ABSTRACT
Purpose
THIOMAB™ drug conjugates (TDCs) with engineered cysteine residues allow site-specific drug conjugation and defined Drug-to-Antibody Ratios (DAR). In order to help elucidate the impact of drug-loading, conjugation site, and subsequent deconjugation on pharmacokinetics and efficacy, we have developed an integrated mathematical model to mechanistically characterize pharmacokinetic behavior and preclinical efficacy of MMAE conjugated TDCs with different DARs. General applicability of the model structure was evaluated with two different TDCs.
Method
Pharmacokinetics studies were conducted for unconjugated antibody and purified TDCs with DAR-1, 2 and 4 for trastuzumab TDC and Anti-STEAP1 TDC in mice. Total antibody concentrations and individual DAR fractions were measured. Efficacy studies were performed in tumor-bearing mice.
Results
An integrated model consisting of distinct DAR species (DAR0-4), each described by a two-compartment model was able to capture the experimental data well. Time series measurements of each Individual DAR species allowed for the incorporation of site-specific drug loss through deconjugation and the results suggest a higher deconjugation rate from heavy chain site HC-A114C than the light chain site LC-V205C. Total antibody concentrations showed multi-exponential decline, with a higher clearance associated with higher DAR species. The experimentally observed effects of TDC on tumor growth kinetics were successfully described by linking pharmacokinetic profiles to DAR-dependent killing of tumor cells.
Conclusion
Results from the integrated model evaluated with two different TDCs highlight the impact of DAR and site of conjugation on pharmacokinetics and efficacy. The model can be used to guide future drug optimization and in-vivo studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Antibody-drug conjugates (ADCs) combine high tumor target specificity of antibodies and potent antitumor activity of cytotoxic agents and have demonstrated convincing efficacy in cancer patients (1,2). ADCs consist of an antibody conjugated to a cytotoxic agent through a linker group that is chemically linked to amino acid residues on the antibody, such as cysteines and lysines (3–5). Cysteine based conjugation often involves controlled reduction of interchain disulphide bonds in the antibody followed by oxidation to produce ADCs (3). For example, humanized IgG antibodies are conjugated to potent cytotoxic agents like monomethyl auristatin E (MMAE) via a protease-labile linker maleimidocaproyl-valine-citrulline-paminobenzyloxycarbonyl through this process (6). However, as a consequence, the resulting ADCs are often produced and administered as a heterogeneous mixture of antibodies with different drug loads or drug-to-antibody ratios (DAR). This heterogeneity leads to complex PK behavior for ADCs, and it has been reported that differences in DAR impact both disposition and efficacy (7,8). To control the heterogeneity of ADCs, a number of site specific conjugation technologies have been developed in recent years, exemplified by a new class of ADCs called THIOMAB™ drug conjugates or TDCs (9,10). TDCs contain engineered cysteine substitutions at positions on light and/or heavy chains, which provide reactive thiol groups for conjugate formation with auristatin class of cytotoxic agents like MMAE (9,11), and allow for the production and purification of homogenous DAR species.
TDCs with uniform DARs afford us the opportunity to characterize the PK properties and biological activities of TDCs with a range of well-defined DARs. Mechanistic modeling dissects the mechanisms involved in ADC disposition and action, and evaluates the relative contributions from deconjugation of the drug from the antibody, site dependent variations in deconjugation rate and differential proteolytic clearance for different DAR species (11). It also helps in identifying the optimal drug load which can maximize in-vivo efficacy.
In this study, we examined the PK characteristics and in-vivo tumor killing efficacy of different DAR variants (DAR1, DAR2 and DAR4) of val-cit (vc)-MMAE based TDCs in mice. We developed an integrated mechanism-based mathematical model to quantitatively understand the complex disposition kinetics and pre-clinical efficacy of TDCs, which will guide further optimization in TDC development.
