Abstract
Purpose
We applied in silico methods to automatically classify drugs according to the Biopharmaceutics Drug Disposition Classification System (BDDCS).
Materials and Methods
Models were developed using machine learning methods including recursive partitioning (RP), random forest (RF) and support vector machine (SVM) algorithms with ChemDraw, clogP, polar surface area, VolSurf and MolConnZ descriptors. The dataset consisted of 165 training and 56 test set molecules.
Results
RF model 3, RP model 1, and SVM model 1 can correctly predict 73.1, 63.6 and 78.6% test compounds in classes 1, 2 and 3, respectively. Both RP and SVM models can be used for class 4 prediction. The inclusion of consensus analysis resulted in improved test set predictions for class 2 and 4 drugs.
Conclusions
The models can be used to predict BDDCS class for new compounds from molecular structure using readily available molecular descriptors and software, representing an area where in silico approaches could aid the pharmaceutical industry in speeding drugs to the patient and reducing costs. This could have significant applications in drug discovery to identify molecules that may have future developability issues.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
INTRODUCTION
Efforts to improve the speed of bringing new drugs to patients are sorely needed, as highlighted by the FDA’s Critical Path Initiative. For example, since its introduction in 1995, the Biopharmaceutics Classification System (BCS) has had a significant impact on the drug regulatory process and practice. For an immediate release orally active dosage form, the rate and extent of its absorption is determined by its aqueous solubility and permeability in the gastrointestinal tract. The BCS therefore represents a new robust model for bioequivalence studies based on physiological parameters and physicochemical properties of drug molecules. The BCS as adopted by the World Health Organization (WHO), classifies the drug molecules listed on the essential medicines list (EML) based on their solubility and permeability characteristics into four different classes (Fig. 1). Accordingly, certain drug classes can be considered for a biowaiver, i.e. approval of products based on their in vitro drug dissolution tests instead of their human bioequivalence data, a costly task for drug manufacturers. Such waivers significantly improve the speed and decrease the cost of bringing orally administered therapeutics to market. Currently, the BCS system allows a waiver of in vivo bioequivalence testing of immediate-release solid dosage forms for class 1 drugs (1). Whereas waivers for class 3 drugs are recommended only based on scientific justifications (2,3).
a The Biopharmaceutics Classification System (BCS) as defined by the FDA after Kasim et al. (4). b Biopharmaceutics Drug Disposition Classification System (BDDCS) proposed by Wu and Benet where major route of elimination (metabolized vs unchanged) serves as the permeability criteria.
Drug classification according to BCS requires knowledge of solubility and permeability data. The determination of drug permeability is typically based on experimental permeability data or well-defined mass balance studies. This information is available only for a small fraction of EML listed drugs (4). A biowaiver currently can be requested for orally active immediate-release dosage forms (≥85% release in 30 min), containing drugs with high solubility over the pH range 1 to 7.5 (dose/solubility ratio <250 ml) and a high permeability (fraction absorbed ≥90%), provided excipients used in the formulation do not interfere with the drug absorption process. Drugs with narrow therapeutic range and drugs designed to be absorbed from the oral cavity may not be considered for biowaivers (5). Thus, the central idea of the BCS classification system is to predict in vivo pharmacokinetic performance of drug products from in vitro drug solubility and permeability characteristics (6).
More recently, Wu and Benet (6) extensively examined about 167 BCS classified drugs. They aptly noticed that pharmacokinetic considerations like effects of food, absorptive transporters, efflux transporters, and routes of elimination (renal/biliary) were important determinants of overall drug absorption and bioavailability for immediate release oral dosage forms. Thus, they suggested that classifying molecules based on the extent of metabolism is less ambiguous as compared to permeability or extent of absorption. This classification may also increase the number of class 1 drugs that would become eligible for biowaivers (7). The BDDCS, like BCS, proposes to classify drug molecules into four classes (Fig. 1), defining the extensive metabolism criterion as ≥50% (±10%) metabolism of an oral dose in vivo in humans. Based on this criterion, a few drugs that were previously BCS class 1 were reclassified as BDDCS class 3 and thus would not be eligible for biowaivers. A study by Takagi et al. (7) observed at least eight drugs in the BDDCS class 1 were eligible for biowaivers. Considering its overall significance, the BDDCS approach could be helpful in successfully classifying drugs in class 1, thereby increasing their eligibility for biowaivers.
A challenge for both BCS and BDDCS is the actual classification of drugs based on the required in vitro data for metabolism, solubility or permeability. However, there has been considerable research over the last decade on computational or in silico methods for prediction of absorption, distribution, metabolism and excretion (ADME) (8,9). The objective of the present study was to enable simple and fast BDDCS classification by developing computational classification models predicting BDDCS class from molecular properties. Computational models were developed based on data for 165 drugs as a training set based on BDDCS data (6). To further test and challenge our models, we have retrieved an additional 56 drugs listed in the WHO EML that were not previously classified under BDDCS but with ample literature data available to enable classification.
