Abstract
The sub-cellular location of enzymes of fatty acid β-oxidation in plants is controversial. In the current debate the role and location of particular thiolases in fatty acid degradation, fatty acid synthesis and isoleucine degradation are important. The aim of this research was to determine the sub-cellular location and hence provide information about possible functions of all the putative 3-ketoacyl-CoA thiolases (KAT) and acetoacetyl-CoA thiolases (ACAT) in Arabidopsis. Arabidopsis has three genes predicted to encode KATs, one of which encodes two polypeptides that differ at the N-terminal end. Expression in Arabidopsis cells of cDNAs encoding each of these KATs fused to green fluorescent protein (GFP) at their C-termini showed that three are targeted to peroxisomes while the fourth is apparently cytosolic. The four KATs are also predicted to have mitochondrial targeting sequences, but purified mitochondria were unable to import any of the proteins in vitro. Arabidopsis also has two genes encoding a total of five different putative ACATs. One isoform is targeted to peroxisomes as a fusion with GFP, while the others display no targeting in vivo as GFP fusions, or import into isolated mitochondria. Analysis of gene co-expression clusters in Arabidopsis suggests a role for peroxisomal KAT2 in β-oxidation, while KAT5 co-expresses with genes of the flavonoid biosynthesis pathway and cytosolic ACAT2 clearly co-expresses with genes of the cytosolic mevalonate biosynthesis pathway. We conclude that KATs and ACATs are present in the cytosol and peroxisome, but are not found in mitochondria. The implications for fatty acid β-oxidation and for isoleucine degradation in mitochondria are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is widely accepted that the complete β-oxidation of medium- and long-chain fatty acids in plants takes place in the peroxisomes (Hooks 2002), as it does in yeast (van Roermund et al. 2003). However, some biochemical evidence suggests that plant mitochondria can also carry out such β-oxidation of fatty acids (Masterson and Wood 2001). It has also become clear recently that plant mitochondria catalyse at least the initial steps in the degradation of branched-chain α-keto acids, derived from leucine, isoleucine and valine, through a branched chain α-keto acid dehydrogenase complex similar to the pyruvate and 2-oxoglutarate dehydrogenase complexes of the TCA cycle (Fujiki et al. 2000; Graham and Eastmond 2002; Taylor et al. 2004). In the case of the leucine carbon skeleton, the later steps of degradation are carried out entirely within the mitochondrion (Graham and Eastmond 2002; Taylor et al. 2004). In contrast, final degradation of valine derivatives requires both mitochondrial and peroxisomal steps (Lange et al. 2004). Meanwhile the complete oxidation of the products from the isoleucine carbon skeleton includes a β-oxidation step that requires a 3-ketoacyl-CoA thiolase (KAT) for the removal of an acetyl-CoA from 2-methylaceto-acetyl CoA to form propionyl-CoA. However, it is unclear if this thiolase catalysed β-oxidation of 2-methylaceto-acetyl CoA occurs in the mitochondrion in plants, as it does in mammals (Fukao et al. 2001), or whether it occurs in the peroxisome in plants akin to the final steps of valine metabolism (Lange et al. 2004).
Comparisons to Saccharomyces cerevisiae are not informative as yeast degrades branched chain amino acids not via the branched chain dehydrogenase complex route in mitochondria, but via the Erhlich pathway involving pyruvate decarboxylase to form the corresponding aldehydes and an aldehyde dehydrogenase to form the corresponding alcohol in the cytosol (Derrick and Large 1993). Thus yeast does not need a thiolase for isoleucine degradation.
Two distinct forms of 3-ketoacyl-CoA thiolase are known. Type 1 3-ketaoacyl-CoA thiolase (KAT; EC 2.3.1.16) is typically involved in the degradative process of fatty acid β-oxidation. The Type II enzyme is an acetoacetyl-CoA thiolase (ACAT; EC 2.3.1.9), typically involved in the mevalonate pathway where it functions in the biosynthetic direction. However, ACATs are not exclusively involved in mevalonate synthesis. In mammals that undertake both fatty acid β-oxidation and isoleucine catabolism in mitochondria, the former is performed by a KAT while the later is performed by an ACAT (Pereto et al. 2005).
In Arabidopsis, thiolase has been reported to be associated with both peroxisomes and mitochondria in sucrose density gradients (Footitt et al. 2002). Kruft et al (2001) and Heazelwood et al (2004) have both claimed the thiolase KAT2 encoded by At2g33150 to be present in purified mitochondria in large-scale proteome analyses. This thiolase has previously been proposed to be a component of isoleucine catabolism in mitochondria (Taylor et al. 2004). However, the thiolase is question is a type I enzyme, while in mammals it is the mitochondrial type II ACATs that have been implicated in isoleucine catabolism (Pereto et al. 2005). The KAT2 thiolase encoded by At2g33150 has a predicted type 2 peroxisomal targeting sequence (PTS2) conforming to the consensus R-(X)6-H/Q-A/L/F with a downstream cysteine residue required for proteolytic cleavage (Baker and Sparkes 2005), and is imported into peroxisomes in vitro (Johnson and Olsen 2003). At2g33150 is well known to be essential for peroxisomal β-oxidation (Germain et al. 2001). However, the protein encoded by At2g33150 is also predicted to be targeted to mitochondria by three different targeting prediction programs (Heazlewood et al. 2004). Furthermore, changing a single amino acid in the peroxisomal targeting signal of the KAT precursor in mammals, a glutamic acid residue to any non-acidic residue, resulted in targeting to both mitochondria and peroxisomes (Tsukamoto et al. 1994). This raises the possibility that the thiolase encoded by At2g33150 is targeted to peroxisomes and mitochondria, representing a dual targeted protein.
This study was carried out to define the subcellular localization of the products from the putative KATs and ACATs in Arabidopsis. This was achieved by identifying all possible thiolase genes in Arabidopsis and comparing their sequences to known-location type I and type II thiolases in yeast, mammals and fungi. We then used transcript sequence data to produce all the possible cDNAs for these gene products. These cDNAs were used with in vivo and in vitro organelle targeting assays to define subcellular localization of type I and II thiolases in Arabidopsis and gene expression profiling data was compared to define likely functional links.
Materials and methods
Identification of genes and cDNAs encoding thiolase
The predicted protein sequence encoded by At2g33150 previously shown to be a thiolase was used to define other thiolase encoding genes in Arabidopsis (Germain et al. 2001), and the sequences from other species as previously published (Pereto et al. 2005). A similarity tree was made using ClustalW multiple sequence alignment and neighbour joining (Thompson et al. 1994, 1997). The gene structures were obtained from The Arabidopsis Information Resource annotation version 6 (TAIR6) and three individual cDNAs were amplified for all possible cDNAs. The cDNAs produced from the genes were defined using the Arabidopsis genome tiling array (Mockler et al. 2005; Yamada et al. 2003). Targeting predictions of the encoded proteins were carried out using a variety of prediction programs: TargetP (Emanuelsson et al. 2000), Mitoprot (Claros and Vincens 1996), Subloc (Hua and Sun 2001), Ipsort (Bannai et al. 2002), Predotar (Small et al. 2004), Mitpred (Kumar et al. 2006) and PeroxP (Emanuelsson et al. 2003). Percentage identity and similarity was calculated using MatGAP v2.02 (Campanella et al. 2003).
Subcellular targeting of predicted thiolase proteins
The coding sequences of the predicted thiolase proteins were cloned in frame with the coding region of GFP in pGEM 3Zf(+) containing the 35S CaMV promoter (Chew et al. 2003). The alternative oxidase (AOX) coding region fused to GFP (Lee and Whelan 2004), and the red fluorescent protein (RFP) fused to a type 1 peroxisomal SRL targeting signal from pumpkin (Pracharoenwattana et al. 2005), were used as mitochondrial and peroxisomal controls respectively. The constructs were used to transform Arabidopsis suspension culture cells by biolistic transformation as previously outlined (Thirkettle-Watts et al. 2003). Fluorescence patterns were obtained 48 h after transformation by visualization under an Olympus BX61 fluorescence microscope and imaged using the CellR imaging software. In vitro protein import assays into mitochondria isolated from Arabidopsis suspension cell cultures were carried out as described in Lister et al. (2004). In vitro mitochondrial uptake assays were performed by adding precursor protein to 100 μg of isolated mitochondria in 200 μl in the presence of respiratory substrate (succinate 5 mM) and ATP (1 mM) and ADP (200 μM) in import buffer (0.3 M sucrose, 50 mM KCl, 10 mM MOPS pH 7.2, 5 mM KH2PO4, 1% (w/v) BSA, 1 mM MgCl2, 1 mM methionine and 5 mM DTT). Reactions were incubated at 24°C for 20 min then divided into two equal aliquots and placed on ice. To one aliquot Proteinase K was added to a final concentration of 40 μg/ml and incubated for 15 min on ice, followed by the addition of PMSF to 2 mM to terminate protease digestion. The mitochondria were pelleted, washed twice in ice-cold import buffer. The final pellet was resuspended in SDS-PAGE sample buffer and proteins separated in 12% (w/v) polyacrylamide gels, then dried. Radiolabelled proteins were visualized by exposing to a BAS TR2040 imaging plate for 24 h and reading on a BAS 2500 Bio imaging analyser (Fuji, Tokyo). Outer membrane ruptured mitochondria (Mit-OM) were prepared after the import assay to test for the intraorganelle location of imported protein. Rupture of the outer membrane allowed access of added protease to intermembrane space components or inner membrane proteins exposed to the intermembrane space. Mit-OM were prepared by resuspending 100 μg of mitochondrial protein in 10 ml SEH buffer (250 mM sucrose, 1 mM EDTA, 10 mM Hepes pH 7.4) and then adding 155 μl of 20 mM Hepes pH 7.4 and incubating on ice for 20 min. To restore osmolarity 25 μl of 2 M sucrose and 10 μl of 3 M KCl was added and mixed, re-pelleted and washed in import buffer. Valinomycin was added to a final concentration of 1 μM where indicated prior to the addition of the precursor protein to mitochondria and commencement of the import assay. AOX was used as a positive control and the small subunit of Ribulose 1, 5-bisphosphate carboxylase/oxygenase (Rubisco SSU) as a negative control. As some precursor proteins displayed protease insensitivity even in the presence of valinomycin the sensitivity of the precursor proteins alone to added protease was tested. Sensitivity of the in vitro synthesized radiolabelled proteins was tested by adding proteinase K to the synthesized protein alone in the absence of mitochondria to ensure that the added protease could digest the protein.
In silico expression analysis
Expression correlation for genes encoding KATs and ACATs was performed using the Expression Angler program on the Botany Array Resource (Toufighi et al. 2005). The Genevestigator Arabidopsis microarray database was used to analyse the response of the genes of interest in this study in a variety of tissue types (Zimmermann et al. 2004). The meta-analyser tool was the function utilized, ATH1 22k array wild type only arrays were chosen and the genes of interest were selected. The data was visualized using a linear scale from a total of 1860 array experiments. TMeV (TIGR Multiple Experiment Viewer) programme was used to cluster the genes and stresses, and Euclidean distance and complete linkage were chosen for the hierarchal clustering (Eisen et al. 1998; Saeed et al. 2003).
Results
The Arabidopsis thiolase gene families
Searches of the Arabidopsis genome identify five loci with sequence similarity to genes encoding known thiolase proteins (Germain et al. 2001). Comparison of predicted amino acid sequences shows that they fall into two classes. Three loci encode the Type I class of enzyme, KAT 1, 2 and 5, typically involved in acetyl-CoA formation in fatty acid β-oxidation by removal of a 2-carbon chain from a 3-ketoacyl-CoA. The other two genes encode the type II class of enzyme, ACAT 1 and 2, typically involved in acetoacetyl-CoA formation from two molecules of acetyl-CoA (Fig. 1A, Supplementary Fig. 1A). The type I genes have previously been annotated as KAT1, KAT2 and KAT5 (3-ketoacyl-CoA thiolase) (Germain et al. 2001), based on the chromosome on which they are found (At1g04710, At2g33150 and At5g48880, respectively). All three are closely related to known peroxisomal located type I thiolases in human, mouse and yeast and this cluster also contains representatives from the fungus Neurospora crassa and the monocot rice (Oryza sativa). The matrix and membrane-bound type I mitochondrial thiolases involved in fatty acid degradation in human, mouse and Drosophila cluster separately and do not contain members from the completed genome sequences of fungi, Arabidopsis or rice.
Classification of thiolase (KAT and ACAT) genes and gene structure in Arabidopsis. (A) A phylogenetic tree was generated using the neighbour joining method, using ClustalW of thiolase proteins from a variety of organisms. KAT = 3-ketoacyl-CoA thiolases, ACAT = acetoacetly-CoA thiolases. (B) Gene structure and predicted proteins encoded by thiolase genes. The differences in the proteins encoded by each locus are indicated in bold where evidence for more than one cDNA exists. The open white boxes indicate exons
The two Arabidopsis type II thiolases (here referred to as ACAT1 and ACAT2, At5g47720 and At5g48230, respectively) cluster with the known cytosolic type II thiolases from yeast and the cytosolic and mitochondrial type II thiolases from human, mouse and Drosophila. The monocot rice also has two type II thiolases that cluster in this set and N. crassa contains a single type II gene that clusters with the yeast cytosolic type II protein.
The sequence divergence of mitochondrial type I thiolases in mammals from the peroxisomal type I KATs in plants, fungi and animals makes the presence of KAT2 in Arabidopsis mitochondria appear unlikely based on phylogenetic evidence if thiolase location is conserved. However, the sequence analysis does not define the location of the type II ACAT proteins in Arabidopsis and leaves open the possibility of a mitochondrial location of at least one of these proteins, especially given the presence of multiple type II thiolases in both plants and mammals.
Definition of the number of products from each Arabidopsis thiolase gene
Analysis of EST sequences and tiling array data shows that KAT1 and KAT2 loci each encode single polypeptide sequences (Fig. 1B) (Mockler et al. 2005; Yamada et al. 2003). In contrast, KAT5 encodes two proteins that differ at the N-terminus due to alternative RNA splicing that generates either 13 (KAT5.1) or 14 (KAT5.2) exons (Fig. 1B). The N-terminal sequences of proteins encoded by KAT1, KAT2 and KAT5.2 include putative PTS2-type sequences, while the protein encoded by KAT5.1 does not (Fig. 1B). Interestingly, proteins encoded by KAT1, KAT2, KAT5.1 and KAT5.2 proteins are predicted to be targeted to mitochondria by at least two of three different programs (Table 1). Analysis of EST sequences and tiling array data shows that ACAT1 and ACAT2 loci also encode three and two proteins respectively (Fig. 1B). Differential RNA splicing results in the protein encoded by ACAT1.1 lacking ten amino acids at the C-terminus relative to the protein encoded by ACAT1.2. The protein ACAT1.3 has 20 different amino acid residues at the N-terminus relative to ACAT1.1. None of the proteins has predicted organelle-targeting information (Table 1). Differential RNA splicing also accounts for the N-terminal 6 amino acid residues of the protein encoded by ACAT2.1 being replaced by 11 different amino acid residues in the case of the protein encoded by ACAT2.2 (Fig. 1B). Neither protein has predicted organelle-targeting information (Table 1).
Targeting of type I and II thiolases in vivo
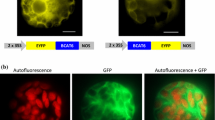
To localize thiolases in vivo, GFP and RFP fusions were employed. GFP and RFP have been used extensively to study protein targeting to mitochondria, peroxisomes and chloroplasts (Heazlewood et al. 2005). To demonstrate specific mitochondrial and peroxisomal targeting in vivo and our ability to distinguish the two, an AOX–GFP construct (Lee and Whelan 2004) and an RFP–PTS1 construct (Pracharoenwattana et al. 2005), were employed. The two gene constructs were co-delivered into Arabidopsis suspension culture cells using a biolistic gene gun (Thirkettle-Watts et al. 2003). After 48 h individual cells expressing both GFP and RFP fluorescence were imaged. The results show that GFP and RFP were targeted to discrete organelles consistent with specific targeting to mitochondria and peroxisomes respectively (Fig. 2).
In vivo targeting ability of thiolases in Arabidopsis. The cDNA coding sequences of thiolase from Arabidopsis were tagged with GFP to assess targeting ability. Each cell shown was transformed with both a GFP construct and with RFP with a type I PTS. Each panel shows the localization of GFP targeted either by the mitochondrial protein alternative oxidase (AOX) or by thiolases (GFP panel). A peroxisomal pattern obtained in the same cell with the RFP with a type I PTS is shown (RFP-SRL panel) together with the merged images (Merged panel)
To examine thiolase targeting we made translational fusions with GFP at the C-terminus since peroxisomal (PTS2) and mitochondrial targeting sequences are both N-terminal. Thiolase cDNAs encoding all nine proteins were linked to the GFP coding region and cloned downstream of the CaMV 35S promoter. They were each delivered into Arabidopsis suspension culture cells together with the RFP–PTS1 construct. After 48 h individual cells expressing both GFP and RFP fluorescence were imaged, and the images merged. The results show that KAT1, KAT2 and KAT5.2 and ACAT 1.3 were targeted to peroxisomes, as indicated by coincidence of GFP and RFP images (Fig. 2). In contrast, ACAT1.1, ACAT1.2, ACAT2.1, ACAT2.2 and KAT5.1 show diffuse fluorescence throughout the cell indicating that no specific targeting to any organelle has occurred, suggesting a cytosolic localization. With peroxisomal targeting of KAT1, KAT2, KAT5.2 and ACAT 1.3, although it was apparent that the patterns of GFP and RFP were essentially identical, the higher intensity of the former means that when merged the green fluorescence was dominant in some cells.
Protein import into isolated mitochondria
None of the thiolases were apparently targeted to mitochondria in vivo. However, it is possible that uptake was prevented by the GFP fusion, or that mitochondria normally take up less thiolase than peroxisomes, such that GFP fluorescence from mitochondria did not reach an intensity to be detected. To test these possibilities we examined the ability of isolated mitochondria to take up all nine thiolases. Each protein was synthesized in a rabbit reticulocyte lysate translation system in the presence of radiolabelled methionine, and then tested for import into mitochondria isolated from Arabidopsis cell cultures. As a control the import and processing of AOX and Rubisco SSU were examined, the former as a positive control for import and the latter as a control to demonstrate the specificity of import into isolated mitochondria (Chew et al. 2003; Chew and Whelan 2004). In this case the AOX precursor protein (36 kDa) was imported and cleaved to a mature protein (32 kDa) as previously demonstrated (Fig. 3, lanes 1 and 2) (Whelan et al. 1995). Addition of protease resulted in the 32-kDa mature form being resistant to protease digestion indicating uptake by mitochondria. This resistance was abolished by the addition of valinomycin that dissipates the inner membrane potential (Fig. 3, lanes 4 and 5) (Tanudji et al. 2001). The phosphate translocator from maize was used as an additional control, after uptake into mitochondria and rupture of the outer membrane protease digestion results in a small cleaved protected fragment of 33 kDa, indicating that the added protease has access to inside the outer membrane (Bathgate et al. 1989; Murcha et al. 2004, 2005; Winning et al. 1992). In contrast to the mitochondrial controls, Rubisco SSU was not protected from protease digestion indicating it was not imported into mitochondria (Fig. 3).
In vitro import of radiolabelled thiolase proteins into mitochondria isolated from Arabidopsis. Lane 1, precursor protein alone. Lane 2, precursor protein incubated with mitochondria under conditions that support import into mitochondria. Lane 3, as lane 2 with proteinase K added after incubation of precursor with mitochondria. Lane 4 and 5, as lane 2 and 3 with valinomycin added to the import assay prior to the addition of precursor protein. Lanes 6–9 as 2–5 except that the mitochondrial outer membrane was ruptured after the incubation period with precursor protein but prior to addition of proteinase K. Apparent mol mass are indicated in kDa. Abbreviations: Mit = mitochondria, Mit-OM = mitochondria with outer membrane ruptured, PK = proteinase K, Val = valinomycin, AOX = alternative oxidase, Pic = phosphate carrier, Rubisco SSU = small subunit of ribulose-1, 5 bisphosphate carboxylase/oxygenase, p = precursor protein band, m = mature protein band
When the nine thiolase proteins were tested for mitochondrial uptake two distinct patterns were observed, KAT1, ACAT 1.1, ACAT 1.2, ACAT 1.3, KAT5.1 and KAT5.2 did not yield any protease protected products upon incubation with mitochondria and thus were deemed not to be imported (Fig. 3). KAT2, ACAT2.1 and ACAT2.2 yielded resistant products upon incubation with mitochondria. Although KAT2 was not proteolytically cleaved by mitochondria, a protease resistant product with a lower mol mass was obtained when mitochondria were treated with Proteinase K (Fig. 3, lanes 1–3). Notably this was also generated in the presence of valinomycin (Fig. 3, lanes 4–5). However, upon rupture of the outer membrane this product was absent (Fig. 3, lanes 6–9). In the case of ACAT2.1 and ACAT2.2 a similar pattern was observed except that the protected protein had the same molecular mass as the protein added to the import assay (Fig. 3, lanes 1–5). Again with rupture of the outer membrane no protease protection was observed (Fig. 3, lanes 6–9).
Although the protease protected fragments produced upon incubation of KAT2, ACAT2.1 and ACAT2.2 may suggest uptake by mitochondria, their presence when valinomycin was added to the import assay and their sensitivity when the outer mitochondrial membrane is ruptured suggests that they may represent protease resistant products in the presence of intact mitochondria. The protease susceptibility of KAT2, ACAT2.1 and ACAT2.2 was tested by the ability of added protease to digest the radiolabelled precursor protein. Incubation of KAT2.1, ACAT2.1 and ACAT2.2 with proteinase K alone indicated that they were resistant to protease digestion; in contrast AOX was completely digested (Fig. 4). Thus it was concluded that there was no uptake of any radiolabelled thiolase proteins into isolated mitochondria, in agreement with the GFP targeting (Fig. 2).
Co-expression analysis
The probable functions of these type I and type II thiolases in their determined subcellular locations was examined by analysis of co-expression of these genes in microarray data from Arabidopsis (Fig. 5). We used the Expression Angler co-expression correlation tool from the Botany Array Resource (Toufighi et al. 2005) to find the most co-expressed genes based on microarray hybridization data on Arabidopsis 22K genechips; the top 25 co-expressed genes are shown in each case (Fig. 5A, Supplementary Table 1). This analysis shows that KAT2 co-expresses (Correlation >0.65–0.79) more highly with a range of peroxisomal fatty acid degradation components in the peroxisome than with any other nuclear genes in Arabidopsis. These included the fatty acid multifunction protein MFP2 (At3g06860), citrate synthase (At2g42790), acyl-CoA oxidases (At5g65110, At3g51840) and enoyl-CoA hydratase (At4g16210) (Fig. 5A). Curiously, Expression Angler showed KAT5 is not co-expressed with β-oxidation enzymes, but instead with a series of flavonoid biosynthesis enzymes (Correlation >0.6–0.71), including flavanone 3-hydroxylase (At3g51240) 4-courmarate CoA ligase (At1g65060), chalcone synthase (At5g13930), chalcone isomerase (At5g05270). Notably this pathway requires short acyl-CoAs for biosynthesis.
In silico expression analysis. (A) The 25 genes co-expressed to the greatest extent with each thiolase gene were determined using the Expression Angler tool from the Botany Array Resource. The annotated genes indicated for KAT2, KAT5 and ACAT2 are those for which the proteins encoded by these genes could function in peroxisomal fatty acid degradation, flavonoid biosynthesis and the mevalonate pathway, respectively. (B) Clustering analysis of co-expressed genes with KAT2, KAT5 and ACAT2 to determine which (KAT or ACAT) branch with co-expressed genes as determined by Expression Angler. Each KAT or ACAT is located in a distinct group in agreement with analysis by Expression Angler, with good bootstrap values supporting the branch points
The gene encoding a type II enzyme ACAT2 (At5g48230) was found by Expression Angler to be co-expressed with a range of genes, but notably, hydroxymethylglutaryl-CoA synthase (At4g11820) and mevalonate diphosphate decarboxylase (At2g38700, At3g54250) were highly co-expressed (Correlation >0.80) (Fig. 5, Supplementary Table 1). This is consistent with the role of type II genes in the cytosolic mevalonate pathway leading to isoprene-containing compounds such as sterols and terpenoids.
The isoleucine catabolism pathway involves the branched chain amino acid dehydrogenase complex (At5g09300, At3g13450, At3g06850), isovaleryl-CoA dehydrogenase (At3g45300), enoyl-CoA hydratase (At4g31810) in mitochondria, and then 3-hydroxy-2-methylbutyryl-CoA dehydrogenases and the fatty acid multifunction proteins (At4g29010, At3g06860, At3g15290), in addition to a thiolase, but these fore-mentioned genes do not appear to be co-expressed with any of the thiolase genes (data not shown).
To confirm the co-expression groups in Fig. 5A, we used Genevestigator (Zimmermann et al. 2004) to cluster KAT2, KAT5 and ACAT2 and their cohort of highlighted co-expressed genes across a series of microarray data based on tissue specific expression (Fig. 5B). This bootstrapped cluster tree showed three separate groupings of genes, confirming the Expression Angler analysis of distinct expression patterns of these three thiolases, correlating with distinct roles in metabolism. Note this type of expression analysis cannot distinguish differential roles for isoforms of proteins resulting from alternative splicing as the probe sets used to determine expression do not distinguish between splice forms.
Discussion
Table 1 summarizes the results of our knowledge on the subcellular localization of thiolase protein in Arabidopsis. Although some thiolase proteins contain a predicted mitochondrial targeting signal both in vitro and in vivo protein localization assays indicate that they are not imported into mitochondria. This conflicts with proteome analysis of isolated mitochondria suggesting a mitochondrial localization for KAT2 (Heazlewood et al. 2004; Kruft et al. 2001). We propose that this is due to contamination by peroxisomal proteins and that KAT2 is not an authentic mitochondrial protein. Heazlewood et al (2004) reported a low level of contamination of their mitochondrial samples with peroxisomes, consistent with the potential for some false positive identifications in this shotgun proteomic study. The apparent requirement of a thiolase for isoleucine catabolism in mitochondria (Taylor et al. 2004) is not a strong argument for the role of KAT2 in mitochondria, as mitochondrial type I enzymes in animals are structurally distinct from Arabidopsis KATs (Fig. 1). The terminal step and isoleucine degradation might be best served by the type II rather than a type I enzyme, and we have now shown convincingly that type II thiolases are in the cytosol and/or peroxisome in Arabidopsis (Fig. 2).
Our data suggests that at least in Arabidopsis, thiolases involved in β-oxidation are not present in mitochondria, despite the fact that some biochemical evidence has suggested this may take place in mitochondria of pea (Masterson and Wood 2001). The results presented here however cannot be definitive for all plant species as it is possible that genes encoding thiolases in other plant species may have mitochondrial targeting ability due to the fact that at least some thiolase genes in Arabidopsis encode proteins that have predicted mitochondrial targeting ability. Thus relatively small changes are likely required to achieve mitochondrial targeting of plant thiolases, as has been observed with some peroxisomal thiolases from other organisms (Danpure et al. 2003; Tsukamoto et al. 1994).
The data on enzymes required for isoleucine degradation from 2-methyl-3-hydroxybutyryl-CoA through to propionyl-CoA increasingly suggests that this part of the biochemical pathway is a non-mitochondrial activity. Of the three 3-hydroxy-2-methylbutyryl-CoA dehydrogenases in Arabidopsis (At4g29010, At3g06860, At3g15290), At3g06860 has been located to peroxisomes by three separate reports using GFP tagging (Cutler et al. 2000; Koh et al. 2005; Tian et al. 2004) and At3g15290 has been located to chloroplasts by mass spectrometry (Kleffmann et al. 2004). The type II ACAT thiolases are all non-mitochondrial (Fig. 2), being present in either the cytosol or peroxisome from our own data. Transport of 2-methyl-3-hydroxybutyryl-CoA out of mitochondria has not been investigated, but the substrate specificity of an array of known mitochondrial carriers from the Mitochondrial Carrier Protein (MCP) family, the Preprotein and Amino acid Transporter (PRAT) family and ATP Binding Cassette (ABC) transporters remain to be studied in Arabidopsis (Pohlmeyer et al. 1997; Rassow et al. 1999; Brugiere et al. 2004; Picault et al. 2004). The distribution of pathways of amino acid biosynthesis and metabolism between organelles and the cytosol is relatively common in plants, but in the case of isoleucine metabolism, although the metabolic enzymes involved are now relatively clear, the transport activities that facilitate this pathway between mitochondria, the cytosol and the peroxisome remain to be elucidated.
Co-expression analysis of transcript data can be a powerful tool to confirm other data or provide leads for further analysis. In this case, the co-expression results for KAT2 are consistent with all our experimental data. This gene encodes a peroxisomal thiolase and is co-expressed with other peroxisomal proteins involved in the same process, namely β-oxidation of fatty acids. For ACAT2 the subcellular location, enzyme class and co-expression also coincide to suggest a role in mevalonate biosynthesis leading to isoprenes. The KAT5 co-expression result was a surprise as this protein was suspected to be involved in β-oxidation based on its enzyme class. However, the different location of KAT5.1 and KAT5.2 (Table 1), the fact that KAT5 does not maintain β-oxidation in seedlings of the KAT knockout but can partially complement for the lack of KAT2 when driven by 35S expression (Germain et al 2001), and the co-expression link with flavonoid biosynthesis rather than β-oxidation genes (Fig. 5), suggests that while KAT5 encodes a thiolase, it has a distinct role to KAT2 in acyl-CoA metabolism in plants.
Abbreviations
- AOX:
-
Alternative oxidase
- ACAT:
-
Acetoacetyl-CoA thiolase
- KAT:
-
3-Ketoacyl-CoA thiolase
- Rubisco SSU:
-
Small subunit of Ribulose 1,5-bisphosphate carboxylase/oxygenase
References
Baker A, Sparkes IA (2005) Peroxisome protein import: some answers, more questions. Curr Opin Plant Biol 8:640–647
Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18:298–305
Bathgate B, Baker A, Leaver CJ (1989) Two genes encode the adenine nucleotide translocator of maize mitochondria. Isolation, characterisation and expression of the structural genes. Eur J Biochem 183:303–310
Brugiere S, Kowalski S, Ferro M, Seigneurin-Berny D, Miras S, Salvi D, Ravanel S, d’Herin P, Garin J, Bourguignon J, Joyard J, Rolland N (2004) The hydrophobic proteome of mitochondrial membranes from Arabidopsis cell suspensions. Phytochemistry 65:1693–1707
Campanella JJ, Bitincka L, Smalley J (2003) MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform 4:29
Chew O, Whelan J (2004) Just read the message: a model for sorting of proteins between mitochondria and chloroplasts. Trends Plant Sci 9:318–319
Chew O, Rudhe C, Glaser E, Whelan J (2003) Characterization of the targeting signal of dual-targeted pea glutathione reductase. Plant Mol Biol 53:341–356
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786
Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97:3718–3723
Danpure CJ, Lumb MJ, Birdsey GM, Zhang X (2003) Alanine:glyoxylate aminotransferase peroxisome-to-mitochondrion mistargeting in human hereditary kidney stone disease. Biochim Biophys Acta 1647:70–75
Derrick S, Large PJ (1993) Activities of the enzymes of the Ehrlich pathway and formation of branched-chain alcohols in Saccharomyces cerevisiae and Candida utilis grown in continuous culture on valine or ammonium as sole nitrogen source. J Gen Microbiol 139:2783–2792
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Emanuelsson O, Elofsson A, von Heijne G, Cristobal S (2003) In silico prediction of the peroxisomal proteome in fungi, plants and animals. J Mol Biol 330:443–456
Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21:2912–2922
Fujiki Y, Sato T, Ito M, Watanabe A (2000) Isolation and characterization of cDNA clones for the e1beta and E2 subunits of the branched-chain alpha-ketoacid dehydrogenase complex in Arabidopsis. J Biol Chem 275:6007–6013
Fukao T, Scriver CR, Kondo N (2001) The clinical phenotype and outcome of mitochondrial acetoacetyl-CoA thiolase deficiency (β-ketothiolase or T2 deficiency) in 26 enzymatically proved and mutation-defined patients. Mol Genet Metab 72:109–114
Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde JP, Bryce JH, Graham IA, Smith SM (2001) Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid β-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J 28:1–12
Graham IA, Eastmond PJ (2002) Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res 41:156–181
Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16:241–256
Heazlewood JL, Tonti-Filippini J, Verboom RE, Millar AH (2005) Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol 139:598–609
Hooks MA (2002) Molecular biology, enzymology, and physiology of ß-oxidation. In: Baker A, Graham I (eds) Plant peroxisomes. Kluwer Academic Publishers, London, pp 19–55
Hua S, Sun Z (2001) Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17:721–728
Johnson TL, Olsen LJ (2003) Import of the peroxisomal targeting signal type 2 protein 3-ketoacyl-coenzyme a thiolase into glyoxysomes. Plant Physiol 133:1991–1999
Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjolander K, Gruissem W, Baginsky S (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14:354–362
Koh S, Andre A, Edwards H, Ehrhardt D, Somerville S (2005) Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J 44:516–529
Kruft V, Eubel H, Jansch L, Werhahn W, Braun HP (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol 127:1694–1710
Kumar M, Verma R, Raghava GP (2006) Prediction of mitochondrial proteins using support vector machine and hidden markov model. J Biol Chem 281:5357–5363
Lange PR, Eastmond PJ, Madagan K, Graham IA (2004) An Arabidopsis mutant disrupted in valine catabolism is also compromised in peroxisomal fatty acid β-oxidation. FEBS Lett 571:147–153
Lee MN, Whelan J (2004) Identification of signals required for import of the soybean F(A)d subunit of ATP synthase into mitochondria. Plant Mol Biol 54:193–203
Lister R, Chew O, Lee MN, Heazlewood JL, Clifton R, Parker KL, Millar AH, Whelan J (2004) A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol 134:777–789
Masterson C, Wood C (2001) Mitochondrial and peroxisomal β-oxidation capacities of organs from a non-oilseed plant. Proc Biol Sci 268:1949–1953
Mockler TC, Chan S, Sundaresan A, Chen H, Jacobsen SE, Ecker JR (2005) Applications of DNA tiling arrays for whole-genome analysis. Genomics 85:1–15
Murcha MW, Elhafez D, Millar AH, Whelan J (2004) The N-terminal extension of plant mitochondrial carrier proteins is removed by two-step processing: the first cleavage is by the mitochondrial processing peptidase. J Mol Biol 344:443–454
Murcha MW, Millar AH, Whelan J (2005) The N-terminal cleavable extension of plant carrier proteins is responsible for efficient insertion into the inner mitochondrial membrane. J Mol Biol 351:16–25
Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16:260–269
Pereto J, Lopez-Garcia P, Moreira D (2005) Phylogenetic analysis of eukaryotic thiolases suggests multiple proteobacterial origins. J Mol Evol 61:65–74
Picault N, Hodges M, Palmieri L, Palmieri F (2004) The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci 9:138–146
Pohlmeyer K, Soll J, Steinkamp T, Hinnah S, Wagner R (1997) Isolation and characterization of an amino acid-selective channel protein present in the chloroplastic outer envelope membrane. Proc Natl Acad Sci USA 94:9504–9509
Pracharoenwattana I, Cornah JE, Smith SM (2005) Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17:2037–2048
Rassow J, Dekker PJ, van Wilpe S, Meijer M, Soll J (1999) The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol 286:105–120
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4:1581–1590
Tanudji M, Dessi P, Murcha M, Whelan J (2001) Protein import into plant mitochondria: precursor proteins differ in ATP and membrane potential requirements. Plant Mol Biol 45:317–325
Taylor NL, Heazlewood JL, Day DA, Millar AH (2004) Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134:838–848
Thirkettle-Watts D, McCabe TC, Clifton R, Moore C, Finnegan PM, Day DA, Whelan J (2003) Analysis of the alternative oxidase promoters from soybean. Plant Physiol 133:1158–1169
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, Rhee SY, Raikhel NV, Citovsky V (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135:25–38
Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The botany array resource: e-Northerns, expression angling, and promoter analyses. Plant J 43:153–163
Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T, Osumi T (1994) Characterization of the signal peptide at the amino terminus of the rat peroxisomal 3-ketoacyl-CoA thiolase precursor. J Biol Chem 269:6001–6010
van Roermund CW, Waterham HR, Ijlst L, Wanders RJ (2003) Fatty acid metabolism in Saccharomyces cerevisiae. Cell Mol Life Sci 60:1838–1851
Whelan J, Hugosson M, Glaser E, Day DA (1995) Studies on the import and processing of the alternative oxidase precursor by isolated soybean mitochondria. Plant Mol Biol 27:769–778
Winning BM, Sarah CJ, Purdue PE, Day CD, Leaver CJ (1992) The adenine nucleotide translocator of higher plants is synthesized as a large precursor that is processed upon import into mitochondria. Plant J 2:763–773
Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, Pham P, Cheuk R, Karlin-Newmann G, Liu SX, Lam B, Sakano H, Wu T, Yu G, Miranda M, Quach HL, Tripp M, Chang CH, Lee JM, Toriumi M, Chan MM, Tang CC, Onodera CS, Deng JM, Akiyama K, Ansari Y, Arakawa T, Banh J, Banno F, Bowser L, Brooks S, Carninci P, Chao Q, Choy N, Enju A, Goldsmith AD, Gurjal M, Hansen NF, Hayashizaki Y, Johnson-Hopson C, Hsuan VW, Iida K, Karnes M, Khan S, Koesema E, Ishida J, Jiang PX, Jones T, Kawai J, Kamiya A, Meyers C, Nakajima M, Narusaka M, Seki M, Sakurai T, Satou M, Tamse R, Vaysberg M, Wallender EK, Wong C, Yamamura Y, Yuan S, Shinozaki K, Davis RW, Theologis A, Ecker JR (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302:842–846
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Acknowledgements
This work was funded through grants from the Australian Research Council (ARC) Centre of Excellence Programme to JW, SS and AHM. AHM is funded as an ARC Queen Elizabeth II Fellow and SS as an ARC Federation Fellow.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Carrie, C., Murcha, M.W., Millar, A.H. et al. Nine 3-ketoacyl-CoA thiolases (KATs) and acetoacetyl-CoA thiolases (ACATs) encoded by five genes in Arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria. Plant Mol Biol 63, 97–108 (2007). https://doi.org/10.1007/s11103-006-9075-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-006-9075-1