MATERIALS AND METHODS
Materials
Antibody/THIOMAB™ Production and MMAE Conjugation
In these studies, we used THIOMAB™ versions of the anti-STEAP1 and anti-HER2 (trastuzumab) antibody that contain single cysteine residues engineered into an IgG heavy or light chain or both. These cysteines serve as exclusive sites for conjugation to monomethyl auristatin E (MMAE) through a linker containing a thiol-reactive maleimide and the protease cleavable val-cit peptide. Methods for construction and production of the THIOMAB™ variant and its drug conjugate (TDC) were reported previously (12). Briefly, a cysteine residue was engineered at A114 position of the heavy chain and/or V205 position of the light chain to produce its THIOMAB™ HC and LC variants, respectively, from which the thio HC and LC MMAE conjugates (referred to as TDC) were generated. The drug-to-antibody ratio (DAR) for each TDC was determined by liquid chromatography-mass spectrometry (LC-MS) analysis as described earlier (13). The DAR of ADC was determined to be approximately 1, 2, or 4 per antibody using the method as described earlier (12).
Animals/In-vivo Studies
All experimental animal studies were conducted according to protocols that were reviewed and approved by the Institutional Animal care and Use Committees (IACUC) of Genentech Laboratory Animal Research (LAR).
Pharmacokinetic Studies in Non-Tumor Bearing Mice
Animals ranging from 6 to 8 weeks old and weighing approximately 18.9-26.6 g at the initiation of the study were randomly assigned to each of the groups. The pharmacokinetics of the trastuzumab unconjugated antibodies and their respective MMAE conjugated variants with various DARs of 1, 2, or 4 at a single dose level of 5 mg/kg were evaluated following a single intravenous (IV) dose in female Nu/Nu female mice (n = 9 per group), while those of anti-STEAP1 were evaluated at dose levels of 2 or 3 mg/kg in male severe combined immunodeficient (SCID) beige mice (n = 12 per group). Blood samples were collected from each animal via the femoral vein for up to 28 days and used to derive plasma for total antibody concentrations by ELISA and relative DAR levels by affinity capture LC-MS (14).
Efficacy Studies in Tumor Bearing Mice
Trastuzumab TDCs were evaluated in female Fo5 mammary tumor bearing CRL nu/nu mice. Animals were approximately 11 weeks old and weighed approximately 25 g each. Female nu/nu mice were implanted with Fo5 tumor fragments (10–20 mm3 in size) at the thoracic mammary fat pad region. When mean tumor size reaches 100–300 mm3, mice were randomly grouped into 7 groups of 8 mice each and given a single IV dose of vehicle or trastuzumab TDCs with DAR1, DAR2, or DAR4 at dose levels of 2.5, 5, or 10 mg/kg. Clinical observations were performed twice per week and any mouse with a tumor volume that was more than 3000 mm3 or when tumors showed signs of impending ulceration were euthanized.
Anti-STEAP1 TDCs were evaluated in male LNCAP-Ner tumor-bearing SCID-beige mice. Animals were approximately 13 weeks old and weighed approximately 23 g each. Male SCID-beige mice were subcutaneously implanted with 12.5 mg testosterone pellets (Innovative Research of America; Sarasota, FL) once every 60 days in the left flank. Two to three days after the first pellet implantation, each mouse was injected with 10 million LNCap-Ner cells subcutaneously in the right flank in a volume of 0.2 mL per mouse. Cells were suspended in HBSS/Matrigel (1:1 ratio). When mean tumor sizes reached 100–300 mm3, mice were randomly grouped into 8 groups of 7 mice each and given a single IV dose of vehicle or anti-STEAP1 TDCs with DAR1 or DAR2 at dose levels of 0.5, 1, 3, or 6 mg/kg. Clinical observations were performed twice per week and any mouse with a tumor volume that was more than 3000 mm3 or when tumors showed signs of impending ulceration were euthanized.
Analytical Measurements
ELISA
The total Anti-STEAP1 antibody concentrations were measured with an ELISA using Anti-STEAP1 anti-ID antibody 5093 for capture and a goat anti-human IgG Fc antibody conjugated to horseradish peroxidase (Bethyl Laboratories, Montgomery, TX) for detection. The total trastuzumab antibody concentrations were determined with an ELISA using NeutrAvidin (Catalog 31000; Thermo Scientific [Pierce]; Rockford, Illinois) as the coat reagent to capture biotinylated sheep anti-human IgG (Cat # AU003CUS01; Binding Site; Birmingham, UK) bridged with conjugated or unconjugated antibody, and goat anti-human IgG horseradish peroxide (HRP) as the detection reagent.
