Abstract
For nearly eight millennia, opium poppy (Papaver somniferum) has been bred and cultivated for therapeutic purposes. The medicinal properties of the plant are conferred by specialized metabolites known as benzylisoquinoline alkaloids (BIAs), comprising the narcotic analgesics morphine and codeine, the antimicrobial agent sanguinarine, and the potential anticancer drug noscapine. In addition, naturally occurring thebaine is used for the semi-synthesis of widely prescribed pain-relievers (e.g., oxycodone and hydrocodone), valuable drugs used in the treatment of opioid addiction (i.e., naltrexone), or antidotes for opioid overdose (i.e., naloxone). The complex stereochemistry of many opiates hinders their chemical synthesis and opium poppy remains the sole commercial source of these important pharmaceuticals. For decades, opium poppy has served as a model plant for research aimed at a comprehensive understanding of BIA metabolism. Recent progress in functional genomics has enabled the discovery of a nearly complete collection of BIA biosynthetic genes, many of which are clustered in the opium poppy genome. Advances in synthetic biology have facilitated the successful reconstitution of several BIA biosynthetic pathways in heterologous hosts such as Saccharomyces cerevisiae and Escherichia coli, although the initially low production levels suggest that commercial scale-up will present additional challenges. This review provides an update of key molecular and biochemical aspects of BIA metabolism in opium poppy, including recent biosynthetic gene discoveries, genomic organization, novel BIA transporters, metabolic regulation, and major efforts in the engineering of pathways in plants and microbes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Opium poppy (Papaver somniferum) is a flowering plant in the Papaveraceae family (order Ranunculales) valued for its ornamental beauty, edible seeds and, above all, significant medicinal properties. Archeological evidence shows that opium poppy was cultivated during the Neolithic and Bronze ages, while references to the beneficial uses of the plant are found in ancient Greek, Egyptian, Indian, Chinese, and Roman texts (Brook et al. 2017). Known as the “joy plant” in Mesopotamian civilization ~ 6000 years BCE, humans have long exploited the therapeutic uses of opium poppy and refined its consumption from the dried milky latex (opium), to the sophisticated extraction of the main bioactive principle (morphine), to the industrial semi-synthesis of widely demanded drugs (oxycodone, naltrexone, and naloxone) in a contemporary billion-dollar market (Presley and Lindsley 2018).
Benzylisoquinoline alkaloids (BIAs), a structurally diverse group of plant specialized metabolites, are responsible for the pharmacological properties of opium poppy (Hagel and Facchini 2013). Notable BIAs found in the plant include (1) the narcotic analgesics morphine and codeine, recognized as essential medicines by the World Health Organization (WHO 2017); (2) thebaine, an important starting material for the semi-synthesis of pain-relievers (oxycodone and hydrocodone), drugs prescribed to treat opioid addiction (naltrexone), and others administered to treat opioid overdose (naloxone); (3) the antimicrobial and prospective anticancer agent sanguinarine; (4) the vasodilator and smooth muscle relaxant papaverine; and (5) the cough suppressant and potential anticancer drug noscapine (Beaudoin and Facchini 2014).
Opium poppy latex or straw is extracted to obtain morphinans for medicinal purposes or for the illicit conversion of morphine to O,O-diacetylmorphine (heroin), the second most widely used recreational drug after cannabis (Dinis-Oliveira 2019). Opioids remain a sensitive topic due to their dual role, both beneficial and harmful, in human health (Presley and Lindsley 2018; Leshner 2019). Safer alternatives to the crop-based production of legitimate medicines derived from opium poppy are desirable. However, the structural complexity of pharmaceutically important BIAs, including the occurrence of several chiral centers, poses a significant challenge to their chemical synthesis at a commercial scale. For example, although more than 30 schemes for the lab synthesis of morphine and its derivatives have been reported, none is currently suitable for the industrial production of the drug (Reed and Hudlicky 2015). Likewise, metabolic engineering approaches in BIA-producing plants or cell cultures have been attempted, but are often hindered due to limitations in the availability of genetic tools and the lengthy development cycles (Diamond and Desgagne-Penix 2016). Both prokaryote and eukaryote microbes have recently emerged as promising platforms to assemble complex pathways leading to BIAs such as noscapine, sanguinarine, morphine, codeine, thebaine, and several derivatives. Nonetheless, although significantly improved in recent years, synthetic biology strategies are not yet feasible for industrial synthesis (Ehrenworth and Peralta-Yahya 2017) and the traditional farming of opium poppy continues to be the sole commercial source of many important phytopharmaceuticals.
The medicinal importance of BIAs has long motivated investigations into their chemistry and biosynthesis, beginning with the groundwork of radiotracer experiments in the 1960s, which established several fundamental metabolic pathways including the general type and sequence of most biochemical reactions (Battersby 1992). To date, ~ 40 BIA biosynthetic genes have been discovered and characterized in opium poppy, expedited by increasing accessibility to metabolite profiling, transcriptome sequencing, recombinant DNA technologies, functional genomics resources, and large-scale sequencing projects such as the recently reported opium poppy genome (Guo et al. 2018).
Comprehensive reviews comprising more than a century of breakthroughs in BIA metabolism (Hagel and Facchini 2013) and its peculiarities in opium poppy plants (Beaudoin and Facchini 2014) have recently been published. Extensive work during the past 5 years has advanced our knowledge of the organization of BIA biosynthetic genes, uncovered novel enzymes leading to the elucidation of branched pathways to major alkaloids such as morphine and noscapine, and increased the spectrum and feasibility of BIA industrial biosynthesis through innovative metabolic engineering strategies. Ongoing research, updated in this review to August 2019, is continuing to provide new and unexpected insight into BIA metabolism in opium poppy.
Biosynthesis of major BIAs in opium poppy
BIAs are mainly found in the order Ranunculales, specifically the Papaveraceae, Ranunculaceae, Berberidaceae, and Menispermaceae families (Hagel and Facchini 2013; Hagel et al. 2015a, b), although several BIAs have also been reported in unrelated families such as the Nelumbonaceae, in the order Proteales (Menendez-Perdomo and Facchini 2018). Biochemical and molecular phylogenetic evidence suggests a monophyletic evolution of BIA biosynthesis in angiosperms, preceding the divergence of the eudicots (Liscombe et al. 2005).
Opium poppy plants are known to produce a collection of BIAs with many structural subgroups (i.e., 1-benzylisoquinolines, morphinans, protoberberines, protopines, benzo[c]phenanthridines, phthalideisoquinolines, and aporphines), with the latex of the aerial organs being the preferred site for alkaloid accumulation (morphine, codeine, thebaine, papaverine, or noscapine, depending on the chemotype), whereas sanguinarine and magnoflorine primarily accumulate in the roots (Hagel and Facchini 2013; Beaudoin and Facchini 2014; Morris and Facchini 2016). Regardless of a marked structural diversity, BIAs share a common biosynthetic origin (Fig. 1).
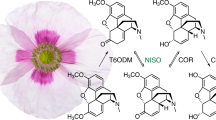
Major BIA biosynthetic pathways in opium poppy leading to (S)-reticuline (pink), morphine (green), sanguinarine (orange), noscapine (blue), magnoflorine (purple), and papaverine (yellow). Red within each alkaloid structure highlights enzyme-catalyzed chemical conversions. Asterisks represent chiral centers in selected structures: the branch point intermediate (S)-reticuline and the end-point alkaloids morphine, noscapine, and magnoflorine. All enzymes shown have been isolated from opium poppy and functionally characterized. 4′OMT, 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase; 6OMT, norcoclaurine 6-O-methyltransferase; AT1, 1,13-dihydroxy-N-methylcanadine 13-O-acetyltransferase; BBE, berberine bridge enzyme; CAS, canadine synthase; CFS, cheilanthifoline synthase; CNMT, coclaurine N-methyltransferase; CODM, codeine O-demethylase; COR, codeinone reductase; CEX1, 3-O-acetylpapaveroxine carboxylesterase; CYP82X1, 1-hydroxy-13-O-acetyl-N-methylcanadine 8-hydroxylase; CYP82X2, 1-hydroxy-N-methylcanadine 13-O-hydroxylase; CYP82Y1, N-methylcanadine 1-hydroxylase; DBOX, dihydrobenzophenanthridine oxidase; MSH, N-methylstylopine 14-hydroxylase; N7OMT, norreticuline 7-O-methyltransferase; NCS, norcoclaurine synthase; NISO, neopinone isomerase; NMCH, N-methylcoclaurine 3′-hydroxylase; NOS, noscapine synthase; OMT2:OMT3, 4′-O-desmethyl-3-O-acetylpapaveroxine 4′-O-methyltransferase; P6H, protopine 6-hydroxylase; REPI, reticuline epimerase; RNMT, reticuline N-methyltransferase; SalAT, salutaridinol 7-O-acetyltransferase; SalR, salutaridine reductase; SalSyn, salutaridine synthase; SanR, sanguinarine reductase; SOMT, scoulerine 9-O-methyltransferase; SPS, stylopine synthase; STOX, tetrahydroprotoberberine oxidase; T6ODM, thebaine 6-O-demethylase; THS, thebaine synthase; TNMT, tetrahydroprotoberberine N-methyltransferase; TYDC, tyrosine decarboxylase; TyrAT, tyrosine aminotransferase. (Color figure online)
(S)-Norcoclaurine constitutes the first committed intermediate from which a wide variety of alkaloid subgroups are generated through functional group modifications and C–C/C–O coupling reactions. These biotransformations are catalyzed by enzymes belonging to different protein families: pathogenesis-related 10 protein (PR10), methyltransferase (MT), cytochrome P450 (CYP), 2-oxoglutarate/Fe(II)-dependent dioxygenase (2-ODD), FAD-linked oxidoreductase (FADOX), acetyltransferase (AT), carboxyltransferase (CXE), short-chain dehydrogenase/reductase (SDR), and aldo–keto reductase (AKR), as recently reviewed (Dastmalchi et al. 2018b). Interestingly, substrate promiscuity is a frequent characteristic of BIA-related enzymes and some of them (e.g., 4′OMT, TNMT, DBOX, CODM, T6ODM, NISO, and COR) are involved in simultaneous pathways (Fig. 1). The restricted number of protein families implicated in the biosynthetic pathways and the monophyletic origin of BIA metabolic enzymes have facilitated empirical gene discovery. To date, most genes involved in BIA biosynthesis in opium poppy have been cloned and the cognate enzymes functionally characterized, providing a solid platform for understanding the complex BIA metabolic network in the plant. The biosynthesis of the central intermediate (S)-reticuline and the end-products of major branch pathways is described below.
(S)-Reticuline
BIA biosynthesis begins with the condensation of two l-tyrosine derivatives, dopamine and 4-hydroxyphenylacetaldehyde (4-HPAA), to form (S)-norcoclaurine (Fig. 1). Despite our comprehensive knowledge of the enzymes catalyzing the conversion of (S)-norcoclaurine to the branch point intermediate (S)-reticuline, as well as each of the downstream reactions leading to complex alkaloids such as morphine, sanguinarine, and noscapine, our current understanding of the enzymes responsible for the shuttling of primary metabolites into the BIA specialized pathway is fairly limited. Early steps involve two transformations of l-tyrosine: (1) decarboxylation to tyramine by a tyrosine decarboxylase (TYDC) and further hydroxylation to dopamine by an uncharacterized hydroxylase (or alternatively, hydroxylation to l-DOPA and decarboxylation to dopamine); and (2) deamination to 4-hydroxyphenylpiruvate by a tyrosine aminotransferase (TyrAT) and subsequent decarboxylation to 4-HPAA by an uncharacterized decarboxylase (Facchini and De Luca 1994; Lee and Facchini 2011). Both TYDC and TyrAT genes have been isolated from opium poppy and the corresponding enzymes functionally characterized, providing considerable biochemical and physiological support for their involvement in the initial stage of BIA biosynthesis (Facchini and De Luca 1994, 1995; Lee and Facchini 2011).