MATERIALS AND METHODS
Drug List
A training set of 165 drugs for computational model building was obtained from the published literature (6). An additional set of 56 drugs, that were not included in the original BDDCS, were retrieved from the WHO EML publication and were used to challenge the computational models (6,10). This collection was subsequently employed as a test set upon classification according to BDDCS criteria. Classification was established based on an extensive literature survey of drug disposition data as well as individual physicochemical parameters described below.
Solubility Definition
Drug solubility data for classification purposes was obtained from standard references (11–13) and expressed in mg/ml. Where solubility data were not available or undefined, guidelines were taken from Kasim et al. (4). Maximum dose strength data was obtained from WHO Essential Medicines core list and expressed in milligrams (10).
Dose Number calculations
The dose number (D 0) was calculated using (14):
Where M 0 is highest dose strength (mg), C s is the solubility (mg/ml) and V 0 is 250 ml.
BDDCS Classification of Compounds
Drug disposition data (Table I) for 56 previously unclassified drugs was obtained from an extensive literature search. Aqueous solubility for each therapeutic drug class was obtained from the Merck Index (14th Edition) and other pertinent literature references (11). Pharmacokinetic data such as plasma half-life, bioavailability, P-glycoprotein (P-gp) affinity, Cytochrome P450 affinity and extent of metabolism were obtained from the literature by MedLine searching using a combination of descriptive keywords and Boolean operators. Additionally, web sources such as DrugBank (15) and http://www.drugs.com were used. Based on this collective information, the drug molecules were assigned to a BDDCS class.
Computational Modeling
Data Collection and Molecule Building
The current dataset comprises 221 drug molecules collected from various literature sources. Molecules were downloaded from PubChem (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) as 2D SMILES strings which were converted to SDF format and imported into Chem3D Ultra (CambridgeSoft, Cambridge, MA) to generate MOL2 files. The molecules were subsequently energy minimized in SYBYL v.7.1 (Tripos Associates, St. Louis, MO) using the Tripos force field (16) and Gasteiger–Hückel charges with distance-dependent dielectrics and the conjugate gradient method with a convergence criterion of 0.001 kcal/mol.
Descriptor Calculations
The number of hydrogen bond donors and hydrogen bond acceptor groups were calculated with ChemDraw Ultra 8.0 (CambridgeSoft, Cambridge, MA); clogP and polar surface area (PSA) were calculated using Sybyl v.7.1. One hundred VolSurf descriptors (17) were calculated from 3D molecular fields using VolSurf 4.0 implemented in SYBYL. Five different probes including water (OH2), carbonyl oxygen atom (O), amphipathic (BOTH), carbonyl oxygen atom (O::), and Sp2N with lone pair (N:=) probes were used for descriptor calculation. Volsurf descriptors include descriptors for size, shape, hydrophilic and hydrophobic regions, interaction energy amongst other descriptors. A total of 149 Molconn-Z descriptors (2D topological) were generated (18), including κ-molecular shape indices, topological state, shape, Wiener and Shannon indices.
Model Building and Validation
Recursive Partitioning (RP) calculations were performed using the rpart module of the R package (19). RP can be used to mine large data sets in order to uncover hidden patterns within data and assign appropriate class. RP attempts to determine the relationship between a set of dependent (X) and independent variables (Y) using the simple mathematical function, Y = f(X). The result of RP is a “tree” or “decision tree” or “graph”. The data is divided (partitioned) into nodes (branches), where data with similar properties tend to occupy the same node. A tenfold cross-validation study was performed on the training set.
The R program was also used for random forest (RF) calculations (20). The total number of trees was set to 1000. The other optimizable parameter in the random forest approach is m try, i.e. the number of descriptors (p) randomly sampled as candidates for splitting at each node. When m try equals the number of descriptors (m try = p) this is commonly termed “bagging.” The number of descriptors was increased systematically with an increment of 5. In general, the so-called “out of bag error” (OBB) estimate can be considered equivalent to a cross-validation study. In OBB, one third of the compounds are randomly selected as a test set and a model is developed from the remaining compounds. The optimum m try was chosen such that %OBB is minimum. Thus, a lower %OBB indicates a higher accuracy of the model.
The Kernlab package in R was used for generating support vector machine (SVM) models. The scaling of the training and test set descriptors avoided domination of any descriptor with a large numerical value in the final SVM model. The two optimizable parameters in the radial basis function (RBF) kernel are C and sigma. The average value obtained from the automated optimal sigma calculation (sigest) method in R was used. The value of C was determined using k-fold cross validation (k = 10). The corresponding value of C with the lowest cross-validation error was then used for modeling. In k-fold cross validation the entire dataset is divided into k subsets of almost equal size. The model is trained using the k − 1 subset and the remaining subset is then used as prediction set. The advantage of k-fold cross validation is that the entire dataset is eventually used for both training and testing.