Affinity Capture Liquid Chromatography Mass Spectrometry Assay
The relative proportions (percentage) of DAR0, DAR1, DAR2 and DAR4 in plasma were determined using a novel affinity capture liquid chromatography mass spectrometric (LC-MS) assay (14). Biotinylated extracellular domain (ECD) of the STEAP1 or HER2 receptor was immobilized onto streptavidin coated paramagnetic beads. This affinity bead system was used to capture anti-STEAP1 or Trastuzumab TDC by incubating with the plasma samples at room temperature. Following affinity capture, bound Anti-STEAP1 or trastuzumab TDC species were deglycosylated on the beads in buffer overnight. Subsequently, the beads were washed extensively and the TDC analytes were eluted by appropriate organic solvent for LC-MS analysis. A proper volume of the sample was injected and separated by a reversed phase capillary LC system run at a 15 μL/min flow rate. Analytes were ionized by electrospray and detected by a Q-Star XL mass spectrometer operated in the TOF-MS mode. Anti-STEAP1 or trastuzumab TDC data was acquired using AB Sciex Analyst QS 1.1 software and the peak deconvolution was performed by AB Sciex BioAnalyst software to obtain the peak area under curve for each component. Relative ratio for the anti-STEAP1 or Trastuzumab TDC components was calculated.
Tumor Volume Measurement
Tumor dimensions were measured using UltraCal-IV calipers (Model 54-10-111, Fred V. Fowler Company; Newton, MA) and volume was calculated according to the following formula: Tumor Volume (mm3) = (length • width2) • 0.5.
Pharmacokinetic/Pharmacodynamic Modeling
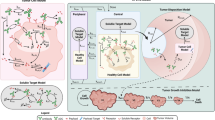
Figure 1 depicts the schematic of the integrated PKPD model developed. The final model consists of 9 distinct species representing TDCs with DAR 0–4 with intact drug conjugation at up to 2 heavy chain (H) and up to 2 light chain (L) sites:: HHLL, HLL, LHH, LL, HH, LH, L, H and DAR0. For each of the 9 species, the model explicitly accounts for drug-deconjugation driven clearance (ie, transition to a lower DAR species) and antibody clearance (ie, elimination of the species). Based on previous in-vitro and in-vivo studies and our understanding of the TDC disposition mechanisms, the rate of deconjugation-driven transition of higher DAR species to lower ones depends on the site of conjugation for MMAE (11). The rate constant kH represents the deconjugation of MMAE from the heavy chain site HC-A114C of the antibody molecule while kL represents deconjugation from the light chain site LC-V205C. Deconjugation-driven clearance CLL (from the light chain site) and CLH (from the heavy chain site) were calculated from their respective deconjugation rate constants as CLL = kL*VC and CLH = kH*VC and are of the units mL/Kg/day.
Schematic of mechanism-based PK-PD model for TDCs in mice. (a) Transition of higher DAR to lower DAR species through site-specific decojugation and DAR dependent antibody clearance in the central compartment, (b) representation of two compartments (central and peripheral) for all DAR species and (c) tumor progression and killing efficacy of TDCs. The model is described by equations in materials and methods section with symbols defined in Table I.
Consistent with PK profiles for ADCs/TDCs (and any monoclonal antibody in general), each species is represented by two compartments, a central and a peripheral compartment (15,16). All the different DAR species share the same parameter values for central volume of distribution (VC), peripheral volume of distribution (VP) and distribution clearance (CLD). Since none of the PK data in the study showed target mediated disposition, a linear clearance of the antibody was used. To account for the impact of DAR on antibody clearance (as opposed to deconjugation-driven clearance) of individual DAR species, the antibody clearance of the different DAR species is defined as:
CLDAR0 represents the antibody clearance of DAR0, A the exponent for DAR dependency for clearance and i, the DAR value. This form was chosen to allow flexibility in capturing the a priori unknown DAR-dependence of the antibody clearance (ie, either a linear or supralinear/concave dependence).
The differential equations describing the central compartments of the different DAR species amounts described in the PK model are:
Equation describing the amounts for different DAR species in the peripheral compartment is:
DARC and DARP represent the amount of that DAR species in the central and the peripheral compartment.
Pharmacodynamic modeling: Consistent with the tumor growth data for both Fo5 mammary tumor model and LNCaP-Ner xenograft tumor model, the tumor growth kinetics were described using a zero order rate constant (kG). The decrease in tumor volumes after trastuzumab TDC treatment in Fo5 mammary tumor model was modeled with a linear killing of tumor by each DAR species, represented by the parameter KKill. The decrease in tumor volumes after anti-STEAP1 TDC treatment in LNCAP-Ner xenograft was modeled as non-linear killing of tumor by each DAR species, represented by the parameters KMax and KC50, where KMax represents maximum killing of tumor and KC50 the concentration of MMAE containing antibody required for 50% tumor killing. The following differential equation represents tumor progression and killing:
where the subscript i ranges over all possible DAR values (0–4) and CDARi represents the total concentration of all species with a DAR of i, regardless of drug conjugation site.
Data Analysis
Model development, data fitting, parameter estimation and simulations were performed in SimBiology® (The MathWorks, Inc., Natick, MA). Data fitting and parameter estimation for PK and PD were performed in two steps. First, total antibody concentrations and fractional DAR distribution at each time point for all DAR0, DAR1 (H), DAR2 (HH) and DAR4 (HHLL) dosing were modeled simultaneously to estimate values for PK parameters. Subsequently, replicate tumor volume data from different DAR variants and different doses were modeled simultaneously to estimate parameters related to tumor progression and killing. Residual variability was modeled using the additive plus proportional variance model. The goodness-of-fit was assessed by model convergence, visual inspection of the fitted curves, and standard error (SE) of the estimated parameters.
RESULTS
The integrated PKPD model development for TDC disposition and efficacy is based on multiple time-course studies performed in mice dosed with either trastuzumab TDC or Anti-STEAP1 TDC. All PK studies were performed in normal mice given a single IV dose of purified DAR0 (LC and HC), DAR1 (H), DAR2 (HH) or DAR4 (HHLL). Measurements of total antibody concentrations and fractional DAR distribution at each time points after dosing for each DAR species were utilized to develop the model. Efficacy studies were performed in tumor-bearing mouse models. A detailed description of the animal studies is provided in the methods section.
TDC Disposition and Pharmacokinetic Model
Total antibody concentration profiles for each purified-DAR dosing experiment (DAR0, DAR1, DAR2 and DAR4 dosing) for both Thio-trastuzumab-vc-MMAE and Thio-anti-STEAP1-vc-MMAE showed multi-exponential disposition over time as shown in Figs. 2 and 3. The fractional DAR distribution data showed a decline of higher DAR species and increase in lower DAR species presumably due to deconjugation driven transition. For example, with the dosing of 5 mg/kg of DAR1 Thio-trastuzumab-vc-MMAE, almost the DAR1 fraction was almost 100% at the time of dosing, but declined over time, more quickly than the total antibody did (Fig. 2c). With the dosing of 5 mg/kg of DAR2 Thio-trastuzumab-vc-MMAE, the DAR2 fraction was almost 100%, and the decrease over time of this fraction corresponded to the simultaneous increase in DAR1 fraction, which started at 0% but increased over time, reaching maximum values at around day 14, beyond which DAR1 fractions started declining too (Fig. 2d). Finally, with the dosing of 5 mg/kg of DAR4 Thio-trastuzumab-vc-MMAE, The DAR4 fraction was almost 100% had at the time of dosing but decreased exponentially over time. Simultaneously, the DAR3 fraction increased, reaching maximum values around day 14, beyond which it started to decline. Fractions of DAR2 and DAR1, started at 0% at the time of dosing, but increased after a lag time (Fig. 2e). Similar fractional DAR distribution profiles were observed with Thio-anti-STEAP1-vc-MMAE dosing studies as shown in Fig. 3. These profiles were consistent with a high-to-low-DAR transition model.
Total antibody concentrations (•) and individual DAR fraction % measurements (*) for thio-anti-Steap1 or thio-anti-Steap1-vc-MMAE given as a single IV dose of (a) 2 mg/kg DAR0 (H), (b) 3 mg/kg DAR0 (H), (c) 3 mg/kg DAR1 (H), (d) 2 mg/kg DAR2 (HH), (e) 3 mg/kg DAR2 (HH) or (f) 2 mg/kg DAR4 (HHLL) in mice. Solid lines represent model fitting.
The pharmacokinetic model (as described in the methods section, Fig. 1) was developed based on known mechanisms of ADC/TDC disposition and data from the animal studies. The model, includes 9 distinct species representing DAR0-4, with each species represented by two compartments, with a distinct deconjugation rate constant for drug at each of two different conjugation sites,, and antibody clearance that increases exponentially with DAR values. This model was successful in describing the total antibody profiles and fractional DAR distributions for both Thio-trastuzumab-vc-MMAE and Thio-anti-STEAP1-vc-MMAE as shown in Figs. 2 and 3. The final parameter estimates are provided in Table I. The values for central volume of distribution (VC) for all the species were estimated to be 47.32 and 57.45 mL/Kg for Thio-trastuzumab-vc-MMAE and Thio-anti-STEAP1-vc-MMAE, respectively. The peripheral volume of distribution (VP) were estimated to be 51.83 and 71.96 mL/Kg and distribution clearance (CLD) values to be 137.39 and 112.2 mL/kg/day for Thio-trastuzumab-vc-MMAE and Thio-anti-STEAP1-vc-MMAE, respectively. The values for distribution volumes and distribution clearances are consistent with PK properties for a typical monoclonal antibody (17). One of the salient features of our pharmacokinetic model is the use of two different rates (or clearance) for deconjugation of MMAE from the different drug conjugation sites. The use of two different deconjugation rates is well justified as observed from the parameter estimates. Values of CLL are estimated to be 1.08 and 0.93 mL/Kg/day and values of CLH are estimated as 4.21 and 4.18 mL/Kg/day for Thio-trastuzumab-vc-MMAE and Thio-anti-STEAP1-vc-MMAE, respectively, suggesting that the deconjugation from the heavy chain site is 4 times faster than deconjugation from the light chain site. Antibody clearance of the different TDC species was assumed to depend exponentially on their DAR value with parameters CL0, the clearance value for the DAR0 species, and A, the exponent for DAR dependency for clearance; the resulting values are estimated to be CL0 = 6.70 and 5.92 mL/Kg/day and A = 0.14 and 0.04 for Thio-trastuzumab-vc-MMAE and Thio-anti-STEAP1-vc-MMAE, respectively. These estimates suggest that although the baseline antibody clearance of DAR0 for both Thio-trastuzumab-vc-MMAE and Thio-anti-STEAP1-vc-MMAE are similar, the DAR dependency shows some modest differences.
Pharmacodynamic Model for Tumor Killing Efficacy
Fo5 mammary tumor model in CRL nu/nu mice was used for testing the efficacy of different DAR species (DAR0-4) of Thio-trastuzumab-vc-MMAE and LNCAP-Ner xenograft tumor model in male SCID beige mice for Thio-anti-STEAP1-vc-MMAE. The tumor volume profiles from the vehicle treatment group provide the growth kinetics behavior for these tumors (Figs. 4a and 5a). Growth kinetics for both the tumor models showed a zero order growth rate. Growth rate constant (kG) values for Fo5 mammary tumor model were estimated as 86.83 mm3/day and growth rate constant for LNCAP-Ner xenograft tumor model were around 5 fold lower than Fo5 mammary tumor with the values estimated as 17.21 mm3/day. Other mechanistic tumor growth kinetics models including the Koch model and Simeoni model were tried, but did not improve data fitting or other model selection criteria (18,19). Thio-trastuzumab-vc-MMAE showed dose-dependent effects in inhibiting the increase in tumor volume of Fo5 mammary tumor model. As shown in Fig. 4, the tumor killing efficacy was higher with dosing of higher DAR species. Similar results were observed for Thio-anti-STEAP1-vc-MMAE efficacy in LNCAP-Ner xenograft model (Fig. 5). Consistent with the mechanism of action for TDCs, the decreases in tumor volumes were modeled as resulting from killing of tumor cells, and concentrations of MMAE attached to antibody were considered to be the driving force for tumor killing for both trastuzumab TDC and Anti-STEAP1 TDC (20). Tumor killing efficacy was directly proportional to DAR values and DAR0 (antibody by itself) did not have any effect on tumor volumes. Linear tumor killing parameter KKill was estimated to be 0.0025 (μg/mL)−1day−1 for trastuzumab TDC. Non-linear tumor killing parameters represented by KMax and KC50 were estimated to be 7 0.13 days−1 and 4.4 μg/mL for Anti-STEAP1 TDC efficacy.
Individual tumor volume (•) over time for Fo5 mammary tumor model in CRL nu/nu mice after (a) vehicle treatment or thio-trastuzumab-vc-MMAE given as a single IV dose of (b) 10 mg/kg DAR1 (H), (c) 5 mg/kg DAR2 (HH), (d) 10 mg/kg DAR2 (HH), (e) 2.5 mg/kg DAR4 (HHLL), (f) 5 mg/kg DAR4 (HHLL) or (g) 10 mg/kg DAR4 (HHLL). Solid lines represent model fitting.
Individual tumor volume (•) over time for LNCAP-Ner xenograft tumor model in male SCID beige mice after (a) vehicle treatment or thio-anti-Steap1-vc-MMAE given as a single IV dose of (b) 0.5 mg/kg DAR1 (H), (c) 1 mg/kg DAR1 (H), (d) 3 mg/kg DAR1 (H), (e) 6 mg/kg DAR1 (H), (f) 0.5 mg/kg DAR2 (HH), (g) 1 mg/kg DAR2 (HH) or (h) 3 mg/kg DAR2 (HH). Solid lines represent model fitting.
DISCUSSION
The development of THIOMAB™ with genetically engineered cysteines for conjugation to drug molecules allows control over drug loading and site of conjugation for the drugs. This provides an opportunity to limit heterogeneity, thereby reducing PK complexities and improving PK properties (21). The target DAR for antibody design is often chosen empirically with little guidance from PK, efficacy or toxicity. On one hand, higher drug load increases the ADC potency, which is evidenced by lower IC50 from in vitro cell killing assays; however, the increased drug load on the antibody disrupts the native structure/hydrophobicity of a mAb and leads to faster clearance of ADC from systemic circulation (8). We utilized purified TDCs with DAR values of 0, 1, 2 or 4 to specifically examine their pharmacokinetic and pharmacodynamic properties and gain quantitative understanding of the interplay of these variables. The integrated modeling approach used here provides insights into mechanisms of TDC disposition and actions and can guide further optimization of TDC and ADC development.
Previous studies have shown that the site of conjugation plays an important role in determining the stability of TDCs (11). These studies compared the stability of different variants of TDCs with the drug conjugation site at the light chain (LC-V205C), heavy chain-Fab (HC-A114C) and Fc region (Fc-S396C) of the antibody. Both in-vitro plasma stability assay and in-vivo time series studies showed that the site of conjugation plays an important role in the stability of drug conjugation with Fc at this amino acid site conjugation as the least stable, while the heavy chain conjugation modestly less stable than the light chain conjugation. It was also shown that these differences in the conjugation stability translated to the in-vivo efficacies with the LC variant being the most efficacious followed by the HC variant, and the Fc variant was the least efficacious in animal tumor models. Solvent accessibility and environment charge distribution were found to likely play a critical roles in determining drug conjugation stability with partial solvent accessibility and positively charged environment as observed in LC were found to form the most stable conjugates. Although we did not study the Fc variant in this study, results from our modeling analysis confirm and quantitate the previous observations that conjugation to the LC-V205C is more stable than HC-A114C conjugation. The deconjugation rate constants from the HC site were estimated to be 4 times faster than from the LC site (Table I). In addition, deconjugation rates, irrespective of the site of conjugation (LC or HC) were found to be linearly dependent on the drug load (DAR values).
Drug loading has been found to affect clearance for vc-MMAE conjugated ADCs with higher DAR species clearing faster compared to lower DAR ones. Hamblett et al. showed that Anti-CD30-Vc-MMAE dosed as a purified DAR8 species cleared 5 fold faster than dosing of the purified DAR2 species and 3 fold faster than the purified DAR4 in mice (8). In-vitro studies show that higher DAR ADCs show better potency compared to lower DAR ADCs (22). However this does not translate to in-vivo efficacies. Our results suggest that this is primarily because of the negative influence of high drug load on PK properties of ADCs and a sweet spot for the most optimal drug load exists at which the maximum in-vivo efficacies are achieved (8). Although the effects of drug loading in determining the PK properties can differ between traditional ADCs and TDCs because the conjugation to engineered cysteines in TDCs preserve the endogenous disulfide bond in the antibody, we did expect some influence on antibody clearance due to potential effects of conjugation on hydrophobicity and other molecular properties of the antibody. As expected, antibody clearance of TDC molecules was found to be DAR dependent (Table I) with higher DAR species clearing faster compared to the lower ones. We tried two different approaches to model the antibody clearance of different DAR species. The first approach was to estimate the clearance of different DAR species independent of each other. The second approach is the one described in the methods section for the final model where the clearance of different DAR species is modeled as an exponential function of its DAR value. As shown in Suppl Fig. 1, both approaches produced similar results for antibody clearance of different DAR species and fitted the data equally well suggesting that the exponential function to describe DAR dependency is reasonable. However, consistent with the relatively low values of the exponential dependence, our results suggest that at least within the range of DAR = 0–4, the antibody clearance can be adequately described by a linear dependence on DAR.
Characterization of in-vivo tumor killing efficacy is an important aspect in understanding the properties of ADCs and TDCs. In-vivo efficacies are affected both by the pharmacology and PK properties of the drug. Because of the heterogeneity in the profiles of both MMAE-based ADCs and TDCs, characterization and prediction of in-vivo efficacies become complicated. In the present study, in addition to characterizing the complex PK profiles of TDCs we were able to link them to in-vivo tumor killing efficacies in appropriate animal models. For both trastuzumab TDC and Anti-STEAP1 TDC, the antibody component just confers target specificity, but does not have any effects on tumor progression in these tumor models. This was confirmed by the absence of tumor killing in response to dosing of the DAR0 TDC species (unconjugated antibody) in these tumor models (data not shown). MMAE conjugated to the antibody confers tumor killing capability and the efficacy is dependent on the drug load. Utilization of mathematical models helps us to delineate this dependence, the tumor growth kinetics, and the PK behavior and pharmacology, thus helping us to understand the contribution of these different components to in-vivo efficacy. Tumor growth kinetics can be different for different tumor models and the use of appropriate tumor progression model is critical for the proper characterization of drug effects. The most common tumor growth kinetics shows initial exponential (or first order) followed by a linear (zero order) growth profile (18,19). However, for both the tumor models utilized in our study, we observe linear growth kinetics over the course of the study and a simple zero order rate constant was sufficient in characterizing the growth profiles. Efforts to incorporate more mechanistic components of tumor progression including transit compartments for tumor growth and killing neither improved fitting nor provided any additional insights into the mechanism of action.
One of the salient features of the study is the integrated approach that we have taken to characterize both PK and efficacy of TDCs. We generated a comprehensive dataset consisting of multiple DAR variants dosing, a variety of analytes including total antibody and different DAR fractions at each individual time points and tumor efficacy data for different DAR variants and doses. Incorporation of known mechanisms of TDC disposition and action along with this rich dataset allowed us to take a systems pharmacology approach to understand and characterize TDCs. Furthermore, testing this model structure with two different TDC molecules targeting two different cancer antigens (Her2 and STEAP1) gives us confidence in its broader applicability and suggests that auristatin MMAE conjugated TDCs shows similar mechanisms of disposition and action, irrespective of the specific antibody molecule or target. In addition, taking an integrated modeling approach will allow us to further refine the model based on evolving information/ data from newer studies and measurements. For example, both TDCs reported in this study did not show a significant target mediated disposition at the dose range studied and hence a linear clearance was sufficient to capture the PK profiles. However, for TDCs showing significant target mediated disposition, a non-linear component for the clearance can be incorporated into the model in addition to the existing clearance mechanisms already represented. Similarly, with relevant data, our current model can be modified to incorporate toxicities caused by TDC dosing.
CONCLUSION
An integrated platform model was developed to describe and predict the complex PK and efficacy profiles of THIOMAB™ ADCs in mice, and can be used to guide future drug optimization and in-vivo studies. Model results suggest that drug deconjugation rates, total antibody clearance, and tumor killing rates increase with DAR and drug deconjugation occurs more readily from the heavy chain (HC-A114C) than the light chain (LC-V205C) sites used on these TDCs, consistent with in vitro and preclinical understanding.
REFERENCES
Senter PD. Potent antibody drug conjugates for cancer therapy. Curr Opin Chem Biol. 2009;13:235–44.
Ballantyneand A, Dhillon S. Trastuzumab emtansine: first global approval. Drugs. 2013;73:755–65.
Singhand R, Erickson HK. Antibody-cytotoxic agent conjugates: preparation and characterization. Methods Mol Biol. 2009;525:445–67. xiv.
Carterand PJ, Senter PD. Antibody-drug conjugates for cancer therapy. Cancer J. 2008;14:154–69.
Schrama D, Reisfeld RA, Becker JC. Antibody targeted drugs as cancer therapeutics. Nat Rev Drug Discov. 2006;5:147–59.
Boswell CA, Mundo EE, Zhang C, Bumbaca D, Valle NR, Kozak KR, et al. Impact of drug conjugation on pharmacokinetics and tissue distribution of anti-STEAP1 antibody-drug conjugates in rats. Bioconjug Chem. 2011;22:1994–2004.
Linand K, Tibbitts J. Pharmacokinetic considerations for antibody drug conjugates. Pharm Res. 2012;29:2354–66.
Hamblett KJ, Senter PD, Chace DF, Sun MM, Lenox J, Cerveny CG, et al. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin Cancer Res. 2004;10:7063–70.
Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotechnol. 2008;26:925–32.
Behrensand, CR, Liu B. Methods for site-specific drug conjugation to antibodies. MAbs. 2013; 6.
Shen BQ, Xu K, Liu L, Raab H, Bhakta S, Kenrick M, et al. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat Biotechnol. 2012;30:184–9.
Junutula JR, Bhakta S, Raab H, Ervin KE, Eigenbrot C, Vandlen R, et al. Rapid identification of reactive cysteine residues for site-specific labeling of antibody-Fabs. J Immunol Methods. 2008;332:41–52.
Alley SC, Benjamin DR, Jeffrey SC, Okeley NM, Meyer DL, Sanderson RJ, et al. Contribution of linker stability to the activities of anticancer immunoconjugates. Bioconjug Chem. 2008;19:759–65.
Xu K, Liu L, Saad OM, Baudys J, Williams L, Leipold D, et al. Characterization of intact antibody-drug conjugates from plasma/serum in vivo by affinity capture capillary liquid chromatography-mass spectrometry. Anal Biochem. 2011;412:56–66.
Chudasama VL, Schaedeli Stark F, Harrold JM, Tibbitts J, Girish SR, Gupta M, et al. Semi-mechanistic population pharmacokinetic model of multivalent trastuzumab emtansine in patients with metastatic breast cancer. Clin Pharmacol Ther. 2012;92:520–7.
Shah DK, Haddish-Berhane N, Betts A. Bench to bedside translation of antibody drug conjugates using a multiscale mechanistic PK/PD model: a case study with brentuximab-vedotin. J Pharmacokinet Pharmacodyn. 2012;39:643–59.
Wang W, Wang EQ, Balthasar JP. Monoclonal antibody pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 2008;84:548–58.
Koch G, Walz A, Lahu G, Schropp J. Modeling of tumor growth and anticancer effects of combination therapy. J Pharmacokinet Pharmacodyn. 2009;36:179–97.
Simeoni M, Magni P, Cammia C, De Nicolao G, Croci V, Pesenti E, et al. Predictive pharmacokinetic-pharmacodynamic modeling of tumor growth kinetics in xenograft models after administration of anticancer agents. Cancer Res. 2004;64:1094–101.
Doronina SO, Toki BE, Torgov MY, Mendelsohn BA, Cerveny CG, Chace DF, et al. Development of potent monoclonal antibody auristatin conjugates for cancer therapy. Nat Biotechnol. 2003;21:778–84.
Bhakta S, Raab H, Junutula JR. Engineering THIOMABs for site-specific conjugation of thiol-reactive linkers. Methods Mol Biol. 2013;1045:189–203.
King HD, Yurgaitis D, Willner D, Firestone RA, Yang MB, Lasch SJ, et al. Monoclonal antibody conjugates of doxorubicin prepared with branched linkers: a novel method for increasing the potency of doxorubicin immunoconjugates. Bioconjug Chem. 1999;10:279–88.
ACKNOWLEDGMENTS AND DISCLOSURES
The authors would like to thank Paul J. Fielder and other PKPD colleagues for their constructive input to this work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 219 kb)
Rights and permissions
About this article
Cite this article
Sukumaran, S., Gadkar, K., Zhang, C. et al. Mechanism-Based Pharmacokinetic/Pharmacodynamic Model for THIOMAB™ Drug Conjugates. Pharm Res 32, 1884–1893 (2015). https://doi.org/10.1007/s11095-014-1582-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-014-1582-1