Papaver somniferum norcoclaurine synthase (PsNCS), a member of the pathogenesis-related 10 protein (PR10) family, catalyzes the Pictet-Spengler enantioselective reaction that yields exclusively (S)-norcoclaurine, the first committed intermediate in BIA biosynthesis (Liscombe et al. 2005; Lee and Facchini 2010). Studies on an ortholog NCS from the related species Thalictrum flavum (TfNCS) showed that the enzyme adopts a homodimeric configuration, with each monomer comprising an active site located between seven antiparallel β-sheets and three α-helices (Ilari et al. 2009). Recent high-resolution X-ray crystallography, computational docking analysis, and site-directed mutagenesis experiments on TfNCS support a reaction mechanism in which dopamine binds first in the active site, followed by 4-HPAA, while Tyr108, Glu110, Lys122, and Asp141 residues facilitate the catalysis (Lichman et al. 2015, 2017). Three active PsNCS isoforms have been characterized to date (Liscombe et al. 2005; Lee and Facchini 2010; Li et al. 2016), one of which (PsNCS3) consists of a fusion protein comprising four tandem repeats of the active domain. PsNCS1 and PsNCS2, as well as each of the four catalytic domains corresponding to PsNCS3, contain the catalytic residues determined for TfNCS (Li et al. 2016). Interestingly, tandem fusions of NCS genes seem to be taxonomically restricted to the Papaveraceae family. Experimental evidence suggests that the detected fusions could contribute to an increase in the characteristically low catalytic efficiency of NCS enzymes (Li et al. 2016).
It is remarkable that the two Pictet-Spenglerases that have been characterized from the plantae kingdom (i.e., norcoclaurine synthase and strictosidine synthase, gatekeepers in the biosynthesis of benzylisoquinoline and monoterpenoid indole alkaloids, respectively) have no structural resemblance based on amino acid sequence identity or protein folding, suggesting a convergent evolution toward their function (Stockigt et al. 2011). In contrast, three enzymes implicated in BIA biosynthesis belonging to the PR10 protein family (i.e., norcoclaurine synthase, thebaine synthase, and neopinone isomerase) share no functional relationship among them (Lee and Facchini 2010; Chen et al. 2018; Dastmalchi et al. 2019a). Interestingly, opium poppy latex is rich in PR10 proteins and, although their functions remain unknown, it could be foreseen that some others may be involved in BIA metabolism (Chen et al. 2018; Dastmalchi et al. 2019a).
(S)-Norcoclaurine is further O- and N-methylated by the enzymes norcoclaurine 6-O-methyltransferase (6OMT) and coclaurine N-methyltransferase (CNMT) to form (S)-N-methylcoclaurine (Facchini and Park 2003; Ounaroon et al. 2003). Crystallographic structures have been solved for T. flavum 6OMT (Tf6OMT) and Coptis japonica CNMT (CjCNMT), increasing our molecular understanding of the methyltransferase reaction mechanism (Robin et al. 2016; Bennett et al. 2018). Tf6OMT catalysis occurs through His256 base-assisted deprotonation of the C6 hydroxyl on the substrate, followed by a nucleophilic attack of the formed phenolate anion on the SAM methyl group (Robin et al. 2016). CjCNMT crystallographic data together with site-directed mutagenesis support a mechanism in which His208 functions as a general base to deprotonate the substrate ammonium ion, generating the nucleophilic free amine required for the methylation step (Bennett et al. 2018). Since Tf6OMT and CjCNMT share more than 60% amino acid sequence identity with opium poppy 6OMT and CNMT, respectively, and the purported catalytic determinants His256, Asp257, and Glu315 (Tf6OMT) or Glu204, His208, and Thr261 (CjCNMT) are conserved in the corresponding orthologs from opium poppy, it is likely that analogous enzymatic mechanisms take place in the plant.
(S)-N-Methylcoclaurine is sequentially converted to (S)-3′-hydroxyl-N-methylcoclaurine and (S)-reticuline via 3′-hydroxylation followed by 4′-O-methylation catalyzed by the enzymes N-methylcoclaurine 3′-hydroxylase (NMCH) and 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase (4′OMT), respectively (Huang and Kutchan 2000; Facchini and Park 2003; Ziegler et al. 2005; Frick et al. 2007). The described enzymatic order from (S)-norcoclaurine to (S)-reticuline has been inferred by the substrate preference of the required enzymes, transcriptional suppression experimental data, and alkaloid profile analyses, although it is expected that, far from the restricted linear arrangement depicted in Fig. 1, the biosynthetic pathway operates through a complex network (Desgagne-Penix and Facchini 2012). In opium poppy (S)-reticuline serves as a central branch point intermediate from which different pathways arise leading to morphinan (e.g., morphine), benzo[c]phenanthridine (e.g., sanguinarine), phthalideisoquinoline (e.g., noscapine) and aporphine (e.g., magnoflorine) alkaloids (Fig. 1).
Morphine
Defined as “God’s own medicine” by the renowned Canadian physician Sir William Osler, morphine is a potent analgesic drug acting on the opioid receptors in the central nervous system. In nature, morphine is exclusively produced by opium poppy and the related species, poppy of Troy (P. setigerum) to a lesser extent. The “principium somniferum” was first isolated from opium poppy latex by Friedrich Sertürner more than two centuries ago, whereas the elucidation of the complex pentacyclic structure was accomplished by Nobel Prize in Chemistry recipient, Sir Robert Robinson in 1925 and later confirmed by chemical synthesis in the 1950s (Brook et al. 2017; Presley and Lindsley 2018). However, the final molecular component involved in the biosynthetic pathway leading to morphine from (S)-reticuline has been determined just this year (Dastmalchi et al. 2019a).
The stereochemical inversion of (S)-reticuline to (R)-reticuline is the gateway reaction to morphinan formation (Fig. 1). A milestone in the complete elucidation of morphine biosynthesis was the discovery of the enzyme catalyzing the required epimerization reaction (Farrow et al. 2015; Winzer et al. 2015). Reticuline epimerase (REPI, also known as STORR) is a unique protein fusion, which consists of an N-terminal cytochrome P450 domain responsible for the oxidation of (S)-reticuline to 1,2-dehydroreticuline and a C-terminal aldo–keto reductase domain that reduces 1,2-dehydroreticuline to (R)-reticuline. Opium poppy plants in which REPI was silenced showed a significant reduction in morphine, codeine, and thebaine levels, but accumulated (S)-reticuline (and derived alkaloids such as noscapine), supporting the key physiological role of the novel enzyme in morphinan biosynthesis (Farrow et al. 2015). Gene family analysis of the closest paralogs of the cytochrome P450 and aldo–keto reductase modules of REPI suggested that the gene fusion might have been the result of a duplication event, followed by an 865 bp deletion between the two tightly linked genes, before the occurrence of opium poppy whole-genome duplication around 7.8 million years ago (Guo et al. 2018). It has been proposed that, besides ensuring co-expression and co-localization of the two domain activities, additional advantages of REPI protein fusion could be an efficient substrate channeling between near catalytic sites and a shielding effect over the unstable 1,2-dehydroreticuline intermediate (Farrow et al. 2015; Winzer et al. 2015). The discovery of REPI fusion, together with the detection of gene tandem fusions for several NCS in opium poppy and other Papaveraceae species (Li et al. 2016), despite some evident functional differences (i.e., combination of two enzymes catalyzing independent conversions versus repeated domains that catalyze the same reaction), highlights the largely unnoticed significance of gene fusions in BIA metabolism (Hagel and Facchini 2017).
(R)-Reticuline undergoes a stereo- and regiospecific C–C phenol-coupling reaction catalyzed by the enzyme salutaridine synthase (SalSyn; CYP719B1) to form salutaridine (Gesell et al. 2009). Salutaridine is successively converted to (7S)-salutaridinol and (7S)-salutaridinol 7-O-acetate by salutaridine reductase (SalR) and salutaridinol 7-O-acetyltransferase (SalAT), respectively (Grothe et al. 2001; Ziegler et al. 2006). The subsequent formation of the first pentacyclic morphinan, thebaine, was long considered to be a spontaneous process (Lenz and Zenk 1995), with limited evidence suggesting an enzymatic step (Fisinger et al. 2007). A recent breakthrough in the thebaine biosynthetic pathway has altered this longstanding belief via the detection and functional characterization of a novel enzyme from opium poppy latex, thebaine synthase (THS). THS is a member of the PR10 protein superfamily with a dimeric quaternary structure (Chen et al. 2018). Two THS active isoforms were confirmed in vivo using an S. cerevisiae heterologous experimental system and a THS transcript suppression in opium poppy plants. Unexpectedly, the search for the THS gene exposed its localization in a cluster containing all genes needed for thebaine formation from (S)-reticuline, consisting of two tandemly repeated REPI and SalSyn genes and two inverted sets of SalAT, SalR, and THS genes (Chen et al. 2018). The recently assembled opium poppy genome (2n = 22 and size nearly 2.7 Gb), included the thebaine biosynthetic gene cluster and a previously reported noscapine biosynthetic gene cluster (Winzer et al. 2012) as part of one larger BIA cluster on chromosome 11, altogether spanning 584 kb (Guo et al. 2018). It is remarkable that 15 genes from BIA biosynthesis in opium poppy account for one of the nearly 30 known plant biosynthetic gene clusters associated with specialized metabolism (Nutzmann et al. 2016; Schlapfer et al. 2017). The clustering of genes strictly associated with the biosynthesis of thebaine (and noscapine) might suggest a selective pressure promoting the co-inheritance of favorable allele combinations and/or a coordinated gene expression at the chromatin level for these BIAs (Nutzmann et al. 2016; Guo et al. 2018).
From thebaine, a bifurcation between major and minor pathways has been proposed to occur for the final steps in morphine biosynthesis. In the major pathway, thebaine is O-demethylated at the C6 to neopinone by thebaine 6-O-demethylase (T6ODM) (Hagel and Facchini 2010). The crystal structure of opium poppy T6ODM has been recently solved (Kluza et al. 2018) and its large substrate binding cavity was purported to account for the previously detected enzyme promiscuity towards several alkaloid subgroups such as 1-benzylisoquinolines, protoberberines, and protopines, with marked differences in backbone structure with respect to the preferred morphinan substrates (Farrow and Facchini 2013). However, the lack of a crystallographic structure of T6ODM combined with BIA substrates precluded any further examination of the catalytic determinants and reaction mechanism of the enzyme. In the minor pathway, thebaine is O-demethylated at the C3 to oripavine by codeine O-demethylase (CODM) and subsequently converted to neomorphinone by T6ODM (Hagel and Facchini 2010). The discovery and functional characterization of T6ODM and CODM, the first known members of the 2-oxoglutarate/Fe(II)-dependent dioxygenase family to catalyze O-demethylation reactions, constituted a milestone in morphine biosynthesis elucidation (Hagel and Facchini 2010). Both enzymes are encoded by genes located as three CODM and five T6ODM (plus partial and pseudogene variants) tandem copies in chromosomes 1 and 2 of opium poppy, respectively (Guo et al. 2018).
Both neopinone and neomorphinone have been proposed to undergo spontaneous isomerization to codeinone and morphinone, respectively. The latest advance in the elucidation of morphine biosynthesis has revolutionised this perspective with the discovery, and biochemical and physiological characterization, of neopinone isomerase (NISO), a novel enzyme responsible for the bidirectional isomerization between neopinone-codeinone and neomorphinone-morphinone (Dastmalchi et al. 2019a). NISO transcripts are significantly abundant in the latex, the major site for the conversion of thebaine to morphine (as well as for alkaloid storage) and virus-induced gene silencing (VIGS) experiments resulted in a significant decrease in morphine levels. Site-directed mutagenesis experiments revealed key domains for NISO activity supporting a proposed catalytic mechanism that involves the stabilization of a dienolate intermediate and subsequent stereoselective proton transfer. Three NISO genes were found clustered together in the opium poppy genome in a scaffold that did not contain other known BIA genes (Dastmalchi et al. 2019a), in agreement with previous reports that T6ODM, COR, and CODM (the other biosynthetic genes involved in the conversion of thebaine to morphine and codeine) are not part of the thebaine biosynthetic gene cluster (Chen et al. 2018; Guo et al. 2018). Similar to THS, NISO is a member of the PR10 protein family that has filled in the catalytic role of a long presumed spontaneous reaction; and the third PR10 protein involved in morphine biosynthesis, stressing the significance of the independent recruitment of members of this superfamily into BIA metabolism.
Codeinone (major pathway) and morphinone (minor pathway) are subsequently converted to codeine and morphine, respectively, by codeinone reductase (COR), an aldo–keto reductase that catalyzes the reversible reactions (Unterlinner et al. 1999). It has been recently demonstrated that COR is also responsible for the irreversible reduction of neopinone to neopine and neomorphinone to neomorphine, structural isomers of codeine and morphine, respectively (Dastmalchi et al. 2018a), although these compounds do not accumulate in opium poppy due to NISO activity (Dastmalchi et al. 2019a). Two inverted copies of COR genes have been detected in chromosome 7 of opium poppy and two other genes have been found in an unplaced scaffold (Guo et al. 2018). Recent site-directed mutagenesis experiments on five COR isoforms suggested that residues Val25, Lys41, Phe129, and Trp279 are crucial for the enzyme stability and catalytic efficiency (Dastmalchi et al. 2018a), although insufficient information regarding the protein structure prevented a more detailed examination of their precise role. A minor but detectable effect of CODM in the conversion of codeinone to morphinone, neopinone to neomorphinone, and neopine to neomorphine has been established, although codeine is the preferred substrate of the enzyme, which undergoes O-demethylation at C3 to yield morphine, the major product of the pathway (Hagel and Facchini 2010; Dastmalchi et al. 2018a, 2019a).