Six different models were generated for each method utilizing different combinations of molecular descriptors. The following descriptor combinations were used: model 1 = ChemDraw (CD) and VolSurf (VS); model 2 = CD, VS, clogP and polar surface area (PSA); model 3 = CD and MolConnZ (MZ); model 4 = CD, MZ, clogP and PSA; model 5 = CD, VS and MZ; model 6 = CD, VS, MZ, clogP and PSA.
Consensus Analysis
The consensus analysis for the test set was performed using predicted data from all three computational modeling methods. Three different rules, namely arithmetic mean, the harmonic mean and the median were applied for the generation of consensus classes. A total of 4 models were generated using each rule. Model 1 = best model from RP, RF and SVM; model 2 = best model from RP, and RF; model 3 = best models from RP and SVM; model 4 = best model from RF and SVM.
Validation Metrics
Three metrics were considered in evaluating model predictive performance. The three metrics are denoted absolute accuracy, consumer’s accuracy, and producer’s accuracy, with the latter two novel metrics attempting to consider the viewpoints of the consumer and the producer of the model. The percent accuracy is the percent of all drugs correctly predicted. In applying percent accuracy to a particular class, absolute accuracy is the percent of all drugs in the class that are correctly predicted to be in that class. Absolute accuracy carries no emphasis for avoiding one type of error over another. However, in practice, it is well appreciated that type I and type II errors represent different categories of inaccuracy. The selection of a final model for each method was based on percent accuracy of the test set. The percent accuracy for each class was calculated using:
Generally, a type I error can be described as a “false positive” error and a type II error as a “false negative” error. Further a type I error occurs when a predicted class is biopharmaceutically more favorable than the true class and was denoted as a “consumer risk.” In order to reflect this perspective in a metric to evaluate model predictive performance, the algorithm was developed to denote “consumer’s accuracy”. In a type II error, a predicted class is biopharmaceutically less favorable than the true class; an error of this type can be denoted as a “producer risk”. In order to reflect this perspective, “producer’s accuracy” was devised.
RESULTS
BDDCS Classification of the Test Set
To validate and challenge the computational models generated from the Wu and Benet data set, a collection of 56 drugs was retrieved from the WHO EML that had not been listed previously under the original BDDCS list (1,6). Drug disposition data was available for 56 drugs after an extensive literature survey (Table I). Interestingly, the collection of 56 drugs extracted from WHO-EML contained 26 compounds that had been classified ambiguously (Table I, column 11) by Lindenberg et al. (5) under BCS. Furthermore, 31 of the drugs in this collection were previously unclassified (Table I, column 12) under BCS by Kasim et al. (4), while 4 drugs were classified ambiguously in this 2004 report. Due to the relatively large number of compounds shared between our test set collection and the compound list classified by Lindenberg and colleagues, we used their BCS classification for comparison against our present BDDCS classification (1,5). Importantly, some of the drugs displayed a shift in BCS class to a new BDDCS class. For example, doxycycline is classified as a BCS class 1 drug, but its drug disposition data, featuring >50% unchanged urinary clearance, and low aqueous solubility would indicate class 4 under BDDCS. This classification is consistent with observations made by Wu and Benet for BDDCS class 4 drugs, which are mainly eliminated, unchanged via biliary or renal routes (6). Eight drugs, namely acetylsalicylic acid, benznidazole, biperidine, methyldopa, nifurtimox, penicillamine, penicillin V and thiamine, that were classified previously as BCS class 3 drugs were reclassified as BDDCS class 1 drugs. As an example, benznidazole has high aqueous solubility at the dose administered, 96% bioavailability and extensive cytochrome P450 metabolism, clearly justifying its classification under BDDCS class 1 (21,22). Similar disposition characteristics rationalize the reclassification of the other seven therapeutics.
Glibenclamide, an oral antidiabetic drug with low solubility (0.01 mg/ml) at the dose administered (5 mg) has been categorized in BCS class 4. However, glibenclamide is a confirmed substrate for both P-gp (23) and cytochrome P450 (24) and should thus be classified within BDDCS class 2. On similar grounds, a BCS 4 to BDDCS 2 class shift can be justified for mercaptopurine (25,26), retinol palmitate (27) and sulfasalazine (28,29). Thus, after extensive literature referencing the 56 test-set drugs from the BCS list were reclassified according to the BDDCS guidelines. This classification was then used to test the computational models that were built using 165 training set drugs obtained from the original BDDCS list (6). The resulting class distribution data for training and test set molecules (Table II) demonstrates equal compound allocation across classes 1–3; however, the percentage of molecules classified within class 4 in both data sets is less than 10%. This low frequency distribution of class 4 compounds is likely to affect both model generation and predictive confidence. Therefore, caution should be used in the interpretation models of the ensuing models with regard to class 4 compounds. The probability of randomly selecting a training set class 1, 2, 3 or 4 drug is 36.4% (60/165), 30.9% (51/165), 25.5% (42/165) and 7.3% (12/165), respectively. In contrast, the random probability for selecting a test set class 1, 2, 3 or 4 drug is 46.5% (26/56), 19.6% (11/56), 25% (14/56) and 8.9% (5/56), respectively.
Model Generation
Recursive Partitioning
To guide splitting criteria and optimize decision tree induction a tenfold cross-validation was performed on the training set data. A total of 6 descriptive models were generated with similar average training class accuracy (67, 70.1, 65.7, 66.1, 68.2, and 68.2% respectively). As expected, prediction of class 4 molecules became the defining criterion for a successful model (Tables III and IV). In fact, only model 1, based on VolSurf descriptors and the number of hydrogen bond donor and acceptor atoms per molecule, was capable of designating appropriate node splitting criteria to determine class 4 compounds. The best model can correctly identify 66.7% (40/60) of the compounds in class 1, 94.1% (48/51) of the compounds in class 2, 73.8% (31/42) of the compounds in class 3, and 33.3% (4/12) of the compounds in class 4 (Table V). However, the average performance (33.1%) on the test set is unsatisfactory.
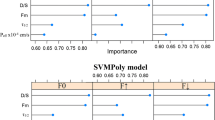
Nevertheless, some simple rules and criteria can be obtained from the decision tree in Fig. 2. The descriptors which are important for BDDCS classification are W1, W3, HB1 and HB7 resulting from an sp2 hybridized nitrogen probe containing one lone pair (N:=), W6 from an sp2 carboxyl oxygen atom probe (O::), and W3 and W6 from a water probe (OH2). In general, W1 and W3 account for polarizability and dispersion forces within a molecule, whereas W6, HB1, and HB7 represent polar and hydrogen bond donor and acceptor regions (recorded with different probe atoms). HB1 and HB7 is calculated as the difference between the hydrophilic volumes between water (OH2) and the (N:=) probe.
Random Forest
Two metrics were used as a measure of prediction accuracy for all models, namely the percent accuracy for predicting external test set molecules and the out-of-bag (OBB) estimate. Again, as in recursive partitioning, six random forest models were developed and the best model (model 3, based on ChemDraw and Molconn-Z descriptors) was selected based on OBB error rate convergence. The overall OBB error ranged from 35.1 to 42.4%. In general, the technique of “bagging” (i.e. m try = p) did not improve the predictive ability of the models (as compared to models with m try < p). Again, predictive ability is poor for class 4 drugs as indicated by 25 and 0% accuracy for training and test set compounds, respectively. However, prediction accuracy for test set molecules in classes 1 and 3 is significantly better than those obtained with recursive partitioning, exhibiting a 1.5 to twofold increase over random selection.
Support Vector Machine
The level of training error tolerated is controlled by the parameter C and models were generated using C values of 0.1, 0.5, 1, and 10 up to 100 (with 10 point increments). The optimum value of C, 40, was determined using k-fold cross-validation studies on the training set data (k = 10). Models 1, 5 and 6 were able to correctly identify all the molecules in their respective classes (100% accuracy); however, based on prediction accuracy on the test set data, the performance of model 1 is superior to that of models 5 and 6, largely due to problematic prediction of class 4 compounds. Models 3–5 failed to predict class 4 compounds altogether in the test set (i.e. 0% accuracy), whereas models 1, 2 and 6 displayed 20% prediction accuracy (data not shown). Overall, the predictive performance of the SVM models is significantly better compared to RP and RF models (Table V). Despite its outstanding internal consistency, the test set prediction for class 1 is actually inferior compared to the other methods, misclassifying 15 out of 26 compounds as class 3. On the other hand, only 3 out of 14 compounds in class 3 were erroneously predicted as class 1. Intrinsic to its algorithm, support vector regression may overfit data within the training set, thereby incurring a performance penalty in predicting test set molecules.
To ascertain the distribution of molecules in descriptor space, a principal component analysis (PCA) was performed on descriptors from the best SVM model, i.e the model including CD and VS descriptors. The PCA score plot provides an estimate of the descriptor space of training and test molecules (Fig. 3). The first three principal components of the training and test set can explain 69.3 and 69.8% of the variance, respectively. Test set molecules such as salbutamol, clomifene, folic acid and cefaxime are outside the descriptor space of training set molecules. Salbutamol, clomifene and folic acid are class 3 drugs and are accurately predicted by the SVM model. However, cefaxime is a class 4 drug predicted to be a class 3 compound by the SVM model. Consequently, PCA analysis provides a convenient method for identifying outliers or molecules that are far removed from the training set descriptor field, thus providing lower confidence in their predictions.
Consensus Analysis of Models
RF, RP, and SVM models can correctly predict 73.1% (19/26), 63.6% (7/11) and 78.6% (11/14) compounds in class 1, class 2 and class 3 respectively. Both RP and SVM models can be used for class 4 prediction however the accuracy is poor possibly due to the limited number of molecules in the training and test set. Along these lines we investigated consensus modeling approaches where combinations of different modeling methods are used (30). The consensus model resulted in significant improvement for predicting class 2 and 4 test set drugs. The prediction accuracy using arithmetic mean model 1 and 3 for class 2 and 4 is 81.8 and 40% respectively (results not shown). However, consensus modeling does not improve prediction accuracy across all classes. The prediction accuracy for class 3 is generally worse than for the individual models. However, the prediction accuracy for class 2 and 4 drugs is higher than individual models, provided a combination of consensus models is used.
Validation Metrics
The three metrics, absolute accuracy, consumer’s accuracy and producer’s accuracy denoted a model’s predictive performance. Percent accuracy is the percent of all drugs correctly predicted (Eq. 2). Absolute accuracy is the percent of all drugs in the class that are correctly predicted to be in that class. Table III identifies the occurrence of type I (false positive error) and II (false negative error), depending on true class. Consumer’s accuracy builds upon the absolute accuracy, but further penalizes type I errors. Correspondingly, consumer’s accuracy de-empasizes type II errors and attenuates the impact of such errors to protect the interest of the consumer. Analogous to consumer’s accuracy, producer’s accuracy attempts to consider a particular viewpoint in assessing model predictions. Consumer’s accuracy emphasizes avoidance of “false positives,” whereas producer’s accuracy emphasizes avoidance of “false negatives”. Each matrix partially de-emphasizes the type of error that it is not focused on. For any real dataset of predictions, consumer’s accuracy and producer’s accuracy can be expected to differ, since type I and type II error rates are generally different. Consumer’s accuracy and producer’s accuracy will span absolute accuracy, except in the case where type I and II errors are identical, where all three metrics will be identical. In general, a drug producer and consumer may have differing needs for accuracy in the prediction of BDDCS classifications. The best SVM model in the present study illustrates that consumer’s accuracy for the test set is lower than its corresponding producer’s accuracy (Table VI), thereby leading to a higher risk of misclassification for the consumer.
DISCUSSION
The BCS has been a helpful guide to classify compounds based on their aqueous solubility and gastrointestinal permeability (31). Wu and Benet (6) emphasize that the clinical impact of efflux transporters in modulating oral absorption and drug pharmacokinetics is most applicable to class 2, and possibly class 4 compounds. For example, high permeability allows facile cellular penetration for class 2 compounds, but low solubility (perhaps mainly due to high lipophilicity) will limit the effective concentration entering the cell, thereby preventing saturation of efflux transporters. Consequently, efflux transport can affect class 2 compounds’ extent of oral bioavailability and their rate of absorption (6). Thus, classification of compounds according to BDDCS guidelines may allow for a scientific basis towards their observed clinical behavior as a result of their interactions with P-gp and other efflux transporters. This, in turn, allows for a deeper understanding of their pharmacokinetic behavior and their potential for drug–drug interactions. Identifying molecules that interact with efflux transporters is important for drug discovery but is also generally reliant on time consuming in vitro and in vivo studies. However, computational models are now available to assist in this process, as we have recently shown for rapidly retrieving substrates or inhibitors for P-gp from commercial databases with in vitro validation (32).
The goal of the current study was to (1) investigate computational methods to produce predictive models; (2) automatically and rapidly classify compounds into BDDCS classes; and (3) use physicochemical properties derived from molecular descriptors alone. In applying these algorithms one can choose from a binary or a quaternary classification system with hard or soft class assignments. A binary system would independently assess the parameters solubility and metabolism for each compound and either uniquely bin the compounds in high/low categories (hard assignment) or use a gradated scale to plot the parameters; class assignment in the latter case would be determined by predefined criteria for low/high solubility and poor/extensive metabolism. The present study chose to apply a unified (quaternary) binning system with unambiguous (hard) class assignment. To accomplish this, we used machine learning methods as these algorithms have been widely used and validated with large datasets and are exceptionally suited to identify important properties and molecular descriptors from diverse arrays of data. In this study, we have captured a range of applicable descriptors for physicochemical properties, including easily interpretable descriptors determined by widely available chemical drawing software (e.g. ChemDraw) or web-based tools (e.g. PubChem or ChemSpider), as well as complex descriptor sets from commercial vendors such as MolconnZ (18) and VolSurf (17).
Among the best models in the current study, SVM model 1 revealed an exceptional level of training and test set prediction accuracy. Interestingly, ChemDraw and VolSurf descriptors are important for classifying class 2, 3 and 4 drugs, whereas a combination of ChemDraw and MolconnZ descriptors are useful for class 1 predictions. Thus, a combination of 2D and 3D descriptors are important for class 2, 3 and 4 drugs, whereas 2D descriptors alone are relevant for class 1 predictions. These observations would suggest that combinations of both models and descriptors may be necessary for optimal prediction of the different BDDCS classes.
Since this is the first report on predictive model development for BDDCS, a direct comparison with previously established computational models is not possible; however, we believe that a critical evaluation with respect to earlier BCS models, especially the study by Bergström et al. (33,34), is warranted. Although the overall prediction accuracies of the training and tests sets between the Bergström study and our models are comparable, there are several important differences that should be highlighted: (1) the Bergström study encompasses a very small number of compounds, presumably with limited chemical space; (2) PTSA is a conformation-dependent property requiring a 3D structure for calculations, thereby reducing the portability of the models between independent laboratories; and (3) solubility and permeability parameters are independently calculated. Interestingly, in the current study, models that included PSA or cLogP as a descriptor generally underperformed compared with models using alternative descriptors. This is in marked contrast to the work by Bergström et al. (34), who determined that PTSA could satisfy both drug solubility and permeability for BCS calculations. In agreement with our present data, however, they determined that cLogP could be excluded as a descriptor without the model losing predictive power. In fact, they found that the molecular surface areas alone contained sufficient information regarding lipophilicity. Analogous to this, it is likely that MolconnZ and VolSurf descriptors alone sufficiently capture lipophilicity to render cLogP redundant as a separate descriptor.
It is important to point out that most difficulty was encountered in predicting class 4 compounds. This was not entirely unexpected, since the number of training set molecules for class 4 compounds is small compared to the other classes. Inevitably, the models were trained primarily for classes 1–3 even when weighting is applied, leading to a disproportionate bias to predicting these classes. However, this may not be a major concern for the pharmaceutical industry because generally the number of compounds under development in classes 3 or 4 is low; for example, fewer than 10% of current compounds in the drug discovery pipeline of GlaxoSmithKline fall within BCS class 3 or 4 (35). Additionally, the bias towards accurately predicting class 1 and 2 compounds can be viewed as favorable in that these drugs may encounter fewer issues during the subsequent drug development process.
Options for improving the confidence in the computational models include the application of a combination of models. Additional computational methods such as k-Nearest Neighbor (kNN), Kohonen and Sammon mapping could be evaluated in the future alongside additional molecular descriptors and an enlarged training and test set with more examples of class 4 compounds. The utilization of Tanimoto similarity, PCA or other graphical mapping tools to assess the distance of a test set molecule from the training set will also aid in improving the confidence in predictions. We also foresee availability of these models to a global audience using web-based applications or their integration into existing database tools.
In summary, the present study represents a new development for rapidly assigning drugs to BDDCS classifications, providing useful additional insight into bioavailability aspects of a drug. This could have significant application in the drug discovery field to a priori identify molecules that may have future developability issues.
Abbreviations
- BCS:
-
Biopharmaceutics Classification System
- BDDCS:
-
Biopharmaceutics Drug Disposition Classification System
- RF:
-
random forest
- RP:
-
recursive partitioning
- SVM:
-
support vector machine
References
WHO. Waiver of in vivo bioavailability and bioequivalence studies for immediate-release solid oral dosage forms based on a biopharmaceutics classification system. FDA guidance for industry, federal drug and food administration, Rockville, MD (2002).
H. H. Blume and B. S. Schug. The biopharmaceutics classification system (BCS): class III drugs—better candidates for BA/BE waiver? Eur. J. Pharm. Sci. 9:117–121 (1999).
J. E. Polli, L. X. Yu, J. A. Cook, G. L. Amidon, R. T. Borchardt, B. A. Burnside, P. S. Burton, M. L. Chen, D. P. Conner, P. J. Faustino, A. A. Hawi, A. S. Hussain, H. N. Joshi, G. Kwei, V. H. Lee, L. J. Lesko, R. A. Lipper, A. E. Loper, S. G. Nerurkar, J. W. Polli, D. R. Sanvordeker, R. Taneja, R. S. Uppoor, C. S. Vattikonda, I. Wilding, and G. Zhang. Summary workshop report: biopharmaceutics classification system-implementation challenges and extension opportunities. J. Pharm. Sci. 93:1375–1381 (2004).
N. A. Kasim, M. Whitehouse, C. Ramachandran, M. Bermejo, H. Lennernas, A. S. Hussain, H. E. Junginger, S. A. Stavchansky, K. K. Midha, V. P. Shah, and G. L. Amidon. Molecular properties of WHO essential drugs and provisional biopharmaceutical classification. Mol. Pharm. 1:85–96 (2004).
M. Lindenberg, S. Kopp, and J. B. Dressman. Classification of orally administered drugs on the World Health Organization Model list of Essential Medicines according to the biopharmaceutics classification system. Eur. J. Pharm. Biopharm. 58:265–278 (2004).
C. Y. Wu and L. Z. Benet. Predicting drug disposition via application of BCS: transport/absorption/ elimination interplay and development of a biopharmaceutics drug disposition classification system. Pharm. Res. 22:11–23 (2005).
T. Takagi, C. Ramachandran, M. Bermejo, S. Yamashita, L. X. Yu, and G. L. Amidon. A provisional biopharmaceutical classification of the top 200 oral drug products in the United States, Great Britain, Spain, and Japan. Mol. Pharm. 3:631–643 (2006).
S. Ekins, C. L. Waller, P. W. Swaan, G. Cruciani, S. A. Wrighton, and J. H. Wikel. Progress in predicting human ADME parameters in silico. J. Pharmacol. Toxicol. Methods. 44:251–272 (2000).
S. Ekins and P. W. Swaan. Computational models for enzymes, transporters, channels and receptors relevant to ADME/TOX. Rev. Comp. Chem. 20:333–415 (2004).
J. B. Ed. by Dressman. Proposal to waive in vivo bioequivalence requirements for the WHO model list of essential medicines immediate release, solid oral dosage forms. World Health Organization (2005).
Merck. The Merck Index, Merck Research Laboratories, Rahway, NJ. The Merck Index, Merck Research Laboratories, Rahway, NJ 13th Edition:(2001).
USP. USP DI Vol III, Approved drug products and legal requirements, 18th Ed., United States Pharmacopoeial Convention Inc., Rockville, MD. 1998. (1998).
USP. The United State Pharmacopoeia, 24th Ed., by authority of the United State Pharmacopoeial convention, Inc., Printed by National Publishing, Philadelphia, PA, 2000. (2000).
D. M. Oh, R. L. Curl, and G. L. Amidon. Estimating the fraction dose absorbed from suspensions of poorly soluble compounds in humans: a mathematical model. Pharm. Res. 10:264–270 (1993).
D. S. Wishart, C. Knox, A. C. Guo, S. Shrivastava, M. Hassanali, P. Stothard, Z. Chang, and J. Woolsey. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34:D668–672 (2006).
M. Clark, R. D. Cramer, and N. van Op den Bosch. Validation of the general purpose Tripos 5.2 force field. J. Comput. Chem. 10:982–1012 (1989).
G. Cruciani, P. Crivori, P.-A. Carrupt, B. Testa. Molecular fields in quantitative structure-permeation relationships:the volSurf approach. THEOCHEM. 503:17–30 (2000).
L. B. Kier and L. H. Hall. Molecular Connectivity in Structure-Activity Analysis. Wiley, Letchworth, Hertfordshire, England, (1986).
T. M. Therneau and E.J. Atkinson. An introduction to recursive partitioning using the RPART routines, Department of health Sciences Research: Mayo clinic, (1997).
A. Liaw and M. Wiener. Classification and regression by random forest. R. News. 2/3:18–22 (2002).
M. I. Walton, C. R. Wolf, and P. Workman. Molecular enzymology of the reductive bioactivation of hypoxic cell cytotoxins. Int. J. Radiat. Oncol. Biol. Phys. 16:983–986 (1989).
P. Workman, R. A. White, M. I. Walton, L. N. Owen, and P. R. Twentyman. Preclinical pharmacokinetics of benznidazole. Br. J. Cancer. 50:291–303 (1984).
P. E. Golstein, A. Boom, J. van Geffel, P. Jacobs, B. Masereel, and R. Beauwens. P-glycoprotein inhibition by glibenclamide and related compounds. Pflugers. Arch. 437:652–660 (1999).
Y. Naritomi, S. Terashita, and A. Kagayama. Identification and relative contributions of human cytochrome P450 isoforms involved in the metabolism of glibenclamide and lansoprazole: evaluation of an approach based on the in vitro substrate disappearance rate. Xenobiotica. 34:415–427 (2004).
R. T. Gilman A. G., Nies, A. S., and P. Taylor. Goodman and Gilman’s The pharmacological basis of therapeutics., Pergamon Press, (1990).
J. Wijnholds, C. A. Mol, L. van Deemter, M. de Haas, G. L. Scheffer, F. Baas, J. H. Beijnen, R. J. Scheper, S. Hatse, E. De Clercq, J. Balzarini, and P. Borst. Multidrug-resistance protein 5 is a multispecific organic anion transporter able to transport nucleotide analogs. Proc. Natl. Acad. Sci. U. S. A. 97:7476–7481 (2000).
T. M. Davis, T. Q. Binh, L. T. Thu, R. Rossi, P. T. Danh, P. H. Barrett, and J. Beilby. Pharmacokinetics of retinyl palmitate and retinol after intramuscular retinyl palmitate administration in severe malaria. Clin. Sci. (Lond). 99:433–441 (2000).
E. Liang, J. Proudfoot, and M. Yazdanian. Mechanisms of transport and structure–permeability relationship of sulfasalazine and its analogs in Caco-2 cell monolayers. Pharm. Res. 17:1168–1174 (2000).
H. Zaher, A. A. Khan, J. Palandra, T. G. Brayman, L. Yu, and J. A. Ware. Breast cancer resistance protein (Bcrp/abcg2) is a major determinant of sulfasalazine absorption and elimination in the mouse. Mol. Pharm. 3:55–61 (2006).
S. E. O’Brien and M. J. de Groot. Greater than the sum of its parts: combining models for useful ADMET prediction. J. Med. Chem. 48:1287–1291 (2005).
G. L. Amidon, H. Lennernas, V. P. Shah, and J. R. Crison. A theoretical basis for a biopharmaceutic drug classification: the correlation of in vitro drug product dissolution and in vivo bioavailability. Pharm. Res. 12:413–420 (1995).
C. Chang, P. M. Bahadduri, J. E. Polli, P. W. Swaan, and S. Ekins. Rapid identification of P-glycoprotein substrates and inhibitors. Drug Metab. Dispos. 34:1976–1984 (2006).
C. A. Bergstrom. In silico predictions of drug solubility and permeability: two rate-limiting barriers to oral drug absorption. Basic Clin. Pharmacol. Toxicol. 96:156–161 (2005).
C. A. Bergstrom, M. Strafford, L. Lazorova, A. Avdeef, K. Luthman, and P. Artursson. Absorption classification of oral drugs based on molecular surface properties. J. Med. Chem. 46:558–570 (2003).
J. Baldoni. Roles of BCS in drug development, AAPS Workshop on BE, BCS and Beyond, Bethesda, MD, (2007).
X. P. Chen, Z. R. Tan, S. L. Huang, Z. Huang, D. S. Ou-Yang, and H. H. Zhou. Isozyme-specific induction of low-dose aspirin on cytochrome P450 in healthy subjects. Clin. Pharmacol. Ther. 73:264–271 (2003).
G. Levy and T. Tsuchiya. Salicylate accumulation kinetics in man. N. Engl. J. Med. 287:430–432 (1972).
S. Nishio, H. Watanabe, K. Kosuge, S. Uchida, H. Hayashi, and K. Ohashi. Interaction between amlodipine and simvastatin in patients with hypercholesterolemia and hypertension. Hypertens. Res. 28:223–227 (2005).
T. Kuzuya, T. Kobayashi, N. Moriyama, T. Nagasaka, I. Yokoyama, K. Uchida, A. Nakao, and T. Nabeshima. Amlodipine, but not MDR1 polymorphisms, alters the pharmacokinetics of cyclosporine A in Japanese kidney transplant recipients. Transplantation. 76:865–868 (2003).
P. A. Meredith and H. L. Elliott. Clinical pharmacokinetics of amlodipine. Clin. Pharmacokinet. 22:22–31 (1992).
R. L. Walsky and R. S. Obach. Validated assays for human cytochrome P450 activities. Drug Metab. Dispos. 32:647–660 (2004).
P. Winstanley, G. Edwards, M. Orme, and A. Breckenridge. The disposition of amodiaquine in man after oral administration. Br. J. Clin. Pharmacol. 23:1–7 (1987).
N. J. White, M. van Vugt, and F. Ezzet. Clinical pharmacokinetics and pharmacodynamics and pharmacodynamics of artemether-lumefantrine. Clin. Pharmacokinet. 37:105–125 (1999).
V. Navaratnam, S. M. Mansor, N. W. Sit, J. Grace, Q. Li, and P. Olliaro. Pharmacokinetics of artemisinin-type compounds. Clin. Pharmacokinet. 39:255–270 (2000).
M. A. van Agtmael, C. A. Van Der Graaf, T. K. Dien, R. P. Koopmans, and C. J. van Boxtel. The contribution of the enzymes CYP2D6 and CYP2C19 in the demethylation of artemether in healthy subjects. Eur. J. Drug Metab. Pharmacokinet. 23:429–436 (1998).
G. Lefevre, S. Looareesuwan, S. Treeprasertsuk, S. Krudsood, U. Silachamroon, I. Gathmann, R. Mull, and R. Bakshi. A clinical and pharmacokinetic trial of six doses of artemether-lumefantrine for multidrug-resistant Plasmodium falciparum malaria in Thailand. Am. J. Trop. Med. Hyg. 64:247–256 (2001).
P. J. de Vries, and T. K. Dien. Clinical pharmacology and therapeutic potential of artemisinin and its derivatives in the treatment of malaria. Drugs. 52:818–836 (1996).
X. Q. Li, A. Bjorkman, T. B. Andersson, L. L. Gustafsson, and C. M. Masimirembwa. Identification of human cytochrome P(450)s that metabolise anti-parasitic drugs and predictions of in vivo drug hepatic clearance from in vitro data. Eur. J. Clin. Pharmacol. 59:429–442 (2003).
P. Reungpatthanaphong, and S. Mankhetkorn. Modulation of multidrug resistance by artemisinin, artesunate and dihydroartemisinin in K562/adr and GLC4/adr resistant cell lines. Biol. Pharm. Bull. 25:1555–1561 (2002).
N. J. White. Clinical pharmacokinetics and pharmacodynamics of artemisinin and derivatives. Trans. R. Soc. Trop. Med. Hyg. 88 Suppl 1:S41–43 (1994).
D. Hornig. Metabolism and requirements of ascorbic acid in man. S. Afr. Med. J. 60:818–823 (1981).
R. P. van Heeswijk, C. L. Cooper, B. C. