, the Ds transposon was inserted into the third exon of At3g62910, within the codon corresponding to the valine residue at amino acid position 100 (Fig. 3A and B, open triangle). APG3 is a single-copy gene, but another homologue of RF1 (DDBJ/EMBL/GenBank Accession No. AAC34223; AGI code, At2g47020) was also found in the Arabidopsis genome. A homology search using the BLASTX program (http://www.ncbi.nlm.nih.gov/blast/) revealed high homology between the APG3 protein and RF1-like proteins from various species such as Synechocystis sp. (strain PCC 6803) (S76914), Clostridium perfringens (NP_563118), Escherichia coli (FCECR1) (Craigen et al. 1985) and mitochondria of Saccharomyces cerevisiae (S28602) (Pel et al. 1992), showing 58, 44, 41, and 30% identity, respectively. Alignments between APG3 and RF1 (E. coli) revealed that APG3 contains additional N-terminal residues, the sequence of which seems to be a transit peptide on the basis of typical characteristics. Position 44 (Fig. 3B, shown by an arrowhead) was predicted to be a transit peptide cleavage site in Arabidopsis using the CHLOROP program (http://www.cbs.dtu.dk/services/chloroP). In contrast, the other homologue of RF1 (At2g47020) does not have a typical transit peptide. The deduced amino acid sequence of this homologue (At2g47020) showed 29, 34, 32, 40, and 45% identity to APG3, and RF1s from Synechocystis sp. PCC6803, S. cerevisiae, C. perfingens, and E. coli, respectively. Thus, At2g47020 is also considered to be a ribosome release factor 1, although its subcellular localization has not yet been elucidated.
The predicted amino acid sequence of APG3 contains a putative tRNA-mimicry region (Fig. 3B, underlined). This region contains the PXT motif, which is responsible for recognition of the UAG stop-codon and determines RF1 specificity in vivo (Ito et al. 2000a). This region also contains the universal GGQ motif, which is conserved in all eubacterial, archabacterial, and eukaryotic release factors, and may mimic the CCA end of tRNA (Mora et al. 2003).
An APG3 rescue temperature-sensitive rf1 mutant of E. coli
On the basis of the significant structural similarity between E. coli RF1 protein (ecRF1) and APG3 (Fig. 3B), we conducted a complementation test of the ecRF1 mutant to determine whether APG3 could functionally substitute RF1 in E. coli. The E. coli mutant, RM695 (rf1ts) bacteria, shows a temperature-sensitive phenotype at 32°C owing to the defect in RF1 (Uno et al. 1996). When APG3 cDNA was overexpressed in RM695 (rf1ts) bacteria, cells could proliferate at the restrictive temperature, although the proliferation rate was slightly less than that of cells expressing the ecRF1 protein (Fig. 4, upper). In order to analyze the specificity of APG3 function, complementation of the temperature-sensitive mutant (RM718), which possesses a mutation in the ecRF2 gene, another type of class 1 release factor gene (Uno et al. 1996), with APG3, was carried out. Neither ecRF1 nor APG3 suppressed the phenotype of RM718 (rf2ts) bacteria at the restrictive temperature, whereas distinct complementation was observed with expression of ecRF2-T246A, in which a mutant of ecRF2 with increased release activity due to a T246A substitution was employed for the positive control of complementation (see ‘Materials and methods’) (Fig. 4, lower). These results indicated that APG3 is a functional orthologue of ecRF1. We, therefore, renamed the APG3 protein AtcpRF1 for A rabidopsis t haliana chloroplast RF1.
E. colirf1 mutant complemented with APG3 encoding Arabidopsis chloroplast RF1 (AtcpRF1). Upper panels: Complementation analysis of the temperature-sensitive rf1 mutant by the control vector pTWV (Vector), pTWV-ecRF1 carrying E. coli RF1 (ecRF1), pTWV-ecRF2 carrying E. coli RF2 (ecRF2 T246A) and pAPG3-lacI carrying APG3 (APG3). Lower panels: Complementation analysis of the temperature-sensitive rf2 mutant (RM718) using the same constructs
The role of the APG3 transit peptide in plastid targeting
The N-terminal 44 amino acid residues of APG3 were predicted to be a transit peptide using the CHLOROP program (Fig. 3B). To analyze this, we constructed two chimeric genes, p35S::APG3tp55-sGFP and p35S::APG3tp40-sGFP, to express the N-terminal 55 or 40 amino acid residues of APG3, respectively, as a fusion protein of sGFP (synthetic green-fluorescent protein) (Chiu et al. 1996). When the p35S::APG3tp55-sGFP chimeric gene was introduced into epidermal cells of Nicotiana tabacum SR1 leaves by particle bombardment (Takechi et al. 2000), GFP fluorescent signals were observed in the chloroplasts (Fig. 5). On the other hand, when a chimeric gene without this N-terminal region was used as a control, GFP signals were dispersed in the cytosol and nuclei (data not shown). The same results were obtained using the p35S::APG3tp40-sGFP construct (data not shown). These results indicate that the N-terminal region of APG3 is a functional transit peptide involved in chloroplast targeting.
Plastid targeting of the APG3tp55-sGFP chimeric protein in tobacco epidermal cells. Tobacco epidermal cells in which the p35S::APG3tp55-sGFP construct was introduced by particle bombardment. Fluorescence from GFP (A) and chlorophyll autofluorescence (B) was detected by laser confocal-scanning microscope. (C) Superimposed pictures of A and B
Expression of APG3 and a homologous gene in various tissues
To understand the effect of photo condition on the expression of RF1s, expression of APG3 and its homologous gene (At2g47020) in etiolated seedlings and various tissues were examined by RT–PCR. Expression of APG3 was barely detected in etiolated seedlings and roots (Fig. 6A). Another homologue of RF1, annotated as At2g47020, showed a similar expression pattern, but its transcript accumulated more than the transcript of APG3, suggesting that this homologue might function to compensate for AtcpRF1 in etiolated seedlings of apg3 plants. In addition, we examined the effect of light on APG3 expression (Fig. 6B). RNA gel blot analysis was carried out using total RNA isolated from leaves harvested at 0, 1, 3, 6, 12, or 24 h after the transfer of 7-day-old dark-grown wild-type plants (Columbia) to a light condition. APG3 was weakly expressed in the dark (0 h), but after transfer of the etiolated plants to the light condition, APG3 mRNA accumulated in a time-dependent manner. This result suggests that the expression of APG3 may be induced by photo signaling pathways. Alternatively, APG3 may be expressed only in differentiated chloroplasts. To confirm whether abnormal chloroplast differentiation could perturb APG3 expression, we analyzed the APG3 mRNA level in another apg mutant, apg2 (Motohashi et al. 2001). The plastids in apg2 plants have similar structures to undifferentiated wild-type proplastids, due to a Ds-insertion in APG2, a TatC homologue of the E. coli ΔpH-dependent protein transporter. As a result, APG3 mRNA was shown to accumulate in apg2 plants as well as in wild-type plants, in the light (data not shown), indicting that the APG3 expression is induced by photo-irradiation rather than chloroplast differentiation.
RNA expression analyses of APG3. (A) Expression of APG3 genes and a homologous gene of APG3 in various tissues. Total RNA was isolated from various tissues (1: etiolated seedlings, 2: seedlings, 3: 21-day-old plants, 4: roots, 5: stems, 6: siliques) and used in RT–PCR experiments with specific primer sets (see ‘Materials and methods’). (B) Expression of the APG3 gene after exposure to light. Total RNA was isolated from 7-day-old dark-grown plants exposed to light for 0, 1, 3, 6, 12, or 24 h. Total RNA was also isolated from 21-day-old Columbia ecotype grown under a cycle of 16-h light/8-h dark at 23°C (lane 7). Each lane was loaded with 20 μg of total RNA. (C) Gene expression of nuclear- and chloroplast-encoded genes in the apg3-1 mutant. Total RNA was prepared from 21-day-old wild-type plants (Nossen), Ds donor plants and apg3-1 mutant plants. The probes used were APG3, APG1, cab, rbcL, 16SrRNA, psaN, rbcS, psbP, and psbA
Expression of nuclear- and chloroplast-encoded genes in the apg3-1 mutant
APG3 expression in apg3-1 plants was examined by RNA gel blot analysis. We detected APG3 mRNA in wild-type and Ds-donor plants, but not in apg3-1 mutants (Fig. 6C). To elucidate the influence of the APG3 deficiency on the expression of photosynthetic genes, we analyzed five nuclear-encoded genes and three chloroplast-encoded genes, namely nuclear genes encoding the small subunit of ribulose-1,5-bisphosphate carboxylase (rbcS; Krebbers et al. 1988), the light-harvesting chlorophyll a/b-binding protein (cab; Leutwiler et al. 1986), the 23-kDa polypeptide of the oxygen-evolving complex of photosystem II (psbP; Kochhar et al. 1996), the N subunit of photosystem I (psaN; Sehnke and Feri 1995) and the 37-kDa protein of the chloroplast inner envelope membrane (APG1; Motohashi et al. 2003), and chloroplast genes encoding the D1 protein of photosystem II (psbA; Liere et al. 1995), the large subunit of ribulose-1,5-bisphosphate carboxylase (rbcL; Zhu et al. 1997), and 16S rRNA (Sato et al. 1999). In apg3-1 plants, the level of rbcL transcript was remarkably decreased, while cab, psbP, psaN, and rbcS transcripts were only slightly decreased. In contrast, APG1, psbA, and 16S rRNA transcripts were detected at similar levels in both wild-type and apg3-1 plants (Fig. 6C). These results implicate APG3 in rbcL expression, although the APG3 defect did not significantly affect expression of the other photosynthetic genes tested.
Chloroplast microarray analysis of apg3
To obtain comprehensive expression profiles of plastid-encoded genes in apg3-1 plants, we generated a DNA microarray equipped with PCR probes for all the plastid genes (Nagashima et al. 2004). Four (a–d) sets of plastid DNA fragments encoding plastid proteins were spotted onto one slide glass, then hybridized with Cy3- and Cy5-labeled cDNA generated from apg3-1 mutant and wild-type RNA, respectively. Moreover, plastid genome microarray analysis of the apg3-2 mutant was also performed. We repeated this experiment four (apg3-1) or five (apg3-2) times using independent RNA preparations, and calculated mean values of the log2(apg3/WT) after global normalization. The plastid gene expression profiles of apg3-1 (Table S1) and apg3-2 (Table S2) plants were essentially similar based on a correlation coefficient of 0.87. It is well known that many plastid-encoded genes are polycistronic, and the levels of each transcript in the same transcript units, such as psaA-psaB-rps14 and psbH-petB-petD-psbB-psbT operons, were quite similar (Tables S1 and S2). These results support the reliability of the microarray data.
In addition to rbcL, a decrease in the expression of genes encoding proteins for photosystems II and I, such as psb and psa, in both apg3-1 and apg3-2 mutants, was also revealed by the microarray (Tables 1, S1, and S2). Furthermore, increased accumulation of plastid gene transcripts was also observed in apg3 mutants, although no genes were upregulated by more than twofold in apg3 mutants compared with wild-type plants. Many of the increased genes, such as rpo, rpl, and rps, are considered to be under the control of NEP (nuclear-encoded plastid RNA polymerase) (Tables S1 and S2). The defects in the UAG/UAA-specific release factor APG3 in mutant plastids gave rise to the hypothesis that the stability of plastid-encoded genes with the UAG/UAA stop codon is decreased in apg3 mutants. However, we could not find a clear correlation between changes in the transcript level of plastid-encoded genes and the type of stop codon (Tables 1, S1, and S2).
Immunoblot analysis of chloroplast proteins in the apg3-1 mutant
The apg3-1 mutant had quite low levels of PSII activity during photosynthesis, whereas decreases in the expression levels of genes participating in PSII were not significant. Therefore, we examined the expression levels of chloroplast proteins involved in photosynthesis, in the apg3-1 mutant, by immunoblot analysis (Fig. 7). The proteins analyzed were soluble Rubisco proteins, LSU and SSU (large and small subunits of ribulose-1,5-bisphosphate carboxylase; encoded by rbcL and rbcS), thylakoid proteins, D1 (encoded by psbA) and LHCII (encoded by cab), and OE23 (encoded by psbP). Among these proteins, Rubisco LSU and D1 are encoded by plastid genes. We could not detect accumulation of Rubisco proteins, D1, or OE23, but the LHCII proteins accumulated slightly in apg3-1 mutants (Fig. 7, lane 3).
Immunoblot analysis of chloroplast proteins in the apg3-1 mutant. Total protein was prepared from 21-day-old wild-type plants (Nossen) (lane 1), a Ds donor line (lane 2) and apg3-1 mutants (lane 3), and separated by SDS–PAGE. For the detection of D1, LHCII, and OE23 proteins, 40 μg of total protein was loaded per lane. For the detection of Rubisco protein, 10 μg of protein was loaded per lane. LSU and SSU refer to the large and small subunits of Rubisco, respectively
Increased polysome association of UGA-containing transcripts in the apg3-1 mutant
The fact that accumulation of rbcL mRNA in apg3-1 mutants was reduced to ∼30% of that in WT plants (Fig. 6C), while the protein level of Rubisco LSU was reduced by less than 5% (Fig. 7), suggested a deficiency of translation in apg3-1 mutants. Therefore, we analyzed chloroplast polysomes to determine whether or not the defects caused by apg3 mutation occur after the initiation of translation. Polysomes were extracted from leaves and fractionated by sucrose gradient centrifugation, prior to being used for the detection, using radio-labeled fragments, of the plastid-encoded genes, rbcL, atpE, psbA, and psbN, and cytosolic rDNA as a control. The transcripts of rbcL and psbN contain a UAG stop codon, which is considered to be recognized by RF1. On the other hand, the transcripts of atpE contain a UGA stop codon, which is considered to be recognized by RF2. In the case of psbA, the transcripts contain a UAA stop codon, which could be recognized by both RF1 and RF2.
The distribution of cytosolic rRNA in the sucrose gradient fraction was shown to be similar between apg3-1 mutants and wild-type plants (Fig. 8). Rapid sedimentation of mRNA in a sucrose gradient, corresponding to a large number of fractions, could be due to the association of mRNA with ribosomes to form polysomes or some other large ribonucleoproteins. The distributions of rbcL and psbN, which contain a UAG stop codon, were shifted toward the bottom fractions of the gradient in extracts from apg3-1 mutants compared with those from wild-type plants, while the distribution of atpE, which contains a UGA stop codon, was not (Figs. 8 and S1). Moreover, the distribution of psbA signals was slightly shifted toward the bottom fractions in extracts from apg3-1 mutants compared with those from wild-type plants. These polysome analyses strongly demonstrated an increased association of RF1-recognized mRNAs with ribosomes in apg3-1 mutants, providing evidence for the function of APG3 in polypeptide release from aminoacyl-t-RNA after translation in plastids.
Polysome analysis of plastid genes in apg3-1 mutants and wild-type leaves. Intact polysomes fractionated by ultracentrifugation were denatured and subjected to RNA gel blot analysis. Fractions 1 and 15 correspond to the top and bottom of the gradient, respectively. The probes used are indicated on the right-hand side. Staining of the membranes before hybridization resulted in equal distribution of the total RNA between WT and apg3-1 mutant plants. Autoradiographic exposure of apg3-1 mutant blots hybridized with the psbN probe was performed for longer than that of wild-type blots hybridized with the same probe, in order to detect the low level of psbN in apg3-1 mutants. These autoradiographs are typical results of the polysome analysis, which was reproduced several times
Discussion
APG3 encodes a ribosome release factor 1 that functions in plastids
We previously isolated 11 albino or pale-green (apg) mutants by screening 2,739 Ds insertion mutants. In this study, we analyzed one of these apg mutants, apg3-1 (Fig. 1), revealing that the mutant is tagged with Ds (Fig. 1D) at the locus annotated as At3g62910. One T-DNA-tagged mutant allele (apg3-2) showed a similar albino phenotype (Fig. 1E), also indicating that a deficiency in APG3 causes an albino phenotype in Arabidopsis (Figs. 1A and 6C).
A database search and multiple alignment of deduced amino acid sequences of APG3 revealed that APG3 belongs to the family of bacterial ribosome release factor 1, which functions in the release of nascent polypeptide chains by recognizing UAA/UAG stop codons during translation termination. The deduced amino acid sequence of APG3 (At3g62910) showed 41% amino acid identity with E. coli RF1, ecRF1 (Craigen et al. 1990). Multiple alignment of APG3 with RF1 homologues from various organisms showed that the APG3 protein had a predicted conserved tRNA-mimicry domain containing the PXT and GGQ motifs. The first and third amino acids in the highly conserved tripeptide PXT motif recognize the adenine and guanine of the UAG stop codon (Ito et al. 2000a), and the GGQ motif is thought to mimic the CCA end of tRNA (Mora et al. 2003). The amino acid sequence of APG3 showed high homology with that of RF1 from cyanobacteria, and the N-terminal region of APG3 was shown to function as a transit peptide involved in chloroplast targeting (Fig. 5). These results indicate that APG3 encodes an Arabidopsis RF1 orthologue, herein named AtcpRF1, with the function of a translation-releasing factor in chloroplasts. AtcpRF1 also showed homology with another type of class 1 RF protein, RF2, in Arabidopsis (AtprfB; Meurer et al. 2002), showing 29% identity, although the homology was especially high around the tRNA-mimicry domain. RF2 has the tripeptide SPF at the corresponding position of the PXT motif in RF1, which functions in the RF2-specific recognition of the UGA stop codon (Ito et al. 2000a). E. coli RF1 and RF2 share 37% identical amino acids (Craigen et al. 1990), which is higher than the identity between AtcpRF1 and AtprfB. A database search also revealed another homologue of RF1, At2g47020, in the Arabidopsis genome. Although the overall identity of the primary structures for AtcpRF1 and the homologue was not high (29%), At2g47020 has the PXT motif as well as the GGQ motif in a putative highly conserved tRNA-mimicry domain (Fig. 3). Thus, the homologue (At2g47020) was predicted to encode another homologue of RF1 in Arabidopsis.
The molecular function of AtcpRF1 in vivo as a ribosome release factor 1 was demonstrated by complementation analysis of the temperature-sensitive (ts) phenotype of E. coli rf1 mutants. AtcpRF1 suppressed the ts phenotype of rf1 mutants, whereas the phenotype of the mutant rf2 was not suppressed by AtcpRF1 expression (Fig. 5). These results indicate that AtcpRF1 could function as RF1 in E. coli, showing specificity of stop codon recognition at a similar extent to ecRF1. Functional complementation by a heterologous RF1 in vivo had not been reported until this study. The chloroplasts of plants and algae are known to have ribosomes whose component RNAs and proteins are strikingly similar to those of eubacteria, consistent with their postulated origin from endosymbiotic cyanobacteria, and the results of this study suggest that plastidial RF1 possesses a common mechanism of interaction with mitochondria and termination of protein synthesis to that in eubacterial RF1.
Taken together with the evidence for the plastid targeting of AtcpRF1 (Fig. 5), we conclude that AtcpRF1 encodes a ribosome release factor 1 that functions in plastids; this is the first report to identify a plastidal RF1 in higher plants.
The AtcpRF1 gene is essential for chloroplast development
We also precisely characterized the phenotype of apg3-1 mutants to analyze the roles of AtcpRF1 in chloroplast development. Although the apg3-1 mutant could grow on medium supplemented with sucrose (Fig. 1B), it showed severe phenotypes: it was seedling-lethal, had small white cotyledons, very pale green true leaves, and a complete loss of PSII activity. A deficiency in RF2 in Arabidopsis, AtprfB, has also been reported to cause severe phenotypes, including seedling-lethality, pale green leaves, and quite low PS II activity (∼10–15% of the value for the wild-type level) (Meurer et al. 1996a, 2002). This mutant, hcf109, has been reported to be able to initiate rudimentary inflorescences, although no fertile flowers develop; apg3-1 mutants cannot even initiate inflorescences (Fig. 1). These differences in phenotypes and the levels of PS II activity between apg3-1 and hcf109 mutants indicate that either of these plastidial RFs may have indispensable roles in the chloroplast development. However, deficiencies in proteins regulated by AtcpRF1 cause more severe defects in chloroplast development than deficiencies in proteins regulated by AtprfB.
The plastids in apg3-1 mutants kept under light contained some internal membranes with few granal structures (Fig. 2), also indicating the important roles of the AtcpRF1 protein in normal development of thylakoid membrane structures during chloroplast biogenesis. The fact that etioplasts in apg3-1 mutants were observed to have structures similar to those in wild-type plants (Fig. 2A and B) strongly supports the importance of AtcpRF1, especially in chloroplast development. Transcripts of AtcpRF1, induced by photo irradiation, were barely detected in etioplasts (Fig. 6A and B), but were abundant in chloroplast-containing photosynthetic organs. These expression profiles of AtcpRF1 are similar to those of AtprfB (Meurer et al. 2002). These results indicate that AtcpRF1, as well as AtprfB, has an important function in the light-regulated activation of translation in chloroplasts.
In chloroplasts, light controls the translation of photosynthesis-related genes at various levels (Gillham et al. 1994; Bruick and Mayfield 1999). Light-regulated translation in chloroplasts has been extensively studied using psbA mRNA encoding the D1 protein of photosystem II, and rbcL mRNA encoding the large subunit of ribulose-bisphosphate carboxylase in higher plants (Bruick and Mayfield 1999; Kim and Mullet 2003). The mRNAs for photosynthesis-related proteins are known to accumulate to quite high levels in plastids, even when the plants are in the dark, while the protein products of these mRNAs are barely detectable. Moreover, by photo-activation, translation of certain chloroplast mRNAs is reported to increase as much as 100-fold, sometimes without significant changes in the amount of mRNA present. Photo-induction of AtcpRF1 (APG3) and AtcpRF2 (AtprfB) at the transcriptional levels is considered to correlate with photo-activated translation during chloroplast development.
AtcpRF1 functions in the regulation of chloroplast translation
The large subunit of ribulose-bisphosphate carboxylase (Rubisco L; rbcL gene product) and the D1 thylakoid protein (psbA gene product) are chloroplast-encoded proteins translated by the plastidial translational system. In apg3-1 mutants, the level of psbA transcripts was shown to be unaffected (Fig. 6C), whereas no D1 protein was detected (Fig. 7). In the case of rbcL, no Rubisco L protein was detected, although rbcL transcript was decreased, but still detected, in apg3-1 mutants. These results indicate that the chloroplast-encoded psbA and rbcL products (D1 and LSU) are not translated from their mRNAs in apg3-1 mutants, which raised the hypothesis that the impaired AtcpRF1 function might disrupt the translation of mRNAs containing UAA/UAG stop codons in apg3-1 mutants.
We suspected that this would lead to an increase in polysome association by UAG-containing transcripts in apg3-1 mutants, which would presumably inhibit the elongation and termination of translation rather than initiation activities. Chloroplast polysome analysis for plastid-coded genes revealed that an increased association between ribosomes and the RF1-recognized mRNAs, rbcL and psbN, in apg3-1 mutants. On the other hand, the RF2-recognized mRNA atpE did not show an increased interaction with ribosomes (Figs. 8 and S1). Moreover, in the case of psbA mRNA with an UAA stop codon, which could be recognized by both RF1 and RF2, the change in polysome association was only slight. In contrast, polysome analysis of hcf109 mutants showed an increased interaction of atpE transcripts with ribosomes, but not of rbcL transcripts (Meurer et al. 2002). These results indicate strict recognition of stop codons by AtcpRF1 and AtprfB in the plastid translation mechanism. In addition, impaired release of ribosomes from mRNAs in the RF mutants was shown to cause defects in translation efficiency in plastids.
RNA gel blot analysis revealed that the transcripts of nuclear-encoded genes (cab, psbP, psaN, and rbcS) were slightly decreased in apg3-1 mutants, whereas their protein levels were markedly decreased (Fig. 7). The loss of nuclear-encoded proteins is an expected outcome of global defects in chloroplast-encoded protein expression because, generally, the stability of the core subunits of each photosynthetic complex decreases markedly when the synthesis of one of core subunits is blocked (Schmidt and Mishkind 1983; Spreltzer et al. 1985). Thus, the absence of large subunits of Rubisco in apg3-1 mutants may have caused the degradation of the small subunits through a loss of the subunits constructing the holoenzyme. Similarly, apg1 mutants (Motohashi et al. 2003), apg2 mutants (Motohashi et al. 2001), and the Ds-tagged mutant of PAC-2 (Grevelding et al. 1996) 54-2038-2 (unpublished) were shown to have abnormal plastids with significantly decreased levels of membrane proteins and slightly decreased levels of soluble proteins (data not shown). The morphological abnormalities of the apg3-1 plastids may correlate with repressed accumulation of membrane proteins.
Translation of chloroplast mRNAs containing the UGA stop codon is impaired, but not inhibited completely, in hcf109 mutants (Meurer et al. 2002). The same might be true for the translation of chloroplast mRNAs containing the UAG stop codon in apg3 mutants. Because the synthesis of photosynthetic proteins is not active under dark conditions, apg3 mutation may have no apparent effect on etioplast development (Fig. 2). Alternatively, it remains a possibility that the gene product of At2g47020, which showed similar tissue-specific expression to AtcpRF1 (Fig. 6A), may function as another homologue of RF1 in plastids, although the molecular function and subcellular localization of At2g47020 remain to be elucidated.
The apg3-1 mutation affects the accumulation of the mRNAs of chloroplast-encoded genes
The decrease in the level of rbcL mRNA in apg3-1 mutants may be attributed to the inhibition ribosome release from the rbcL mRNA, owing to the deficiency in AtcpRF1. Thus, most of the rbcL mRNA is accumulated in stroma, destabilized as a consequence of its decreased association with ribosomes, and degraded rapidly. It has been reported that defects in polysome assembly can be corrected by altered metabolism of rbcL mRNA (Barkan 1993). Therefore, changes in the chloroplast translation machinery during chloroplast development, such as a defect in rRNA processing and lesions in genes encoding ribosome proteins or initiation factors, might be one factor modulating the stability of a subset of chloroplast mRNAs (Barkan 1993). Similarly, hcf109, the mutant of AtcpRF2, shows a defect in translational termination. Although UGA-containing transcripts accumulated to ∼50% of wild-type levels in this mutant, their protein levels were reduced to less than 10% of wild-type levels (Meurer et al. 2002). Interestingly, some proteins of UGA-containing transcripts were decreased but still detectable in hcf109 mutants by immunoblot analysis (Meurer et al. 2002). These results show that the translation of UGA-containing transcripts is not completely terminated by the deficiency in AtprfB in hcf109 mutants. In contrast, none of the plastid proteins tested could be detected in apg3-1 mutants (Fig. 7), at least in this study, a finding that may correlate with the fact that apg3-1 mutants show a more severe phenotype than hcf109 mutants.
RF1 and RF2 have been shown to recognize UAA/UAG and UAA/UGA stop codons, respectively, in E. coli (Craigen et al. 1990). In hcf109 mutants, plastid mRNAs containing the stop codons UAA (28 analyzed out of 47) and UAG (11 analyzed out of 18) were unaffected in size and abundance, but the levels of all 12 UGA stop codon-containing plastid transcripts were decreased (Meurer et al. 1996a, b, 2002). In contrast, in apg3 mutants, microarray analysis showed that the levels of all 18 UAG stop codon-containing plastid transcripts were not always decreased. However, a number of chloroplast transcripts for proteins involved in photosynthesis were significantly decreased (Tables 1, S1, and S2).
In higher plants, transcription of chloroplast genes is controlled by two RNA polymerases, NEP (nuclear-encoded RNA polymerase) and PEP (plastid-encoded RNA polymerase) (Stern et al. 1997; Hess and Börner 1999). The subunits of the E. coli-like RNA polymerase PEP are encoded by the plastid genome in higher plants, and PEP initiates transcription from E. coli σ7-type promoters in the plastid genome. The existence of a second RNA polymerase, NEP, in photosynthetic higher plants, was determined by deleting the genes encoding the essential β subunit of the tobacco PEP rpoB (Allison et al. 1996). PEP is composed of proteins encoded by rpoA, rpoB, rpoC1, and rpoC2, and transcribes most of the photosynthesis-related genes (Hajdukiewicz et al. 1997). In tobacco, mutants of these rpo genes showed albino phenotypes without pigments, due to the loss of stacked thylakoid membranes. The level of transcripts of the major photosynthesis-related genes encoded on plastid DNA is decreased in rpo mutants (Allison et al. 1996; Hajdukiewicz et al. 1997; Serino and Maliga 1998; De Santis-MacIossek et al. 1999; Krause et al. 2000; Legen et al. 2002). In PEP-deficient plastids, the transcription of atpA, psbC, psaA, petB, and petC was decreased (Legen et al. 2002), but accD, rpl2, ycf2, rps15, and rps18 exhibited upregulation (Allison et al. 1996; Hajdukiewicz et al. 1997; De Santis-Maciossek et al. 1999; Legen et al. 2002). Because rpoA (containing the stop codon UAG), rpoB (UAA), rpoC1 (UAA), and rpoC2 (UAA) mRNAs can be recognized by RF1, transcripts of rpo genes may not be translated efficiently in apg3-1 mutants, possibly causing repression of the transcription of many photosynthesis-related genes (Tables S1 and S2). Among them, the mRNA level of psbA was not exceptionally decreased (Fig. 6A). Transcripts of psbA have been reported to be very stable and abundant in the chloroplasts of light-grown plants (Baumgartner et al. 1993; Kim et al. 1993).
In contrast to the many photosynthesis-related genes, transcripts of rpo, rpl, and rps, housekeeping genes transcribed by NEP, were unaffected or increased in apg3-1 mutants (Tables S1 and S2). Such upregulation of NEP-dependent housekeeping genes has been reported in PEP-deficient plastids. The upregulation of rpo, rpl, and rps might be due to a feedback mechanism mediated by signals from defective chloroplast proteins transferred to the nucleus. Such a signal, a ‘plastid factor’, thought to be an intermediate of the tetrapyrrole biosynthetic pathway, has been reported (Vinti et al. 2000; Mochizuki et al. 2001; Strand et al. 2003). apg3 mutants showed an albino phenotype and decreased transcript levels of photosystem I- and II-related genes, which is much the same state as plants with PEP-deficient plastids. Thus, the accumulation of transcripts might not be related to whether the stop codon is UAG or UGA; however, on the contrary, it may be strongly dependent on the RNA polymerase (NEP or PEP).
The mutant of AtcpRF1, apg3-1, showed a more severe albino phenotype than the mutant of AtprfB, hcf109 (Meurer et al. 2002). One of the possible reasons for this difference is a deficiency in the translation of rpoA, which has a UAG stop codon, in apg3 mutants. It has been reported that the knockout mutants of genes encoding PEP subunits, such as rpoA, shows severe phenotypes concomitant with a decrease in the transcription levels of genes involved in photosystems I and II (Allison et al. 1996; Santis-Maciossek et al. 1999; Legen et al. 2002). On the other hand, other subunits of the PEP complex, such as rpoB, rpoC1, and rpoC2, have a UAA stop codon, which could be regulated by both RF1 and RF2. Thus, mutation in AtcpRF1 (APG3), which causes a decrease in PEP-regulated gene expression, results in a more severe phenotype than that due to mutation in AtcpRF2 (hcf109 mutants). This hypothesis is partially supported by evidence obtained from microarray analysis (Tables 1, S1, and S2). Among the genes affected by the mutation in APG3, most of the PEP-regulated genes were downregulated.
In conclusion, using the apg3 mutant, we demonstrated that a ribosome release factor 1 homologue is essential for chloroplast development in Arabidopsis. APG3 protein is an orthologue of E. coli RF1, and has been renamed AtcpRF1. AtcpRF1 functions as part of the chloroplast translation machinery and is important in chloroplast development and thylakoid biogenesis.
Materials and methods
Construction of Ds insertion lines
We made crosses between a transgenic line expressing Ac transposase (Nae Ac380-16) as the female parent and Ds-GUS-T-DNA lines (Ds2 389-13 and Ds4 391-20) as pollen parents (Fedoroff and Smith 1993; Smith et al. 1996). Selection of the transposed lines was described previously (Ito et al. 1999, 2000b).
Plant growth conditions
Seeds sterilized with 70% ethanol and 1% Antiformin were sown on GM agar plates (Valvekens et al. 1988) containing 1% sucrose and kept under 16-h light (illumination of about 70 μmol photons m−2 s−1)/8-h dark conditions at 22°C.
Chlorophyll fluorescence measurements
The chlorophyll fluorescence of leaves was measured at room temperature using a pulse-amplitude-modulated (PAM) fluorometer (TEACHING-PAM, Walz, Effeltrich, Germany) and a photosynthesis yield analyzer (MINI-PAM, Walz, Effeltrich, Germany). Before the chlorophyll fluorescence measurements, plants grown normally for 3 weeks were dark-adapted for 20 min. The results represent the mean values of at least four measurements.
Isolation of Ds-flanking sequences by TAIL-PCR, isolation of APG3 cDNA, and DNA gel blot analysis
TAIL-PCR, cloning of APG3 cDNA, and DNA gel blot analysis were carried out as described by Motohashi et al. (2003).
Electron microscopic analysis
Leaves were fixed, ultrathin sections were cut, sections were stained, and electron microscope (Jeol, Tokyo, Japan) observations were made as described by Motohashi et al. (2001).
Complementation analysis
Construction of the expression plasmid harboring APG3 was conducted using essentially the same procedure previously reported (Uno et al. 1996; Ito et al. 1998). For addition of the Shine-Dalgarno sequence into APG3, we amplified the NdeI–EcoRI cDNA fragment of APG3 by PCR using the primers 5Nde-AtRF and 3Sac-AtRF (5Nde-AtRF: 5′-TTTCATATGATGGCCGAACCTTACCTCATTAGG-3′ and 3Sac-AtRF: 5′-TTTGAGCTCTTATCCAGAAGTAGCAGAAGAAGC-3′) and cloned it into the NdeI–EcoRI sites of the pET30a vector (Novagen). To construct a lac promoter-regulated expression vector, pAPG3-lacI, the XbaI–EcoRI fragment of the pET-APG3 vector was introduced into the pTWV229 vector (TaKaRa, Tokyo, Japan), followed by introduction of an EcoRI fragment of lacI q (Uno et al. 1996) for high repression. The resulting plasmids, pAPG3-lacI, pTWV229, pTWV-ecRF1 harboring RF1 of E. coli, and pTWV-ecRF2 harboring the RF2 mutant of E. coli T246A, were introduced into the mutant E. coli strains, rf1ts (RM695) and rf2ts (RM718) (Ito et al. 1998) and grown at 42°C on agar medium containing 0.1 mM IPTG (isopropyl-1-thio-β-d-galactoside). It has been reported that E. coli RF2 terminates translation very weakly at UAA codons, while Salmonella RF2 decodes this signal efficiently. Moreover, an excess of E. coli RF2 was toxic to cells, whereas an excess of Salmonella RF2 was not. The residue at position 246 is solely responsible for these two phenotypes. Upon substituting an Ala for Thr-246 of E. coli RF2, the protein acquired increased release activity for UAA codons as well as for UGA codons (Uno et al. 1996). To avoid these inconvenient effects caused by overexpression of wild-type RF2, the RF2 mutant T246A was employed in this analysis. RM695 (rf1ts) was cultured in LB medium containing 1% bacto-tryptone, 0.5% yeast extract, 0.5% NaCl (Miller 1972) and ampicillin (50 μg/ml). RM718 (rf2ts) was cultured in YT medium containing 1% bacto-tryptone, 0.1% yeast extract, 0.25% NaCl and ampicillin (50 μg/ml) (Nakamura et al. 1986).
Transient assay by particle bombardment
The putative short transit peptide region (orange underline in Fig. 3A) of APG3 (40 amino acids) was amplified by PCR using the primers 1F and 1R (1F: 5′-TTTTCTAGAATGAATAGCTCGATGACGACG-3′, 1R: 5′-TTTGGATCCAGGCGGCCACGAGACTAGACA-3′). The putative long transit peptide region (green underline) from APG3 (55 amino acids) was amplified by PCR using the primers 1F and 2R (2R: 5′-TTTGGATCCAGGTTCGGCCATACAAACGAG-3′). The PCR-amplified fragments, coding the 40- and 55-amino-acid N-terminal regions of APG3, were cloned into the XbaI–BamHI sites of p35S-sGFP, consisting of a synthetic green fluorescent protein (GFP) under the control of cauliflower mosaic virus (CaMV) 35S promoter (Chiu et al. 1996). The resulting fusion constructs were named p35S::APG3tp40-sGFP and p35S::APG3tp55-sGFP, respectively. These constructs and the p35S-sGFP construct were introduced into the leaves of N. tabacum SR1 by particle bombardment. Particle bombardment and observation of GFP signals were carried out as described by Motohashi et al. (2001).
Reverse transcriptase–polymerase chain reaction
RT–PCR was performed according to the protocol of the mRNA Selective PCR kit ver.1.1 (TaKaRa, Japan). First-strand cDNA was synthesized from 1 μg of total RNA. PCR was performed with primers specific for the 5′ end of APG3 (5′-TTTTCTAGAATGAATAGCTCGATGACGACG-3′) and the 3′ end of APG3 (5′-TTTGGATCCAGGTTCGGCCATACAAACGAG-3′), the 5′ end of At2g47020 (5′-CGATATTGCTCTAGAGGTACGC-3′) and the 3′ end of At2g47020 (5′-CAAATCATTGATCAGCAACATTG-3′). Reactions comprised 30 cycles of 1 min at 80°C, 1 min at 50°C, and 1 min at 72°C.
RNA gel blot analysis
RNA gel blots were prepared as described by Motohashi et al. (2003). The probes were generated by PCR amplification of the large (DDBJ/EMBL/GenBank Accession Nos. U91966; 471–1,247) and small (X13611; 791–1,614) subunits of Rubisco, psaN (U32176; 31–529), psbA (X79898; 281–1,045), psbP (X98108; 290–1,537), 16S rRNA (AP000423; 136,147–137,637), APG1 (AB054257; 1–1,014), and the chlorophyll a/b-binding protein (X03909; 201–934) of Arabidopsis. After hybridization, the filters were washed twice with 0.1 × SSC/0.1% SDS at 65°C for 15 min and analyzed by autoradiography.
Microarray analysis
Microarray preparation and analysis were conducted as reported previously (Nagashima et al. 2004). For fluorescent probe preparation, 20 μg of each RNA sample was mixed with 35 ng of λ -RNA as internal controls. Q-RNA was synthesized by Riboprobe System-SP6 (Promega Co. Ltd., Madison, WI, USA) from a Q-DNA fragment containing an SP6 promoter and a poly-T tract. Preparation of Q-DNA was performed as described previously (Nagashima et al. 2004). This RNA solution was then added to a primer mixture containing 0.5 pmol of each gene-specific antisense primer (Nagashima et al. 2004), and reverse-transcribed in the presence of Cy3-dUTP or Cy5-dUTP (Amersham Pharmacia, Piscataway, NJ, USA). After array hybridization at 50°C for 16 h, the cover glass was removed from the microarray in 2 × SSC, washed twice with 0.1 × SSC containing 0.1% SDS for 5 min, and then twice in 0.1 × SSC for 5 min.
For microarray data analysis, image analysis and signal quantification were performed with QuantArray version 2.0 (GSI Lumonics, Oxnard, CA, USA). We normalized the data according to the global normalization method (Yang et al. 2002).
Immunological detection of plastid proteins
Soluble proteins for immunological detection of the Rubisco L/S complex were prepared by homogenizing leaves in a microcentrifuge tube on ice in a solution containing 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 10% (v/v) glycerol, 1.4 mM 2-mercaptoethanol, and 1 mM dithiothreitol. Total proteins for immunological detection of thylakoid proteins D1, OE23, and LHCII were prepared as described previously (Takeda et al. 1990; Nakajima et al. 1996).
Proteins were separated by SDS–PAGE (5% acrylamide stacking gel and 13% acrylamide separating gel) as described by Laemmli (1970). After electrophoresis, proteins were stained with Coomassie brilliant blue, or transferred electrophoretically to nitrocellulose filters in a solution of 48 mM Tris, 39 mM glycine, 0.037% (w/v) SDS, and 20% (v/v) methanol, at 15 V for 1 h. After blocking for 1 h in PBS (80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl, pH 7.5) buffer containing 5% non-fat dried milk (Yukijirushi, Sapporo, Japan) at room temperature, the membrane was incubated in PBS buffer with polyclonal antibodies (Rubisco L/S complex 1:5,000 dilution, LHCII 1:5,000 dilution, D1 1:500 dilution, OE23 1:1,000 dilution) for 2 h at 4°C. After washing in PBS buffer, the blots were then incubated with horseradish peroxidase-conjugated secondary antibody (Amersham, Little Chalfont, Buckinghamshire, UK), and the complexes were made visible by ECL (enhanced chemiluminescence; Amersham) following the manufacturer’s instructions. A polyclonal antibody against the tobacco Rubisco L/S complex was provided by F. Sato of Kyoto University and T. Nakano of RIKEN. A polyclonal antibody against spinach chlorophyll a/b-binding protein (LHCII) was provided by T. Masuda and K. Takamiya of the Tokyo Institute of Technology. A polyclonal antibody against spinach D1 protein was provided by M. Ikeuchi of Tokyo University. A polyclonal antibody against pea OE23 protein was provided by T. Endo of Nagoya University.
Polysome analysis
Polysome isolation from 3-week-old plants (1 g) was performed as described previously (Barkan 1998). Leaf tissue was ground to a fine powder in liquid nitrogen to which polysome extraction buffer was then added. The polysome extraction buffer including leaf powder was centrifuged to remove debris and the supernatant was incubated to solubilize membranes. Aliquots (1.4 mL) of polysomes were layered onto 15–55% sucrose gradients (8 mL per experiment) and centrifuged for 65 min at 39,000 rpm at 4°C in a SW41Ti rotor (Beckman, Munich, Germany). Fractions (0.7 mL) were collected by gentle pipetting from the top of the gradient. RNA was extracted from each fraction and analyzed by Northern hybridization.
References
Akkaya MS, Welsch PL, Wolfe MA, Duerr BK, Becktel WJ, Breinberger CA (1994) Purification and N-terminal sequence analysis of pea chloroplast protein synthesis factor EF-G. Arch Biochem Biophys 308:109–117
Albrecht V, Ingenfeld A, Apel K (2006) Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol Biol 60:507–518
Allison LA, Simon LD, Maliga P (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15:2802–2809
Barkan A (1993) Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5:389–402
Barkan A (1998) Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol 297:38–57
Baumgartner BJ, Rapp JC, Mullet JE (1993) Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development (evidence for selective stabilization of psbA mRNA). Plant Physiol 101:781–791
Bruick RK, Mayfield SP (1999) Light-activated translation of chloroplast mRNAs. Trends Plant Sci 4:190–195
Campos F, Garcia-Gomez BI, Solorzano RM, Salazar E, Estevez J, Leon P, Alvarez-Buylla ER, Covarrubias AA (2001) A cDNA for nuclear-encoded chloroplast translational initiation factor 2 from a higher plant is able to complement an infB Escherichia coli null mutant. J Biol Chem 276:28388–28394
Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6:325–330
Craigen WJ, Cook RG, Tate WP, Caskey CT (1985) Bacterial peptide chain release factors: conserved primary structure and possible frame shift regulation of release factor 2. Proc Natl Acad Sci USA 82:3616–3620
Craigen WJ, Lee CC, Caskey CT (1990) Recent advances in peptide chain termination. Mol Microbiol 4:861–865
De Santis-MacIossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, Rudiger W, Koop HU, Herrmann RG (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J 18:477–489
Fedoroff NV, Smith DL (1993) A versatile system for detecting transposition in Arabidopsis. Plant J 3:273–289
Gillham NW, Boynton JE, Hauser CR (1994) Translational regulation of gene expression in chloroplasts and mitochondria. Annu Rev Genet 28:71–93
Grevelding C, Suter-Crazzolara C, von Menges A, Kemper E, Masterson R, Schell J, Reiss B (1996) Characterisation of a new allele of pale cress and its role in greening in Arabidopsis thaliana. Mol Gen Genet 251:532–541
Hajdukiewicz PTJ, Allisom LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16:4041–4048
Hernandez-Torres J, Breitenberger CA, Spielmann A, Stutz E (1993) Cloning and sequencing of a soybean nuclear gene coding for a chloroplast translation elongation factor EF-G. Biochim Biophys Acta 1174:191–194
Hess W, Börner T (1999) Organellar RNA polymerases of higher plants. Int Rev Cytol 190:1–59
Hirose T, Ideue T, Wakasugi T, Sugiura M (1999) The chloroplast infA gene with a functional UUG initiation codon. FEBS Lett 445:169–172
Ito K, Uno M, Nakamura Y (2000a) A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403:680–684
Ito K, Uno M, Nakamura Y (1998) Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc Natl Acad Sci USA 95:8165–8169
Ito T, Kim G-T, Shinozaki K (2000b) Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J 22:257–264
Ito T, Motohashi R, Kuromori T, Mizukado S, Sakurai T, Kanahara H, Seki M, Shinozaki K (2002) A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol 129:1695–1699
Ito T, Motohashi R, Kuromori T, Noutoshi Y, Seki M, Kamiya A, Mizukado S, Sakurai T, Shinozaki K (2005) A resource of 5814 Dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol 46:1149–1153
Ito T, Seki M, Hayashida N, Shibata D, Shinozaki K (1999) Regional insertional mutagenesis of genes on Arabidopsis thaliana chromosome V using the Ac/Ds transposon in combination with a cDNA scanning method. Plant J 17:433–444
Kim M, Christopher DA, Mullet JE (1993) Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol Biol 22:447–463
Kim J, Mullet JE (2003) A mechanism for light-induced translation of the rbcL mRNA encoding the large subunit of ribulose-1,5-bisphosphate carboxylase in barley chloroplasts. Plant Cell Physiol 44:491–499
Klein HA, Capecchi MR (1971) Polypeptide chain termination: Purification Of The Release Factors, R1 and R2, from Escherichia Coli. J Biol Chem 246:1055–1061
Kochhar A, Khurana JP, Tyagi AK (1996) Nucleotide sequence of the psbP gene encoding precursor of 23-kDa polypeptide of oxygen-evolving complex in Arabidopsis thaliana and its expression in the wild-type and a constitutively photomorphogenic mutant. DNA Res 3(5):277–285
Krause GH, Grafflage S, Rumich-Bayer S, Somersalo S (1988) Effects of freezing on plant mesophyll cells. Symp Soc Exp Biol 42:311–327
Krause K, Maier RM, Kofer W, Krupinska K, Herrmann RG (2000) Disruption of plastid-encoded RNA polymerase genes in tobacco: expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Mol Gen Genet 263:1022–1030
Krebbers E, Seurinck J, Herdies L, Cashmore AR, Timko MP (1988) Four genes in two diverged subfamilies encode the ribulose-1,5-bisphosphate carboxylase small subunit polypeptides of Arabidopsis thaliana. Plant Mol Biol 11:745–759
Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K (2004) A collection of 11,800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J 37:897–905
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lee CC, Kohara Y, Akiyama K, Smith CL, Craigen WJ, Caskey CT (1988) Rapid and precise mapping of the Escherichia coli release factor genes by two physical approaches. J Bacteriol 170:4537–4541
Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31:171–188
Leutwiler LS, Meyerowitz EM, Tobin EM (1986) Structure and expression of three light-harvesting chlorophyll a/b binding protein genes in Arabidopsis thaliana. Nucl Acids Res 14:4051–4064
Liere K, Kestermann M, Müller U, Link G (1995) Identification and characterization of the Arabidopsis thaliana chloroplast DNA region containing the genes psbA, trnH and rps19′. Curr Genet 28(2):128–130
Liu Y-G, Whittier RF (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681
Matsumura K, Ito K, Kawazu Y, Mikuni O, Nakamura Y (1996) Suppression of temperature-sensitive defects of polypeptide release factors RF-1 and RF-2 by mutations or by an excess of RF-3 in Escherichia coli. J Mol Biol 258:588–599
Meurer J, Lezhneva L, Amann K, Gödel M, Bezhani S, Sherameti I, Oelmüller R (2002) A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell 14:3255–3269
Meurer J, Berger A, Westhoff P (1996a) A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid, psbB, psbD/C, ndhH, and hdhC operons. Plant Cell 8:1193–1207
Meurer J, Meierhoff K, Westhoff P (1996b) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterization by spectroscopy, immunoblotting and northern hybridization. Planta 198:385–396
Millen RS, Olmstead RG, Adama KL, Palmer JD, Lao NT, Heggie L, Kavanagh TA, Hibberd JM, Gray JC, Morden CW, Calie PJ, Jermiin LS, Wolfe KH (2001) Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13:645–658
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98:2053–2058
Mora L, Heurgue-Hamard V, Champ S, Ehrenberg M, Kisselev LL, Buckingham RH (2003) The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol Microbiol 47:267–275
Motohashi R, Ito T, Kobayashi M, Taji T, Nagata N, Asami T, Yoshida S, Yamaguchi-Shinozaki K, Shinozaki K (2003) Functional analysis of the 37-kDa inner envelope membrane polypeptide in chloroplast biogenesis, using a Ds-tagged Arabidopsis pale green mutant. Plant J 34:719–731
Motohashi R, Nagata N, Ito T, Takahashi S, Hobo T, Yoshida S, Shinozaki K (2001) An essential role of a TatC homologue of ΔpH-dependent protein transporter in thylakoid membrane formation during chloroplast development in Arabidopsis thaliana. Proc Natl Acad Sci USA 98:10499–10504
Nagashima A, Hanaoka M, Fujiwara M, Motohashi R, Seki M, Shinozaki K, Kanamaru K, Takahashi H, Tanaka K (2004) DNA microarray analysis of plastid gene expression in an Arabidopsis mutant deficient in a plastid transcription factor sigma, SIG2. Biosci Biotech Biochem 68:694–704
Nakahigashi K, Kubo N, Narita S, Shimaoka T, Goto S, Oshima T, Mori H, Wada C, Inokuchi H (2002) HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc Natl Acad Sci USA 99:1473–1478
Nakajima Y, Yoshida S, Inoue Y, Ono T (1996) Occupation of the QB-binding pocket by a photosystem II inhibitor triggers dark cleavage of the D1 protein subject to brief preillumination. J Biol Chem 271:17383–17389
Nakamura Y, Ito K, Isaksson LA (1996) Emerging understanding of translation termination. Cell 87:147–150
Nakamura Y, Ito K, Matsumura K, Kawazu Y, Ebihara K (1995) Regulation of translation termination: conserved structural motifs in bacterial and eukaryotic polypeptide release factors. Biochem Cell Biol 73:1113–1122
Nakamura Y, Mizusawa S, Court DL, Tsugawa A (1986) Regulatory defects of a conditionally lethal nusAts mutant of Escherichia coli: positive and negative modulator roles of NusA protein in vivo. J Mol Biol 189:103–111
Pannekoek Y, Heurgue-Hamard V, Langerak A, Speijer D, Buckingham R, Ende A (2005) The N5-glutamine S-adenosyl-l-methionine-dependent methyltransferase PrmC/HemK in Chlamydia trachomatis methylates class 1 release factors. J Bacteriol 187:507–511
Pel HJ, Maat C, Rep M, Grivell LA (1992) The yeast nuclear gene MRF1 encodes a mitochondrial peptide chain release factor and cures several mitochondrial RNA splicing defects. Nucleic Acids Res 20:6339–6346
Rochaix JD (1996) Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol Biol 32:327–341
Rolland N, Janosi L, Block MA, Teyssier E, Miége C, Chéniclet C, Carde J-P, Kaji A, Joyard J (1999) Plant ribosome recycling factor homologue is a chloroplastic protein and is bactericidal in Escherichia coli carrying temperature-sensitive ribosome recycling factor. Proc Natl Acad Sci USA 96:5464–5469
Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res 6:283–290
Schmidt G, Mishkind N (1983) Rapid degradation of unassembled ribulose 1,5-bisphosphate carboxylase small subunits in chloroplasts. Proc Natl Acad Sci USA 80:2632–2636
Schoof H, Ernst R, Nazarov V, Pfeifer L, Mewes HW, Mayer KF (2004) MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource for plant genomics. Nucleic Acids Res 1:32, 373–376
Scolnick E, Tompkins R, Caskey T, Nirenberg M (1968) Release factors differing in specificity for terminator codons. Proc Natl Acad Sci USA 61:768–774
Sehnke PC, Ferl RJ (1995) Nucleotide sequence of an Arabidopsis thaliana cDNA clone (Accession No. U32176) encoding the complete precursor for a homologue to the barley extrinsic thylakoid lumenal polypeptide PSI-N. Plant Physiol 109:1126
Seki M, Ito T, Shibata D, Shinozaki K (1999) Regional insertional mutagenesis of specific genes on the CIC5F11/CIC2B9 locus of Arabidopsis thaliana chromosome 5 using the Ac/Ds transposon in combination with the cDNA scanning method. Plant Cell Physiol 40:624–639
Serino G, Maliga P (1998) RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol 117:1165–1170
Smith D, Yanai Y, Liu Y-G, Ishiguro S, Okada K, Shibata D, Whittier RF, Fedoroff NV (1996) Characterization and mapping of Ds-GUS-T-DNA lines for targeted insertional mutagenesis. Plant J 10:721–732
Spreltzer RJ, Goldschmidt-Clermont M, Rahire M, Rochalx J-D (1985) Nonsense mutations in the Chlamydomonas chloroplast gene that codes for the large subunit of ribulose bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA 82:5460–5464
Stern DB, Higgs DC, Yang J (1997) Transcription and translation in chloroplasts. Trends Plant Sci 2:308–315
Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrin IX. Nature 421:79–83
Sugita M, Sugiura M (1996) Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol 32:315–326
Sugiura M (1992) The chloroplast genome. Plant Mol Biol 19:149–168
Takechi K, Sodmergen Murata M, Motoyoshi F, Sakamoto W (2000) The YELLOW VARIGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol 41:1334–1346
Takeda S, Sato F, Ida K, Yamada Y (1990) Characterization of polypeptides that accumulate in cultured Nicotiana tabacum cells. Plant Cell Physiol 31:215–221
Uno M, Ito K, Nakamura Y (1996) Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie 78:935–943
Valvekens D, Van Montagu M, Van Lijsebettens M (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85:5536–5540
Vinti G, Hills A, Campbell S, Bowyer JR, Mochizuki N, Chory J, Lopez-Juez E (2000) Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signaling. Plant J 24:883–894
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Nagai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucl Acids Res 30:e15
Zerges W (2000) Translation in chloroplasts. Biochimie 82:583–601
Zhu G, Jensen RG, Bohnert HJ (1997) DNA sequence of ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit from Arabidopsis thaliana. Plant Physiol 114:395
Acknowledgements
We thank I. Furukawa and S. Kawamura of RIKEN for their skillful technical assistance; R. Yoshida and T. Amano for their helpful comments; F. Sato of Kyoto University and T. Nakano of RIKEN for the Rubisco L/S complex antibody; M. Ikeuchi of Tokyo University for the D1 antibody; and T. Masuda and K. Takamiya of Tokyo Institute of Technology for the LHCII antibody. This work was supported by a grant for Genome Research from RIKEN to K.S. and a Grant-in-Aid for Scientific Research and NISSAN Science foundation to R.M.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material
Rights and permissions
About this article
Cite this article
Motohashi, R., Yamazaki, T., Myouga, F. et al. Chloroplast ribosome release factor 1 (AtcpRF1) is essential for chloroplast development. Plant Mol Biol 64, 481–497 (2007). https://doi.org/10.1007/s11103-007-9166-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9166-7