Sanguinarine
The biosynthesis of sanguinarine (Fig. 1), a planar and achiral benzo[c]phenanthridine, begins with the introduction of a protoberberine ring system in the (S)-reticuline backbone via a C2-C2′ stereospecific coupling reaction catalyzed by the berberine bridge enzyme (BBE) to form (S)-scoulerine (Winkler et al. 2008; Hagel et al. 2012). In opium poppy, the protoberberine (S)-scoulerine is a common intermediate in the formation of two other BIA structural subgroups: phthalideisoquinolines (i.e., noscapine) if O-methylated and benzo[c]phenanthridines (i.e., sanguinarine) if oxidized. Two BBE genes have been isolated from opium poppy and the corresponding enzymes functionally characterized (Facchini et al. 1996; Hagel et al. 2012). Interestingly, BBE isoforms are differentially expressed in the stem and roots, which could be related to a potential dual role of the enzymes in noscapine and sanguinarine biosynthesis (Hagel et al. 2012).
In the pathway leading to benzo[c]phenanthridine alkaloids, (S)-scoulerine undergoes two consecutive methylenedioxy bridge formations, catalysed by the cytochrome P450s cheilanthiofoline synthase (CFS; CYP719A25) and stylopine synthase (SPS; CYP719A20) to yield cheilanthiofoline and stylopine, respectively (Fossati et al. 2014). In opium poppy, both enzymes are exclusively or predominantly expressed in the roots (Fossati et al. 2014), consistent with sanguinarine organ-specific accumulation in the plant. Subsequently, (S)-cis-N-methylstylopine is formed through N-methylation of (S)-stylopine, catalyzed by the enzyme tetrahydroprotoberberine cis-N-methyltransferase (TNMT). Opium poppy TNMT is a promiscuous enzyme and its preference for stylopine and canadine (among other protoberberines) matches the shared role in the biosynthesis of sanguinarine and noscapine (Liscombe and Facchini 2007). Recent crystallographic work, combined with the functional characterization of the Glaucium flavum ortholog (GfTNMT), has enlightened our understanding of the structural differences that distinguish TNMTs from other N-methyltransferases involved in BIA metabolism (i.e., CNMT, PavNMT, and RNMT) (Lang et al. 2019). Notably, GfTNMT possesses a hydrophobic l-shaped substrate recognition pocket that exclusively accepts the bent conformation of cis-protoberberine substrates, explaining the cis-isomer stereospecificity. Site-directed mutagenesis suggested that residues Tyr81, Glu204, Glu207, and His208, surrounding the amino (BIA) and methyl (SAM) groups, are key for catalysis. Due to the significant amino acid sequence identity (> 80%) between PsTNMT and GfTNMT and the absolute conservation of the purported catalytic determinants, a similar reaction mechanism can be proposed for PsTNMT.
Next, hydroxylation by the root-specific (S)-cis-N-methylstylopine 14-hydroxylase (MSH; CYP82N4) causes ring opening to yield protopine and the further hydroxylation by protopine 6-hydroxylase (P6H; CYP82N3) leads to spontaneous ring rearrangement to form dihydrosanguinarine (Beaudoin and Facchini 2013). Finally, the enzyme dihydrobenzophenanthridine oxidase (DBOX) catalyzes a two-electron oxidation (completing the ring aromatization) to yield sanguinarine (Hagel et al. 2012). Although DBOX is also involved in the last step of papaverine biosynthesis (Hagel et al. 2012), the enzyme has shown a higher substrate turnover for dihydrosanguinarine compared to tetrahydropapaverine and it is exclusively expressed in opium poppy roots, where benzophenanthridine biosynthesis occurs. Several opium poppy sanguinarine reductase (SanR) isoforms have been isolated and characterized, catalyzing a DBOX opposite reaction with exclusive activity towards sanguinarine, which has been suggested as a mechanism to reduce the cytotoxic effects of sanguinarine in the plant (Beaudoin 2015).
Noscapine
The phthalideisoquinoline alkaloid noscapine was discovered by the French pharmacist Jean-François Derosne in 1803, but despite its early isolation most advances regarding its biosynthetic genes and enzymes have been accomplished during the last decade (Chen et al. 2015). Eleven enzymatic steps are required to convert (S)-reticuline to noscapine (Park et al. 2018) and notably, 10 out of the 12 biosynthetic genes implicated in the pathway are contained in the noscapine cluster (Winzer et al. 2012), recently determined to be part of the more complex BIA biosynthetic gene cluster (Guo et al. 2018). Interestingly, the noscapine cluster excludes two genes (BBE and TNMT) coding for enzymes that are not exclusively involved in noscapine formation. It has been proposed that BBE and TNMT, also implicated in sanguinarine biosynthesis, may have been subject to selective pressure to remain out of the cluster, most likely because of the different spatial biosynthesis of sanguinarine (roots) versus noscapine (aerial organs) in opium poppy (Guo et al. 2018).
Noscapine biosynthesis in opium poppy has been extensively reviewed (Chen et al. 2015) however, new experimental evidence (Li and Smolke 2016; Park et al. 2018) has prompted a revision and adjustment of the previously proposed biosynthetic outline. The branched pathway leading to noscapine begins with the cyclization reaction catalyzed by BBE to form (S)-scoulerine (Fig. 1). The sequential transformation of (S)-scoulerine to noscapine has been determined largely based on opium poppy VIGS experimental data combined with a thorough biochemical characterization of the candidate enzymes. The successive reactions include (S)-scoulerine O-methylation by scoulerine 9-O-methyltransferase (SOMT) (Dang and Facchini 2012; Winzer et al. 2012), closure of the isoquinoline moiety via methylenedioxy bridge formation by canadine synthase (CAS; CYP719A19) (Winzer et al. 2012; Dang and Facchini 2014a), and N-methylation by TNMT to yield (S)-N-methylcanadine (Liscombe and Facchini 2007). Two consecutive hydroxylation reactions catalyzed by N-methylcanadine 1-hydroxylase (CY82Y1) and 1-hydroxy-N-methylcanadine 13-O-hydroxylase (CYP82X2) are followed by the addition of a protective acetyl group by 1,13-dihydroxy-N-methylcanadine 13-O-acetyltransferase (AT1), preceding another hydroxyl group incorporation by 1-hydroxy-13-O-acetyl-N-methylcanadine 8-hydroxylase (CYP82X1), spontaneous ring opening via cleavage of the N7-C8 bridge, and 4′-O-desmethyl-3-acetylpapaveroxine formation (Winzer et al. 2012; Dang and Facchini 2014b; Dang et al. 2015).
A further 4′-O-methylation was known to be key in noscapine formation, but both the enzyme responsible for the activity and its substrate were challenging to come across. VIGS experiments showed that OMT2 (encoded in the noscapine cluster) had a significant role in noscapine biosynthesis (Winzer et al. 2012), but attempts to corroborate its function in vitro (Dang and Facchini 2012) and in vivo using yeast (Li and Smolke 2016) were unable to detect the expected activity. Remarkably, when OMT2 was co-expressed with either OMT3 (also encoded in the noscapine cluster) or 6OMT (participating in the BIA common pathway to (S)-reticuline) in a yeast strain capable of producing noscapine, the catalytic 4′OMT activity was detected (Li and Smolke 2016). This work demonstrated, for the first time, the capability of plant O-methyltransferases to form functional heterodimers, though it incorrectly pointed to narcotoline as the enzyme–substrate. This statement was promptly corrected by extensive experimental work involving in vitro assays with opium poppy stem protein extracts and purified recombinant heterodimers (OMT2:OMT3 and OMT2:6OMT) and all six possible intermediates in the noscapine biosynthetic pathway, unambiguously demonstrating that the unique substrate for the heterodimeric enzyme was 4′-O-desmethyl-3-acetylpapaveroxine (not previously tested) to form 3-O-acetylpapaveroxine and that narcotoline was formed most likely as the result of a pathway bifurcation before the 4′-O-methylation occurred (although narcotoline might also be converted to noscapine via a non-characterized mechanism) (Park et al. 2018). In addition, site-specific mutagenesis experiments together with VIGS analysis, established that both OMT2:OMT3 and OMT2:6OMT heterodimers are physiologically relevant but the OMT2 monomer is the key subunit needed for dimerization and subsequent 4′-O-methylation (Park et al. 2018).
Lastly, 3-O-acetylpapaveroxine undergoes ester hydrolysis by 3-O-acetylpapaveroxine carboxylesterase (CXE1) and noscapine synthase (NOS) catalyzes the dehydrogenation of narcotine hemiacetal to yield noscapine (Winzer et al. 2012; Chen and Facchini 2014; Dang et al. 2015).
Magnoflorine
Recent work has also shed light on the biosynthesis of magnoflorine (Fig. 1), an aporphine alkaloid found in opium poppy roots (Morris and Facchini 2016). Two metabolic routes are possible starting from reticuline, although the stereochemistry of magnoflorine from opium poppy plants (and hence, the implication of (R)- and/or (S)-reticuline) remains to be determined. In the first route, a C8–C2′ coupling reaction is catalyzed by the enzyme corytuberine synthase (CTS; CYP80G2) to yield corytuberine, followed by its N-methylation to form the quaternary amine magnoflorine. The alternative path entails the same reactions in the inverse order, involving the 1-benzylisoquinoline tembetarine (instead of corytuberine) as the intermediate. Although C. japonica CTS has been functionally characterized (Ikezawa et al. 2008), an ortholog from opium poppy has not yet been isolated. Instead, a cDNA encoding a novel reticuline N-methyltransferase (RNMT) preferentially expressed in the roots was recently isolated from opium poppy plants and comprehensively characterized (Morris and Facchini 2016). Taking into account that (1) RNMT accepted corytuberine as substrate, whereas C. japonica CTS does not accept tembetarine (Ikezawa et al. 2008) and (2) RNMT transcript suppression through VIGS experiments significantly reduced magnoflorine levels and increased corytuberine accumulation (while tembetarine and reticuline were not affected), it has been suggested that the major route to magnoflorine in opium poppy is via corytuberine rather than tembetarine formation (Morris and Facchini 2016).
Papaverine
Papaverine is a 1-benzylisoquinoline optically-inactive alkaloid that accumulates preferentially in the aerial organs of opium poppy (Farrow and Facchini 2015). Discovered in the nineteenth century by the son of the Merck Corporation’s founder, first synthesized by Amé Pictet in 1909, and commercially produced by chemical synthesis half a century ago, papaverine is a widely established antispasmodic drug (Ricci et al. 2016; Presley and Lindsley 2018). However, due to limited information regarding key biosynthetic enzymes, the pathway leading to papaverine has not been completely elucidated to date (Fig. 1). Nonetheless, two alternative routes have been proposed involving N-methylated (NCH3 pathway) or N-desmethylated (NH pathway) intermediates (Beaudoin and Facchini 2014).
The NCH3 pathway implies that papaverine biosynthesis, as for all other major BIAs, occurs via (S)-reticuline. This route has been supported by isotope labeling experiments using opium poppy seedlings, in which (S)-coclaurine and (S)-reticuline, as well as (S)-laudanine and (S)-laudanosine, were partially incorporated into papaverine (Han et al. 2010). According to this hypothesis, (S)-reticuline is subject to two consecutive O-methylations, first by reticuline 7-O-methyltransferase (7OMT) to (S)-laudanine (Ounaroon et al. 2003) and then by an uncharacterized 3′OMT to (S)-laudanosine, further N-demethylated to (S)-tetrahydropapaverine by a purported N-demethylase. The alternative NH pathway involves (S)-coclaurine as the gateway intermediate, which is hydroxylated by an uncharacterized 3′-hydroxylase to form (S)-6-O-methylnorlaudanosoline and further O-methylated by the consecutive action of 4′OMT, norreticuline 7-O-methyltransferase (N7OMT), and an uncharacterized 3′OMT to form (S)-norreticuline, (S)-norlaudanine, and (S)-tetrahydropapaverine, respectively (Ziegler et al. 2005; Pienkny et al. 2009). Decisive evidence pointing at the NH pathway as the major route contributing to papaverine biosynthesis in opium poppy plants was provided through VIGS experiments targeting N7OMT (strictly related to the NH pathway), which showed a significant decrease in papaverine accumulation, whereas the silencing of CNMT (exclusively involved in the NCH3 pathway) produced a substantial increase in papaverine content (Desgagne-Penix and Facchini 2012). No effect on papaverine levels was observed by the suppression of NMCH and 7OMT transcripts, although the corresponding enzymatic products are required for the formation of (S)-reticuline and the purported NCH3 pathway intermediate (S)-laudanine, respectively. The detection of norreticuline and norlaudanine (proposed intermediates in the NH pathway) in opium poppy latex, together with comparative transcriptome studies showing up-regulation of N7OMT and down-regulation of 7OMT in a high-papaverine cultivar, further supported the NH pathway as the main route in the plant (Desgagne-Penix and Facchini 2012; Pathak et al. 2013).
In this regard, it has been previously established that opium poppy NMCH does not accept coclaurine as a substrate for 3′-hydroxylation (Huang and Kutchan 2000) and hence, a redundant enzyme is likely responsible for this particular biotransformation. In addition, although silencing of the opium poppy SOMT1 isoform (primarily involved in noscapine biosynthesis) significantly affected papaverine accumulation due to residual 3′- and 7-O-methyltransferase activity of the enzyme on (S)-norreticuline and/or (S)-reticuline (Dang and Facchini 2012), a more efficient 3′OMT exclusively implicated in papaverine biosynthesis remains to be characterized. Though a recent study predicted the involvement of a novel 3′OMT enzyme in the conversion of (S)-norlaudanine to (S)-tetrahydropapaverine by in silico modeling and in planta silencing experiments (Agarwal et al. 2019); in vitro experimental data supporting the enzyme catalytic activity with physiologically significant substrates (e.g., (S)-norlaudanine, (S)-6-O-methylnorlaudanosoline, or (S)-laudanine) is still required for a conclusive functional characterization of this purported 3′OMT.
The last step in papaverine biosynthesis is catalyzed by DBOX via a two double bond formation and ring aromatization of (S)-tetrahydropapaverine (Hagel et al. 2012). The enzyme has been thoroughly characterized in vitro and in vivo, supporting its dual physiological role in papaverine and sanguinarine biosynthesis (Hagel et al. 2012). The observed restriction of DBOX transcripts to the roots could imply that papaverine biosynthesis does not necessarily occur in the aerial organs in which it accumulates (Hagel et al. 2012), although certain opium poppy cultivars also accumulate papaverine in the roots, at a lesser extent (Farrow and Facchini 2015). A novel 2-oxoglutarate/Fe(II)-dependent dioxygenase from opium poppy named papaverine 7-O-demethylase (P7ODM), which catalyzes the 7-O-demethylation of papaverine to pacodine, has also been isolated and characterized, adding new information about papaverine metabolism in the plant (Farrow and Facchini 2015). It is expected that, with the recent publication of the opium poppy genome and the well-established platforms for BIA biosynthetic gene discovery, the completion of the papaverine pathway is imminent.
Regulation of alkaloid metabolism
Cellular localization and transport
Studies on BIA biosynthetic gene expression, enzymatic transformations, and alkaloid accumulation in opium poppy revealed a complex arrangement interconnecting three cell types: companion cells, sieve elements, and laticifers (Bird et al. 2003; Samanani et al. 2006; Onoyovwe et al. 2013). The sieve elements, where most BIA biosynthetic reactions are proposed to occur, are incomplete cells unable to carry out gene expression and protein biosynthesis, functions that are taken over by the companion cells. Hence, it has been suggested that BIA-related enzymes must be translocated from the companion cells to the sieve elements, and then the biosynthetic products transported to the laticifers associated with the plant phloem. The major site for BIA accumulation in opium poppy plants is the latex (i.e., the laticifer cytoplasm) (Beaudoin and Facchini 2014). However, laticifers also constitute a significant site for gene expression and catalytic transformations, mostly related to the latter biosynthetic steps of major end-products (Onoyovwe et al. 2013; Chen and Facchini 2014; Dang et al. 2015; Dastmalchi et al. 2019a).
All enzymes implicated in the common biosynthetic pathway (NCS, 6OMT, CNMT, NMCH, and 4′OMT) have been exclusively detected in the sieve elements of the phloem, whereas most of the corresponding transcripts have been localized in the companion cells, confirming the specific role of different cell types in the biosynthesis of BIAs (Bird et al. 2003; Samanani et al. 2006; Lee and Facchini 2010; Onoyovwe et al. 2013). Interestingly, in specialized pathways such as those leading to morphine and noscapine, the enyzmes associated with early biosynthetic steps have also been reported in the sieve elements (e.g., SalSyn and BBE), whereas those implicated in latter steps have been detected in both the sieve elements and the laticifers (e.g., SalR and SalAT), or predominantely/solely in the laticifers (e.g., THS, T6ODM, NISO, COR, CODM, CXE1, and NOS) (Onoyovwe et al. 2013; Chen and Facchini 2014; Chen et al. 2018; Dastmalchi et al. 2019a). These findings suggest that both the sieve elements and the laticifers host critical catalytic steps of partitioned pathways, requiring the transport of BIA intermediates between these cells. The export of BIA intermediates from the sieve elements to the apoplast and its uptake by the laticifers could occur through a symplastic transport via plasmodesmata and/or apoplastic movement facilitated by transporters (Onoyovwe et al. 2013).
Until recently, the mechanisms underlying BIA intercellular transport in opium poppy plants remained unknown. The latest breakthrough in BIA metabolism arose from research involving purine permease-type sequences linked to the previously reported BIA biosynthetic gene clusters (Winzer et al. 2012; Chen et al. 2018), which led to the discovery of a unique family of transporters specific for BIAs, designated as benzylisoquinoline alkaloid uptake permeases (BUPs) (Dastmalchi et al. 2019b). Six active isoforms (denoted as BUP1-6) were functionally characterized in engineered S. cerevisiae strains, revealing a broad but distinctive substrate acceptance profile for each transporter. For instance, expression of BUP1 in yeast consistently facilitated opiate production from precursors (e.g., l-DOPA) or pathway intermediates (e.g., (S)-reticuline and thebaine), improving the end-product titres and increasing the uptake and intracellular concentration of a wide range of supplemented BIAs such as norcoclaurine, reticuline, thebaine, codeine, and morphine (but not noscapine and papaverine); whereas other isoforms showed a marked preference for certain substrates. Curiously, BUP1 was the only detected gene linked to the BIA (noscapine) biosynthetic gene cluster (in addition to a BUP1 pseudogene linked to the thebaine gene cluster). Further characterization of the BUP1 transporter showed it was localized in the plasma membrane and predominantly expressed in opium poppy latex (as well as BUP4 and BUP6), whereas BUP5 was mostly expressed in the root, BUP2 showed low expression in the stem, and BUP3 transcripts were not detected. Although VIGS-mediated suppression of several BUP transcripts did not significantly affect alkaloid accumulation in opium poppy plants, analysis of both gene expression and substrate acceptance overlapping profiles suggests a redundant role for the transporters that might justify the low impact of their silencing on plant alkaloid levels. The discovery of BUPs constitutes the first (and so far only) report related to the cellular transport of BIAs in opium poppy.
Subcellular compartmentalization and trafficking
It has been suggested that the subcellular compartmentalization of BIAs might constitute a self-defence mechanism used by opium poppy plants to sequester toxic metabolites. A model for the subcellular localization of 12 enzymes involved in the biosynthesis of the cytotoxic alkaloid sanguinarine (which accumulates in the vacuoles of elicitor-treated opium poppy cell cultures) has been proposed (Alcantara et al. 2005; Hagel and Facchini 2012). Accordingly, NCS, BBE, and DBOX are presumably localized in the lumen of the endoplasmic reticulum (ER), cytochrome P450 enzymes (NMCH, CFS, SPS MSH, and P6H) are anchored in the cytosolic side of the ER membrane, whereas methyltransferases (6OMT, CNMT, 4′OMT, and TNMT) are contained in the cell cytosol (Alcantara et al. 2005; Lee and Facchini 2010; Hagel and Facchini 2012). Seemingly, the direct precursors of BIAs, dopamine and 4HPAA, need to be translocated to the ER lumen for the NCS-catalyzed condensation to occur, whereas the reaction product (S)-norcoclaurine will be exported to the cytosol to be converted into (S)-reticuline. The presence of BBE in the ER lumen indicates that (S)-reticuline is imported into the ER for (S)-scoulerine biosynthesis, which is then exported to the cytosol for further conversion to dihydrosanguinarine, and next internalized into the ER for the final DBOX-catalyzed reaction to form sanguinarine. This last step might favor sanguinarine sequestration in vesicles to be transported to (and stored in) the vacuole. However, there is not direct experimental evidence supporting the proposed trafficking of sanguinarine biosynthetic intermediates, and future work is still needed to confirm this elegant model.
Less information is known regarding the subcellular compartmentalization of other BIA-related enzymes, although analogies can be inferred for enzymes with similar functions. For instance, RNMT, implicated in magnoflorine biosynthesis in opium poppy roots, lacks subcellular targeting signal sequences, which might suggest that, similar to other MTs involved in BIA metabolism, the enzyme catalyzes the reaction in the cytosol (Morris and Facchini 2016).
Hormonal and transcriptional regulation
Phytohormones are often used as elicitors to study BIA biosynthetic gene response in opium poppy. For instance, application of methyl jasmonate to opium poppy cell cultures induced the expression of NMCH and BBE, but not COR, in agreement with the observed accumulation of sanguinarine but no morphine (Huang and Kutchan 2000). Likewise, methyl jasmonate-treated opium poppy capsules showed a fourfold increase in thebaine, linked to the induction of several genes from the BIA common biosynthetic pathway, as well as SalSyn and SalR (exclusive to the thebaine branch) (Gurkok et al. 2015). In addition, research directed to customize and/or enhance alkaloid levels in opium poppy has also relied on hormonal treatments. Application of gibberellic acid in combination with triacontanol (a naturally occurring plant growth promoter) produced higher opium yield as well as morphine content in opium poppy plants (Khan et al. 2007). In an opposite approach, application of the anti-gibberellic acid and plant growth retardant trinexapac-ethyl (alone or in combination with methyl jasmonate) provoked thebaine and oripavine accumulation at the expense of morphine (Cotterill 2013). However, a detailed molecular examination of the role of phytohormones in controlling BIA metabolism still needs to be addressed.
Only a few transcription factors (TFs) involved in BIA biosynthesis have been identified in opium poppy, from which WRKY is the most represented gene family. A WRKY gene was first identified in opium poppy by the analysis of thebaine-accumulating T-DNA insertion mutants (Kawano et al. 2012). A wound-induced WRKY protein highly expressed in opium poppy capsules, was proven to interact in vitro with a W-box present in promoter regions of BIA biosynthetic genes, whereas a yeast-one-hybrid assay confirmed its binding to (and transcriptional activation of) a TYDC promoter, suggesting a regulatory role in the early steps of BIA biosynthesis (Mishra et al. 2013). Another WRKY and a MYB gene (preferentially expressed in the leaves) have also been isolated from opium poppy plants (Kakeshpour et al. 2015). Remarkably, since several WRKY and MYB motifs are present in the promoter region of each of the 10 genes comprising the opium poppy noscapine biosynthetic gene cluster, it has been hypothesized that co-regulation of the pathway-related genes might occur via these TFs, at least in part (Winzer et al. 2012; Kakeshpour et al. 2015). In addition, correlational analysis between TF expression levels and alkaloid profiles among 10 opium poppy cultivars suggested the implication of a CH3 gene in BIA biosynthesis regulation (likely related to thebaine and oripavine accumulation), but the lack of a comprehensive functional characterization of the protein limits a further understanding of its possible regulatory role (Agarwal et al. 2016). Despite mounting evidence suggesting a key role for TFs in BIA metabolism, their functions have not yet been thoroughly explored in opium poppy.
Additionally, it has been suggested that endophytes located in opium poppy capsules are able to modulate BIA accumulation (Pandey et al. 2016; Ray et al. 2019). For instance, a low-alkaloid cultivar inoculated with SM1B (Acinetobacter spp.) increased morphine (> 1000%) via upregulation of TYDC, NCS, 6OMT, CNMT, NMCH, SalSyn, SalR, SalAT, and COR genes expression (Pandey et al. 2016), whereas when SM1B was combined with SM3B (Marmoricola spp.), which is capable of upregulating T6ODM and CODM gene expression, morphine content was augmented more than 2000% compared to the non-inoculated plants (Ray et al. 2019). The precise molecular mechanism by which endophytes (individually or in combination) might regulate opium poppy BIA metabolism is not clear and needs further investigation.
Experimental evidence also suggests a post-transcriptional micro-RNA-mediated regulation of BIA metabolism in opium poppy, as has been detected for several medicinal plants (Sabzehzari and Naghavi 2019). Micro-RNAs, first detected in opium poppy nearly a decade ago (Unver et al. 2010), are endogenous noncoding small RNAs that regulate gene expression in eukaryotes, either by targeted-mRNA cleavage or by affecting translation. Several micro-RNA have been predicted to target TYDC, 4′OMT2, 7OMT, BBE, SalAT, and COR transcripts (Boke et al. 2015). Although correlational analysis regarding organ-specific expression of micro-RNA and their target genes indicated a plausible regulatory role (e.g., pso-miR13 and its expected target 7OMT), future investigations are needed to gain a better insight into the role of these new components in BIA biosynthesis.
Metabolic engineering
After thousands of years of exploitation in both traditional and modern medicine, opium poppy continues to be the sole commercial source for most BIAs of pharmaceutical significance. Opioid analgesics, such as morphine, constitute essential medicines widely administered for pain management, while thebaine is the preferential feedstock for the semi-synthesis of highly demanded painkillers and drugs prescribed to treat opioid overdose or addiction. Although the total chemical synthesis of morphine has been accomplished (Reed and Hudlicky 2015), the structural complexity of this pentacyclic molecule containing five stereocenters (5R, 6S, 9R, 13S, 14R) still renders the process commercially unfeasible. Likewise, de novo chemical synthesis of thebaine is hindered by the occurrence of three chiral centers in its structure. Other opiates like the antimicrobial sanguinarine and the cough suppressant noscapine are currently under extensive research regarding their potential anticancer properties (Rida et al. 2015; Galadari et al. 2017). However, the scaffolds of natural intermediates are challenging to replicate (e.g., (S)-scoulerine is formed by BBE through a reaction that cannot be simulated in synthetic chemistry). Noscapine has been chemically synthesized using different methodologies, however, two chiral centers in the complex molecular backbone make this process non-competitive with respect to the natural product extraction from the plant (Chen et al. 2015). Only the muscle relaxant achiral papaverine is produced synthetically for industrial purposes (Hagel and Facchini 2013). However, the susceptibility of opium poppy farming to environmental factors (e.g., diseases and climate conditions), the desired safer opiate sources, and the unstable relationships between countries growing the plant and those in which the drugs are manufactured, constitute driving forces in the search for practicable synthetic alternatives. Recent progress on this topic using genetically altered plants and microbes is discussed below.
Plants
Papaver somniferum has been genetically manipulated to modulate BIA composition and yields. In principle, by altering the expression of certain genes in the BIA pathway, unwanted metabolites can be reduced (or totally removed) using different approaches such as RNA interference (RNAi), virus-induced gene silencing (VIGS) or the novel CRISRP/Cas9 system. Likewise, it is possible to increase the amounts of particular alkaloids of pharmaceutical importance that naturally exist in low quantities (e.g., by overexpressing a rate-limiting enzyme).
A major goal in opium poppy metabolic engineering is to manipulate morphinan levels in the plant. One of the first attempts to manipulate BIA-producing poppies used a COR-specific RNAi to silence the expression of this aldo–keto reductase, key for codeine and morphine formation (Allen et al. 2004). Although the transformed poppies showed decreased levels of morphinans, unexpected accumulation of (S)-reticuline (7 steps upstream intermediate) was also detected. A decade later, this intriguing result was finally clarified as the consequence of off-target co-suppression of a COR paralog, the reductase domain in the fusion protein REPI, catalyzing the (S)-to-(R)-reticuline stereochemical inversion (Farrow et al. 2015; Winzer et al. 2015). VIGS-mediated transcript suppression is a transient reverse/forwards genetics technique that has become a valuable functional genomics tool for elucidating or confirming gene function related to the common BIA biosynthetic pathway (Lee and Facchini 2010; Desgagne-Penix and Facchini 2012), morphinans (Hagel and Facchini 2010; Wijekoon and Facchini 2012; Farrow et al. 2015; Chen et al. 2018; Dastmalchi et al. 2019a), benzo[c]phenanthridines (Hagel et al. 2012), phthalideisoquinolines (Dang and Facchini 2012; Winzer et al. 2012; Dang et al. 2015; Park et al. 2018), and other major alkaloids from opium poppy (Gurkok et al. 2016; Morris and Facchini 2016). However, VIGS technique impacts gene expression at the post-transcriptional level, which decreases (but does not totally abolish) gene function, and some effects can be masked when using this technique. Recently, gene editing through CRISPR/Cas9 technology has been used to knockout opium poppy 4′OMT2 gene using synthetic and viral-based delivery systems (Alagoz et al. 2016). 4′OMT2 has a critical role for both papaverine and (S)-reticuline biosynthesis (Facchini and Park 2003; Ziegler et al. 2005; Desgagne-Penix and Facchini 2012), and its knockout affected (although not completely) the levels of all major BIAs in the leaves, including (S)-reticuline, papaverine, laudanosine, morphinans, and noscapine (Alagoz et al. 2016). This system can be exploited in the future to obtain stable transgenic plants devoid of undesired traits concerning specific alkaloid levels or general profile.
On the other hand, gene overexpression has also been useful to study the biosynthetic function of enzymes and ultimately, to manipulate BIA metabolism in opium poppy plants. For instance, overexpression of the NMCH gene led to an expected overall increase in latex alkaloid content, with little impact on BIA profile (Frick et al. 2007), whereas transgenic plants overexpressing COR (Larkin et al. 2007) or SalAT (Allen et al. 2008) showed accumulation of morphinan alkaloids.
Although considerable work on plant metabolic engineering has the potential to increase the yields (and customize the profile) of medicinally significant BIAs, the difficulties faced when working with a complex organism harboring many endogenous pathway regulatory mechanisms (still not fully understood) and the absence of high-throughput plant transformation and screening technologies, make the engineering of plants a time-consuming effort and a slow-moving field (Ehrenworth and Peralta-Yahya 2017).
Microbes
Synthetic biology is emerging as a promising alternative to traditional harvesting, chemical synthesis, or plant metabolic engineering for the industrial production of significant BIAs. During the past decade, noteworthy efforts have enabeled the integration of complex pathways into microbial hosts to produce 1-benzylisoquinolines, morphinans, benzo[c]phenanthridines, phthaleidoisoquinolines, and aporphine alkaloids (Table 1). Lab workhorses such as Escherichia coli and Saccharomyces cerevisiae, have resulted in the most convenient, cost-effective, and speedy heterologous systems (Narcross et al. 2016). Beyond question, the reconstruction of biosynthetic pathways in microbes raises numerous challenges due to poor gene expression, loss of intermediates, intrinsic metabolic regulation, and variable enzymatic performance (low catalytic efficiency, promiscuity, unstable proteins, etc.). However, as knowledge regarding BIA metabolism continues to build, more sophisticated and efficient systems are becoming available.
The first reported microbial system for the production of BIAs (Minami et al. 2008) used E. coli expressing monoamine oxidase (MAO) for dopamine deamination to 3,4-dihydroxyphenylacetaldehyde and C. japonica NCS to mediate the condensation of both precursors to norlaudanosoline (a synthetic C3′-OH analogue of norcoclaurine). This strategy avoided the inclusion of NMCH, a CYP for which expression usually constitutes a bottleneck in heterologous systems. The integration of three more well-characterized enzymes from C. japonica led to a reticuline-producing culture (11 mg/l), that could be further co-cultivated with engineered yeast for the production of other complex BIAs (i.e., protoberberine and aporphine) (Minami et al. 2008). Further modification of this system by increasing NCS expression levels (Kim et al. 2013) improved reticuline titres by nearly five-fold and the latter combination with in vitro enzymatic steps augmented the efficiency for (S)-reticuline synthesis another 12-fold, to nearly 600 ml/l (Matsumura et al. 2017). Extensive work by Nakagawa and others made possible the optimization of bacterial fermentation for the highly desired de novo biosynthesis of norlaudanosoline, reticuline, and morphinans (Table 1), although still not at a commercially feasible-scale (Nakagawa et al. 2011, 2012, 2014, 2016).
The de novo production of BIAs from simple precursors supplied by the host metabolism has been a major breakthrough for developing a practical microbial-based process. Enhancing the cell overproduction of aromatic amino acids is a critical strategy to guarantee that the primary metabolite that feeds BIA biosynthesis (l-tyrosine) is not a limitation for the efficiency of the system (Narcross et al. 2016). However, not all the catalytic enzymes participating in the conversion of l-tyrosine to (S)-norcoclaurine have been characterized and the use of non-BIA-producing-plant heterologous genes is usually required to perform the upstream steps (Table 1). For instance, chromosomal integration of a tyrosine hydroxylase (CYP76AD1) from sugar beet, engineered to enhance its catalytic activity to increase l-DOPA yields, in combination with the expression of bacterial DOPA-specific decarboxylase (DODC) for the conversion of supplemented l-DOPA to dopamine, and five other enzymes from BIA-producing plants allowed the first reported de novo production of (S)-norcoclaurine (104.6 µg/l) and (S)-reticuline (80.6 µg/l) in S. cerevisiae (DeLoache et al. 2015). In an alternative approach, yeast endogenous gene modification (to increase the metabolic flux towards l-tyrosine and 4-HPAA biosynthesis), the introduction of a complex array of heterologous genes coding for the mammalian feedback inhibition-resistant tyrosine hydroxylase (TyrHM), and additional enzymes required for the replenishment of its cosubstrate tetrahydrobiopterin, was used to increase the de novo production of l-DOPA, and the subsequent formation of norcoclaurine and reticuline (Trenchard et al. 2015). Both fermentative systems produced BIAs at a comparable, yet low yields (DeLoache et al. 2015; Trenchard et al. 2015).
A common bottleneck for norcoclaurine production (and henceforth, for the downstream pathways) is the low catalytic efficiency of NCS proteins. In a screening of NCS orthologs in transcriptome databases among BIA-producing plant species aimed to detect more efficient variants in engineered S. cerevisiae it was found that, while Papaver bracteatum NCS5 (PbNCS5) produced the highest titres of (S)-reticuline (11 mg/l), the catalytic performance of each candidate positively correlated with the number of repeated domains (i.e., four-domain PsNCS3 was more efficient than three-domain PbNCS5) (Li et al. 2016). This discovery could have significant implications for the improvement of current synthetic biology platforms. In addition, a novel synthetic DNA landing pad system designed for CRISPR/Cas9-mediated precise control of multicopy gene integration in S. cerevisiae was recently used to screen NCS homologs from eight BIA-accumulating species, in order to assess the effect of gene copy number on (S)-norcoclaurine de novo production (Bourgeois et al. 2018). In most cases, the authors detected a positive correspondance between the increase in gene copy number and product yields (e.g., integration of up to four gene copies corresponding to catalytically efficient NCS doubled the amount of product formed when compared to the respective single copy strains). This approach could help to circumvent bottlenecks related to NCS catalytic low efficiency by facilitating a precise gene copy number tuning for efficient NCS variants.
The microbial biosynthesis of morphinan alkaloids is challenging for several reasons, including the number of side-products that can be generated, the broad substrate specificity of enzymes, and combinatorial order in which the reactions can occur (Narcross et al. 2016). Nonetheless, morphine has been produced in engineered yeast from thebaine (Thodey et al. 2014) or codeine (Fossati et al. 2015) and morphinan-derived drugs (hydrocodone, oxycodone, hydromorphone, and dihydrocodeine) have been microbially produced from thebaine (Thodey et al. 2014; Dastmalchi et al. 2019a) or from simple carbon sources as in the case of hydrocodone (Galanie et al. 2015). Other semi-synthesized pharmaceuticals such as buprenorphine and naltrexone are chemically synthesized from thebaine (Ehrenworth and Peralta-Yahya 2017), which has already been succesfully made by de novo synthesis in a sophisticated pathway reconstruction that required the complex assembly of 17 heterologous genes from BIA-producing plants, mammals, and bacteria (Galanie et al. 2015). Even though this innovative platform introduced numerous modifications to redirect carbon flux through tyrosine to (S)-reticuline; overexpressed bottleneck enzymes such as TyrHM, 4′OMT, and NCS; corrected N-terminal sorting and glycosylation of SalSyn by protein engineering; and deleted/overexpressed yeast native genes, the thebaine yields ultimately produced (< 0.001 mg/l) still need to be increased by more than five orders of magnitude to become an efficient alternative to opium poppy farming (Galanie et al. 2015). Additional protein engineering approaches can further improve morphine biosynthesis, such as the swapping of amino acid regions between T6ODM and CODM, which was shown to produce a CODM variant that selectively demethylates codeine, avoiding thebaine shuttling through the minor pathway (Runguphan et al. 2012).
Recently, two PR10 proteins related to the biosynthesis of morphinans have been discovered in opium poppy, filling in the catalytic role for previously alleged spontaneous reactions. Thebaine synthase (THB) is responsible for the conversion of (7S)-salutaridinol 7-O-acetate to thebaine (Chen et al. 2018), while neopinone isomerase (NISO) catalyzes the conversion of neopinone/neomorphinone to codeinone/morphinone (Dastmalchi et al. 2019a). The inclusion of THB (integrated and/or episomal) in engineered yeast containing another eight chromosomally integrated BIA-biosynthetic genes (Table 1) substantially increased thebaine titers by nearly 25-fold when racemic norlaudanosoline was administered in the culture (Chen et al. 2018). This strategy could be implemented to improve previously reported systems for opioid fermentation (Fossati et al. 2015; Galanie et al. 2015) in which thebaine was only produced in low yields by spontaneous intramolecular cyclization of the unstable precursor (7S)-salutaridinol 7-O-acetate. NISO also has an eminent potential for the perfection of current synthetic biology approaches, which usually suffer from carbon loss in alternate pathways (Fossati et al. 2015; Galanie et al. 2015). The formation of unwanted by-products such as neopine and neomorphine by the irreversible action of COR, -which can account for nearly half of alkaloid products in engineered yeast (Thodey et al. 2014), but occur at trace levels or not at all in opium poppy plants (Dastmalchi et al. 2018a), could be circumvented by the inclusion of NISO in yeast strains expressing T6ODM and COR (and CODM), a strategy that has already been proven to increase the conversion of thebaine to codeine (and morphine) (Dastmalchi et al. 2019a). Experimental evidence based on NISO and COR gene expression under promoters with different strengths suggests that the ratio between both enzymes should be tuned in future optimizations of engineered yeast systems for codeine production (Dastmalchi et al. 2019a), similar to the previous observation regarding T6ODM:COR:CODM (2:1:3) optimal gene copy number ratio for the efficient production of morphine in yeast (Thodey et al. 2014). In addition, substitution of COR by morphinone reductase (MorB), a bacterial flavoprotein oxidoreductase that catalyzes the conversion of codeinone to hydrocodone, has facilitated the yeast conversion of thebaine to this highly demanded semi-synthetic drug (Dastmalchi et al. 2019a).
The recent discovery of BUP importers localized in the laticifer plasma membrane represents a promising advance for the microbial production of valuable opiates (Dastmalchi et al. 2019b). The inclusion of BUPs in available synthetic biology platforms could significantly counteract current low yields of desirable end-products related to the efflux of pathway intermediates to the culture media. For instance, S. cerevisiae strains expressing BUPs showed considerable improvements in uptake for both early intermediates (300-fold for dopamine) and pathway intermediates (10-fold for (S)-reticuline). Moreover, a modular co-culture consisting of three engineered yeast strains, each expressing BUP1 as the interconnecting component that facilitated the uptake of intermediates, resulted in the bioconversion of supplemented l-DOPA to approximately 3 mg/l of codeine.
The de novo biosynthesis of thebaine has also been reported in an optimized E. coli stepwise fermentation (Nakagawa et al. 2016) resulting in a 300-fold increase compared with de novo thebaine biosynthesis in yeast (Galanie et al. 2015). Interestingly, different approaches for the functional expression of REPI in the prokaryotic platform were not successful due to low catalytic activity and (R,S)-reticuline was used instead for the downstream R enantioselective reaction, preventing a higher efficiency (Nakagawa et al. 2016). Further studies aimed at manipulating REPI through protein engineering might allow the inclusion of this key component in the morphinan biosynthetic pathway in bacterial cultures to build up its productivity.
The advances in the fermentative biosynthesis of morphinans could lead to inexpensive, less addictive, and more effective pharmaceuticals. Although still far from feaseable and routine commercialization (Galanie et al. 2015; Nakagawa et al. 2016), microbial-production of opioids has raised concerns related to the illicit abuse of the so-called poppy yeast (Ehrenberg 2015; Oye et al. 2015; Rinaldi 2015). Thus, future developments in this emerging field will also require engineered “safety features” accompanied by strict regulations (Narcross et al. 2016).
The biosynthesis of benzo[c]phenanthridines (e.g., sanguinarine), protoberberines and protopines have only been accomplished in eukaryotic systems (S. cerevisiae and Pichia pastoris), suitable for the functional expression of membrane-bound tailoring enzymes such as cytochrome P450s (Fossati et al. 2014; Trenchard and Smolke 2015; Hori et al. 2016). In 2014, dihydrosanguinarine was produced starting from (R,S)-norlaudanosoline via the reconstruction of three catalytic blocks integrated in the genome of S. cerevisiae for the consecutive formation of (R,S)-reticuline, (S)-stylopine, and dihydrosanguinarine, which could be further converted to sanguinarine by the ex vivo activity of a DBOX enzyme (Fossati et al. 2014). This complex multienzymatic system allowed a 1.5% yield in dihydrosanguinarine production, mostly affected by enzyme promiscuity (i.e., TNMT activity towards scoulerine and cheilanthifoline intermediates) and complexities associated with the co-expression of several P450s. Further improvement of yeast-based systems by screening plant species for highly efficient enzyme variants (and combinations), genetic manipulation, and culture-condition optimization allowed the production of (S)-cheilanthifoline, (S)-stylopine, (S)-N-methylstylopine, protopine, and sanguinarine at the highest titres reported to date (Table 1) (Trenchard and Smolke 2015).
The noscapine pathway has also been fully reconstructed in yeast (Table 1) (Li and Smolke 2016; Li et al. 2018). Early attempts were limited to the production of protoberberine intermediates (e.g., scoulerine, tetrahydrocolumbamine, and canadine) due to the largely unknown BIA biosynthetic genes (Hawkins and Smolke 2008; Minami et al. 2008; Galanie and Smolke 2015). With the discovery and functional characterization of all the enzymes implicated in the transformation of (S)-reticuline to noscapine (Chen et al. 2015; Li and Smolke 2016) it has been possible to produce the medicinal phthalideisoquinoline by synthetic biology. An initial approach integrated 16 plant enzymes into S. cerevisiae to transform racemic norlaudanosoline to noscapine (Li and Smolke 2016). A recently reported upgraded pathway based on previous work (Galanie and Smolke 2015; Trenchard et al. 2015; Li and Smolke 2016) allowed noscapine biosynthesis de novo (Li et al. 2018). Noscapine titres have been enhanced by engineering rate-limiting enzymes (i.e., NCS N-terminal truncation and TyrHM codon-optimization), boosting the NADPH supply, fundamental for P450 function (i.e., increasing copy number of NADPH regenerating genes from yeast), and optimizing culture growth conditions, leading to an over 18,000-fold system improvement (Li et al. 2018). It is expected that an additional 102–103 fold increase will be required for the scale-up of noscapine microbial-biosynthesis to be competitive (Li et al. 2018), a goal that might be reached before long.
Synthetic biology aimed at aporphine production (Table 1) has been limited by lacking information about key biosynthetic genes in BIA-producing plants. Although early efforts were able to produce magnoflorine from dopamine by mixing reticuline-producing E.coli with CNMT- and CTS-expressing S. cerevisiae cell cultures (Minami et al. 2008), new available information on RNMT, an enzyme involved in magnoflorine production in opium poppy (Morris and Facchini 2016) might be exploited to increase the aporphine yields. Although papaverine microbial biosynthesis has not been accomplished, it has been demonstrated that a simple system such as E. coli expressing Glaucium flavum OMT2, is able to produce tetrahydropapaverine (papaverine inmediate precursor) when racemic norlaudanosoline is fed (Chang et al. 2015). In addition, several FDA-approved BIA-derived drugs, such as apomorphine, used to treat Parkinson’s disease, the skeletal muscle relaxant cisatracurium, and the analgesics levorphanol and butorphanol, could also benefit from current advances in synthetic biology to improve their industrial manufacture (Ehrenworth and Peralta-Yahya 2017).
The levels of microbially synthesized drugs achieved so far are still not commercially viable but constitute an undoubtable proof of concept waiting for future improvements. The ability to assemble multistep pathways in a manageable system benefits both research and industrial applications. Gene discovery could be facilitated through a “plug-and-play” strategy using engineered microbes with the ability to perform individual sections of BIA metabolism, to circumvent low-throughput in vitro analysis of recombinant enzymes, limitations in the availability of authentic BIA substrates, and in planta strategies such as VIGS, which could be challenging to implement in some species (Morris et al. 2016). The more enzyme variants that are unveiled and characterized (i.e., novel catalytic mechanisms, substrate specificity, and/or kinetic parameters), the greater the potential for continuing to develop rational engineered platforms. In addition, when combined with combinatorial biochemistry, synthetic biology will offer a major opportunity to design an almost unlimited variety of novel pharmaceutical compounds (Facchini et al. 2012; Hagel and Facchini 2013).
Future prospects
During the past few years, extensive research on opium poppy BIA metabolism has resulted in impressive gene discoveries, including those encoding unique protein fusions (REPI), unusual heterodimers (OMT2:OMT3), enzymes filling in for long presumed spontaneous reactions (THS and NISO), and a novel family of BIA transporters. Researchers have unveiled new information about the genomic organization of opium poppy BIA biosynthetic genes, such as the occurrence of NCS tandem gene fusions (coding for the gateway enzyme of BIA biosynthesis) and the clustering of genes involved in thebaine and noscapine formation. In-depth functional and physiological characterization of new genes and their cognate enzymes has expedited a comprehensive (and presumably complete) elucidation of major biosynthetic routes leading to valuable pharmaceuticals such as morphine, codeine, thebaine, and noscapine. In addition, the readiness of whole BIA pathways at the genetic level will improve current (and enable new) metabolic engineering strategies, particularly those related to the sophisticated gene assembly in heterologous hosts for the de novo production of critical medicines. Regardless, the challenging task of increasing BIA production to industrial-scale levels in microbial systems will be facilitated by the availability of an upgraded “toolbox” of enzyme variants, together with a better understanding of BIA metabolic networks and regulatory mechanisms.
Increased access to modern technologies from the post-genomics era has transformed our approach to study BIA metabolism through functional genomics tools, including VIGS-mediated analyses, transcriptomics, proteomics, and large-scale sequencing projects such as the opium poppy genome. However, despite the tremendous advances in gene discovery, the enzymatic steps in the transformation of l-tyrosine to (S)-norcoclaurine or the branch leading to papaverine remain unknown or controversial and research aimed to fill these gaps is largely overdue. The recent discovery of opium poppy BUP cellular importers (the first opium poppy transporters known to operate on BIA metabolism) constitutes a crucial advance in our understanding of alkaloid biosynthesis in the plant and a valuable tool for improving the titres of desired end-products when mimicking the biosynthetic processes in engineered microorganisms. Additional studies addressing the cellular and subcellular localization of BIA biosynthetic enzymes will provide a thorough knowledge of metabolic channelling and transportation of BIAs. Likewise, further assessment of pathway regulatory mechanisms at different levels is still required.
After thousands of years of opium poppy exploitation for therapeutic purposes, the refined extraction of its multiple bioactive components, a detailed understanding of most BIA biosynthetic gene functions, and contemporary promising attempts to simulate plant BIA metabolism in engineered microbial factories, human efforts and ingenuity remains challenged by “nature’s own pharmacy”.
Abbreviations
- 2-ODD:
-
2-Oxoglutarate-dependent dioxygenase
- 4-HPAA:
-
4-Hydroxyphenylacetaldehyde
- 4′OMT:
-
3′-Hydroxy-N-methylcoclaurine 4′-O-methyltransferase
- 6OMT:
-
Norcoclaurine 6-O-methyltransferase
- 7OMT:
-
Reticuline 7-O-methyltransferase
- AKR:
-
Aldo-keto reductase
- AT:
-
Acetyltransferase
- AT1:
-
1,13-Dihydroxy-N-methylcanadine 13-O-acetyltransferase
- ATR:
-
Arabidopsis thaliana P450 reductase
- BBE:
-
Berberine bridge enzyme
- BIA:
-
Benzylisoquinoline alkaloid
- BUP:
-
BIA uptake permease
- CAS:
-
Canadine synthase
- Cas9:
-
CRISPR-associated protein 9
- CFS:
-
Cheilanthifoline synthase
- CNMT:
-
Coclaurine N-methyltransferase
- CODM:
-
Codeine O-demethylase
- COR:
-
Codeinone reductase
- CPR:
-
Cytochrome P450 reductase
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- CTS:
-
Corytuberine synthase
- CXE:
-
Carboxylesterase
- CXE1:
-
3-O-acetylpapaveroxine carboxylesterase
- CYP:
-
Cytochrome P450
- CYP2D6:
-
Human cytochrome P450
- CYP76AD1:
-
Tyrosine hydroxylase double mutant W13L F309L
- CYP82X1:
-
1-Hydroxy-13-O-acetyl-N-methylcanadine 8-hydroxylase
- CYP82X2:
-
1-Hydroxy-N-methylcanadine 13-O-hydroxylase
- CYP82Y1:
-
N-methylcanadine 1-hydroxylase
- DBOX:
-
Dihydrobenzophenanthridine oxidase
- DHFR:
-
Dihydrofolate reductase
- DODC:
-
l-DOPA-specific decarboxylase
- DOPA:
-
l-3,4-Dihydroxyphenylalanine
- ER:
-
Endoplasmic reticulum
- FADOX:
-
Flavin adenine dinucleotide-linked oxidoreductase
- HPLC:
-
High performance liquid chromatography
- MAO:
-
Monoamine oxidase
- morB:
-
Morphinone reductase
- MS:
-
Mass spectrometry
- MSH:
-
N-methylstylopine 14-hydroxylase
- MT:
-
Methyltransferase
- N7OMT:
-
Norreticuline 7-O-methyltransferase
- NADPH:
-
Nicotinamide adenine dinucleotide phosphate
- NCS:
-
Norcoclaurine synthase
- NISO:
-
Neopinone isomerase
- NLDS:
-
Norlaudanosoline
- NMCH:
-
N-methylcoclaurine 3′-hydroxylase
- NOS:
-
Noscapine synthase
- OMT:
-
O-methyltransferase
- OMT2:OMT3:
-
4′-O-desmethyl-3-O-acetylpapaveroxine 4′-O-methyltransferase
- P6H:
-
Protopine 6-hydroxylase
- P7ODM:
-
Papaverine 7-O-demethylase
- PCD:
-
Pterin-4-alpha-carbinolamine dehydratase
- PR10:
-
Pathogenesis-related 10 protein
- PTPS:
-
6-Pyruvoyltetrahydropterin synthase
- QDHPR:
-
Quinonoid dihydropteridine reductase
- REPI:
-
Reticuline epimerase
- RNAi:
-
RNA interference
- RNMT:
-
Reticuline N-methyltransferase
- SalAT:
-
Salutaridinol 7-O-acetyltransferase
- SalR:
-
Salutaridine reductase
- SalSyn:
-
Salutaridine synthase
- SanR:
-
Sanguinarine reductase
- SDR:
-
Short-chain dehydrogenases/reductase
- SDR-DRR:
-
1,2-Dehydroreticuline synthase-1,2-dehydroreticuline reductase
- SepR:
-
Sepiapterin reductase
- SOMT:
-
Scoulerine 9-O-methyltransferase
- SPS:
-
Stylopine synthase
- STORR:
-
S to R Reticuline
- STOX:
-
Tetrahydroprotoberberine oxidase
- T6ODM:
-
Thebaine 6-O-demethylase
- TF:
-
Transcription factor
- THS:
-
Thebaine synthase
- TNMT:
-
Tetrahydroprotoberberine N-methyltransferase
- TYDC:
-
Tyrosine decarboxylase
- TYR:
-
Tyrosinase
- TyrAT:
-
Tyrosine aminotransferase
- TyrHM :
-
Feedback inhibition-resistant tyrosine hydroxylase triple mutant R37E R38E W166Y
- VIGS:
-
Virus-induced gene silencing
References
Agarwal P, Pathak S, Lakhwani D et al (2016) Comparative analysis of transcription factor gene families from Papaver somniferum: identification of regulatory factors involved in benzylisoquinoline alkaloid biosynthesis. Protoplasma 253(3):857–871
Agarwal P, Pathak S, Kumar RS et al (2019) 3′-O-methyltransferase, Ps3′OMT, from opium poppy: involvement in papaverine biosynthesis. Plant Cell Rep. https://doi.org/10.1007/s00299-019-02439-5
Alagoz Y, Gurkok T, Zhang B et al (2016) Manipulating the biosynthesis of bioactive compound alkaloids for next-generation metabolic engineering in opium poppy using CRISPR-Cas 9 genome editing technology. Sci Rep 6:30910
Alcantara J, Bird DA, Franceschi VR et al (2005) Sanguinarine biosynthesis is associated with the endoplasmic reticulum in cultured opium poppy cells after elicitor treatment. Plant Physiol 138(1):173–183
Allen RS, Millgate AG, Chitty JA et al (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22(12):1559–1566
Allen RS, Miller JA, Chitty JA et al (2008) Metabolic engineering of morphinan alkaloids by over-expression and RNAi suppression of salutaridinol 7-O-acetyltransferase in opium poppy. Plant Biotechnol J 6(1):22–30
Battersby AR (1992) Probing nature’s pathway to alkaloids. Curr Contents 42:10
Beaudoin GA (2015) Characterization of oxidative enzymes involved in the biosynthesis of benzylisoquinoline alkaloids in opium poppy (Papaver somniferum). Dissertation, University of Calgary
Beaudoin GA, Facchini PJ (2013) Isolation and characterization of a cDNA encoding (S)-cis-N-methylstylopine 14-hydroxylase from opium poppy, a key enzyme in sanguinarine biosynthesis. Biochem Biophys Res Commun 431(3):597–603
Beaudoin GA, Facchini PJ (2014) Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta 240(1):19–32
Bennett MR, Thompson ML, Shepherd SA et al (2018) Structure and biocatalytic scope of coclaurine N-methyltransferase. Angew Chem Int Ed Engl 57(33):10600–10604
Bird DA, Franceschi VR, Facchini PJ (2003) A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15(11):2626–2635
Boke H, Ozhuner E, Turktas M et al (2015) Regulation of the alkaloid biosynthesis by miRNA in opium poppy. Plant Biotechnol J 13(3):409–420
Bourgeois L, Pyne ME, Martin VJJ (2018) A highly characterized synthetic landing pad system for precise multicopy gene integration in yeast. ACS Synth Biol 7(11):2675–2685
Brook K, Bennett J, Desai SP (2017) The chemical history of morphine: an 8000-year journey, from resin to de-novo synthesis. J Anesth Hist 3(2):50–55
Chang L, Hagel JM, Facchini PJ (2015) Isolation and characterization of O-methyltransferases involved in the biosynthesis of glaucine in Glaucium flavum. Plant Physiol 169(2):1127–1140
Chen X, Facchini PJ (2014) Short-chain dehydrogenase/reductase catalyzing the final step of noscapine biosynthesis is localized to laticifers in opium poppy. Plant J 77(2):173–184
Chen X, Dang TT, Facchini PJ (2015) Noscapine comes of age. Phytochemistry 111:7–13
Chen X, Hagel JM, Chang L et al (2018) A pathogenesis-related 10 protein catalyzes the final step in thebaine biosynthesis. Nat Chem Biol 14(7):738–743
Cotterill P (2013). Method of altering the alkaloid composition in poppy plants. International patent. WO/2005/107436
Dang TT, Facchini PJ (2012) Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol 159(2):618–631
Dang TT, Facchini PJ (2014a) Cloning and characterization of canadine synthase involved in noscapine biosynthesis in opium poppy. FEBS Lett 588(1):198–204
Dang TT, Facchini PJ (2014b) CYP82Y1 is N-methylcanadine 1-hydroxylase, a key noscapine biosynthetic enzyme in opium poppy. J Biol Chem 289(4):2013–2026
Dang TT, Chen X, Facchini PJ (2015) Acetylation serves as a protective group in noscapine biosynthesis in opium poppy. Nat Chem Biol 11(2):104–106
Dastmalchi M, Chang L, Torres MA et al (2018a) Codeinone reductase isoforms with differential stability, efficiency and product selectivity in opium poppy. Plant J. https://doi.org/10.1111/tpj.13975
Dastmalchi M, Park MR, Morris JS et al (2018b) Family portraits: the enzymes behind benzylisoquinoline alkaloid diversity. Phytochem Rev 17:249–277
Dastmalchi M, Chen X, Hagel JM et al (2019a) Neopinone isomerase is involved in codeine and morphine biosynthesis in opium poppy. Nat Chem Biol 15(4):384–390
Dastmalchi M, Chang L, Chen R et al (2019b) Purine permease-type benzylisoquinoline alkaloid transporters in opium poppy. Plant Physiol 181(3):916–933
DeLoache WC, Russ ZN, Narcross L et al (2015) An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat Chem Biol 11(7):465–471
Desgagne-Penix I, Facchini PJ (2012) Systematic silencing of benzylisoquinoline alkaloid biosynthetic genes reveals the major route to papaverine in opium poppy. Plant J 72(2):331–344
Diamond A, Desgagne-Penix I (2016) Metabolic engineering for the production of plant isoquinoline alkaloids. Plant Biotechnol J 14(6):1319–1328
Dinis-Oliveira RJ (2019) Metabolism and metabolomics of opiates: a long way of forensic implications to unravel. J Forensic Leg Med 61:128–140
Ehrenberg R (2015) Engineered yeast paves way for home-brew heroin. Nature 521(7552):267–268
Ehrenworth AM, Peralta-Yahya P (2017) Accelerating the semisynthesis of alkaloid-based drugs through metabolic engineering. Nat Chem Biol 13(3):249–258
Facchini PJ, De Luca V (1994) Differential and tissue-specific expression of a gene family for tyrosine/dopa decarboxylase in opium poppy. J Biol Chem 269(43):26684–26690
Facchini PJ, De Luca V (1995) Phloem-specific expression of tyrosine/DOPA decarboxylase genes and the biosynthesis of isoquinoline alkaloids in opium poppy. Plant Cell 7(11):1811–1821
Facchini PJ, Park SU (2003) Developmental and inducible accumulation of gene transcripts involved in alkaloid biosynthesis in opium poppy. Phytochemistry 64(1):177–186
Facchini PJ, Penzes C, Johnson AG et al (1996) Molecular characterization of berberine bridge enzyme genes from opium poppy. Plant Physiol 112(4):1669–1677
Facchini PJ, Bohlmann J, Covello PS et al (2012) Synthetic biosystems for the production of high-value plant metabolites. Trends Biotechnol 30(3):127–131
Farrow SC, Facchini PJ (2013) Dioxygenases catalyze O-demethylation and O, O-demethylenation with widespread roles in benzylisoquinoline alkaloid metabolism in opium poppy. J Biol Chem 288(40):28997–29012
Farrow SC, Facchini PJ (2015) Papaverine 7-O-demethylase, a novel 2-oxoglutarate/Fe2+-dependent dioxygenase from opium poppy. FEBS Lett 589:2701–2706
Farrow SC, Hagel JM, Beaudoin GA et al (2015) Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy. Nat Chem Biol 11(9):728–732
Fisinger U, Grobe N, Zenk MH (2007) Thebaine synthase: a new enzyme in the morphine pathway in Papaver somniferum. Nat Prod Commun 2(3):249–253
Fossati E, Ekins A, Narcross L et al (2014) Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat Commun 5:3283
Fossati E, Narcross L, Ekins A et al (2015) Synthesis of morphinan alkaloids in Saccharomyces cerevisiae. PLoS ONE 10(4):e0124459
Frick S, Kramell R, Kutchan TM (2007) Metabolic engineering with a morphine biosynthetic P450 in opium poppy surpasses breeding. Metab Eng 9(2):169–176
Galadari S, Rahman A, Pallichankandy S et al (2017) Molecular targets and anticancer potential of sanguinarine—a benzophenanthridine alkaloid. Phytomedicine 34:143–153
Galanie S, Smolke CD (2015) Optimization of yeast-based production of medicinal protoberberine alkaloids. Microb Cell Fact 14:144
Galanie S, Thodey K, Trenchard IJ et al (2015) Complete biosynthesis of opioids in yeast. Science 349(6252):1095–1100
Gesell A, Rolf M, Ziegler J et al (2009) CYP719B1 is salutaridine synthase, the C–C phenol-coupling enzyme of morphine biosynthesis in opium poppy. J Biol Chem 284(36):24432–24442
Grothe T, Lenz R, Kutchan TM (2001) Molecular characterization of the salutaridinol 7-O-acetyltransferase involved in morphine biosynthesis in opium poppy Papaver somniferum. J Biol Chem 276(33):30717–30723
Guo L, Winzer T, Yang X et al (2018) The opium poppy genome and morphinan production. Science 362(6412):343–347
Gurkok T, Turktas M, Parmaksiz I et al (2015) Transcriptome profiling of alkaloid biosynthesis in elicitor induced opium poppy. Plant Mol Biol Rep 33:673–688
Gurkok T, Ozhuner E, Parmaksiz I et al (2016) Functional characterization of 4′OMT and 7OMT genes in BIA biosynthesis. Front Plant Sci 7:98
Hagel JM, Facchini PJ (2010) Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat Chem Biol 6(4):273–275
Hagel JM, Facchini PJ (2012) Subcellular localization of sanguinarine biosynthetic enzymes in cultured opium poppy cells. Vitro Cell Dev Biol Plant 48:233–240
Hagel JM, Facchini PJ (2013) Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol 54(5):647–672
Hagel JM, Facchini PJ (2017) Tying the knot: occurrence and possible significance of gene fusions in plant metabolism and beyond. J Exp Bot 68(15):4029–4043
Hagel JM, Beaudoin GA, Fossati E et al (2012) Characterization of a flavoprotein oxidase from opium poppy catalyzing the final steps in sanguinarine and papaverine biosynthesis. J Biol Chem 287(51):42972–42983
Hagel JM, Mandal R, Han B et al (2015a) Metabolome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol 15:220
Hagel JM, Morris JS, Lee EJ et al (2015b) Transcriptome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol 15:227
Han X, Lamshoft M, Grobe N et al (2010) The biosynthesis of papaverine proceeds via (S)-reticuline. Phytochemistry 71(11–12):1305–1312
Hawkins KM, Smolke CD (2008) Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat Chem Biol 4(9):564–573
Hori K, Okano S, Sato F (2016) Efficient microbial production of stylopine using a Pichia pastoris expression system. Sci Rep 6:22201
Huang FC, Kutchan TM (2000) Distribution of morphinan and benzo[c]phenanthridine alkaloid gene transcript accumulation in Papaver somninferum. Phytochemistry 53(5):555–564
Ikezawa N, Iwasa K, Sato F (2008) Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C–C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J Biol Chem 283(14):8810–8821
Ilari A, Franceschini S, Bonamore A et al (2009) Structural basis of enzymatic (S)-norcoclaurine biosynthesis. J Biol Chem 284(2):897–904
Kakeshpour T, Nayebi S, Rashidi Monfared S et al (2015) Identification and expression analyses of MYB and WRKY transcription factor genes in Papaver somniferum L. Physiol Mol Biol Plants 21(4):465–478
Kawano N, Kiuchi F, Kawahara N et al (2012) Genetic and phenotypic analyses of a Papaver somniferum T-DNA insertional mutant with altered alkaloid composition. Pharmaceuticals (Basel) 5(2):133–154
Khan R, Khan MMA, Singh M et al (2007) Gibberellic acid and triacontanol can ameliorate the opium yield and morphine production in opium poppy (Papaver somniferum L.). Acta Agric Scandinavica B 57:307–312
Kim JS, Nakagawa A, Yamazaki Y et al (2013) Improvement of reticuline productivity from dopamine by using engineered Escherichia coli. Biosci Biotechnol Biochem 77(10):2166–2168
Kluza A, Niedzialkowska E, Kurpiewska K et al (2018) Crystal structure of thebaine 6-O-demethylase from the morphine biosynthesis pathway. J Struct Biol 202(3):229–235
Lang DE, Morris JS, Rowley M et al (2019) Structure-function studies of tetrahydroprotoberberine N-methyltransferase reveal the molecular basis of stereoselective substrate recognition. J Biol Chem 294(40):14482–14498
Larkin PJ, Miller JA, Allen RS et al (2007) Increasing morphinan alkaloid production by over-expressing codeinone reductase in transgenic Papaver somniferum. Plant Biotechnol J 5(1):26–37
Lee EJ, Facchini P (2010) Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. Plant Cell 22(10):3489–3503
Lee EJ, Facchini PJ (2011) Tyrosine aminotransferase contributes to benzylisoquinoline alkaloid biosynthesis in opium poppy. Plant Physiol 157(3):1067–1078
Lenz R, Zenk MH (1995) Acetyl coenzyme A: salutaridinol-7-O-acetyltransferase from Papaver somniferum plant cell cultures. The enzyme catalyzing the formation of thebaine in morphine biosynthesis. J Biol Chem 270(52):31091–31096
Leshner AI (2019) Integrating tactics on opioids. Science 363(6434):1367
Li Y, Smolke CD (2016) Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat Commun 7:12137
Li J, Lee EJ, Chang L et al (2016) Genes encoding norcoclaurine synthase occur as tandem fusions in the Papaveraceae. Sci Rep 6:39256
Li Y, Li S, Thodey K et al (2018) Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc Natl Acad Sci USA 115(17):E3922–E3931
Lichman BR, Gershater MC, Lamming ED et al (2015) ‘Dopamine-first’ mechanism enables the rational engineering of the norcoclaurine synthase aldehyde activity profile. FEBS J 282(6):1137–1151
Lichman BR, Sula A, Pesnot T et al (2017) Structural evidence for the dopamine-first mechanism of norcoclaurine synthase. Biochemistry 56(40):5274–5277
Liscombe DK, Facchini PJ (2007) Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J Biol Chem 282(20):14741–14751
Liscombe DK, MacLeod BP, Loukanina N et al (2005) Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 66(20):2501–2520
Matsumura E, Nakagawa A, Tomabechi Y et al (2017) Laboratory-scale production of (S)-reticuline, an important intermediate of benzylisoquinoline alkaloids, using a bacterial-based method. Biosci Biotechnol Biochem 81(2):396–402
Menendez-Perdomo IM, Facchini PJ (2018) Benzylisoquinoline alkaloids biosynthesis in sacred lotus. Molecules 23(11):2899
Minami H, Kim JS, Ikezawa N et al (2008) Microbial production of plant benzylisoquinoline alkaloids. Proc Natl Acad Sci USA 105(21):7393–7398
Mishra S, Triptahi V, Singh S et al (2013) Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS ONE 8(1):e52784
Morris JS, Facchini PJ (2016) Isolation and characterization of reticuline N-methyltransferase Involved in biosynthesis of the aporphine alkaloid magnoflorine in opium poppy. J Biol Chem 291(45):23416–23427
Morris JS, Dastmalchi M, Li J et al (2016) Plug-and-play benzylisoquinoline alkaloid biosynthetic gene discovery in engineered yeast. Methods Enzymol 575:143–178
Nakagawa A, Minami H, Kim JS et al (2011) A bacterial platform for fermentative production of plant alkaloids. Nat Commun 2:326
Nakagawa A, Minami H, Kim JS et al (2012) Bench-top fermentative production of plant benzylisoquinoline alkaloids using a bacterial platform. Bioeng Bugs 3(1):49–53
Nakagawa A, Matsuzaki C, Matsumura E et al (2014) (R,S)-tetrahydropapaveroline production by stepwise fermentation using engineered Escherichia coli. Sci Rep 4:6695
Nakagawa A, Matsumura E, Koyanagi T et al (2016) Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat Commun 7:10390
Narcross L, Fossati E, Bourgeois L et al (2016) Microbial factories for the production of benzylisoquinoline alkaloids. Trends Biotechnol 34(3):228–241
Nutzmann HW, Huang A, Osbourn A (2016) Plant metabolic clusters—from genetics to genomics. New Phytol 211(3):771–789
Onoyovwe A, Hagel JM, Chen X et al (2013) Morphine biosynthesis in opium poppy involves two cell types: sieve elements and laticifers. Plant Cell 25(10):4110–4122
Ounaroon A, Decker G, Schmidt J et al (2003) (R,S)-reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum—cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J 36(6):808–819
Oye KA, Lawson JC, Bubela T (2015) Regulate ‘home-brew’ opiates. Nature 521(7552):281–283
Pandey SS, Singh S, Babu CS et al (2016) Endophytes of opium poppy differentially modulate host plant productivity and genes for the biosynthetic pathway of benzylisoquinoline alkaloids. Planta 243(5):1097–1114
Park MR, Chen X, Lang DE et al (2018) Heterodimeric O-methyltransferases involved in the biosynthesis of noscapine in opium poppy. Plant J 95(2):252–267
Pathak S, Lakhwani D, Gupta P et al (2013) Comparative transcriptome analysis using high papaverine mutant of Papaver somniferum reveals pathway and uncharacterized steps of papaverine biosynthesis. PLoS ONE 8(5):e65622
Pienkny S, Brandt W, Schmidt J et al (2009) Functional characterization of a novel benzylisoquinoline O-methyltransferase suggests its involvement in papaverine biosynthesis in opium poppy (Papaver somniferum L). Plant J 60(1):56–67
Presley CC, Lindsley CW (2018) DARK Classics in chemical neuroscience: opium, a historical perspective. ACS Chem Neurosci 9(10):2503–2518
Ray T, Pandey SS, Pandey A et al (2019) Endophytic consortium with diverse gene-regulating capabilities of benzylisoquinoline alkaloids biosynthetic pathway can enhance endogenous morphine biosynthesis in Papaver somniferum. Front Microbiol 10:925
Reed JW, Hudlicky T (2015) The quest for a practical synthesis of morphine alkaloids and their derivatives by chemoenzymatic methods. Acc Chem Res 48(3):674–687
Ricci JA, Koolen PG, Shah J et al (2016) Comparing the outcomes of different agents to treat vasospasm at microsurgical anastomosis during the papaverine shortage. Plast Reconstr Surg 138(3):401e–408e
Rida PC, LiVecche D, Ogden A et al (2015) The noscapine chronicle: a pharmaco-historic biography of the opiate alkaloid family and its clinical applications. Med Res Rev 35(5):1072–1096
Rinaldi T (2015) “Poppy” yeast. EMBO Rep 16(11):1410
Robin AY, Giustini C, Graindorge M et al (2016) Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids. Plant J 87(6):641–653
Runguphan W, Glenn WS, O’Connor SE (2012) Redesign of a dioxygenase in morphine biosynthesis. Chem Biol 19(6):674–678
Sabzehzari M, Naghavi MR (2019) Phyto-miRNAs-based regulation of metabolites biosynthesis in medicinal plants. Gene 682:13–24
Samanani N, Alcantara J, Bourgault R et al (2006) The role of phloem sieve elements and laticifers in the biosynthesis and accumulation of alkaloids in opium poppy. Plant J 47(4):547–563
Schlapfer P, Zhang P, Wang C et al (2017) Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol 173(4):2041–2059
Stockigt J, Antonchick AP, Wu F et al (2011) The Pictet–Spengler reaction in nature and in organic chemistry. Angew Chem Int Ed Engl 50(37):8538–8564
Thodey K, Galanie S, Smolke CD (2014) A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat Chem Biol 10(10):837–844
Trenchard IJ, Smolke CD (2015) Engineering strategies for the fermentative production of plant alkaloids in yeast. Metab Eng 30:96–104
Trenchard IJ, Siddiqui MS, Thodey K et al (2015) De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab Eng 31:74–83
Unterlinner B, Lenz R, Kutchan TM (1999) Molecular cloning and functional expression of codeinone reductase: the penultimate enzyme in morphine biosynthesis in the opium poppy Papaver somniferum. Plant J 18(5):465–475
Unver T, Parmaksiz I, Dundar E (2010) Identification of conserved micro-RNAs and their target transcripts in opium poppy (Papaver somniferum L.). Plant Cell Rep 29(7):757–769
WHO (2017) WHO model list of essential medicines, 20th list. Genova WHO technical report series, no. 1006
Wijekoon CP, Facchini PJ (2012) Systematic knockdown of morphine pathway enzymes in opium poppy using virus-induced gene silencing. Plant J 69(6):1052–1063
Winkler A, Lyskowski A, Riedl S et al (2008) A concerted mechanism for berberine bridge enzyme. Nat Chem Biol 4(12):739–741
Winzer T, Gazda V, He Z et al (2012) A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336(6089):1704–1708
Winzer T, Kern M, King AJ et al (2015) Plant science. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science 349(6245):309–312
Ziegler J, Diaz-Chavez ML, Kramell R et al (2005) Comparative macroarray analysis of morphine containing Papaver somniferum and eight morphine free Papaver species identifies an O-methyltransferase involved in benzylisoquinoline biosynthesis. Planta 222(3):458–471
Ziegler J, Voigtlander S, Schmidt J et al (2006) Comparative transcript and alkaloid profiling in Papaver species identifies a short chain dehydrogenase/reductase involved in morphine biosynthesis. Plant J 48(2):177–192
Acknowledgements
This work was supported by financial contributions from a Natural Sciences and Engineering Research Council of Canada Discovery Grant to PJF (Grant No. 183573). IMMP is the recipient of the Alberta Innovates Technology Futures Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
PJF owns stock in, serves on the Board of Directors of, and is provided compensation by Willow Biosciences Inc. IMMP owns stock in Willow Biosciences Inc.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Singh, A., Menéndez-Perdomo, I.M. & Facchini, P.J. Benzylisoquinoline alkaloid biosynthesis in opium poppy: an update. Phytochem Rev 18, 1457–1482 (2019). https://doi.org/10.1007/s11101-019-09644-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11101-019-09644-w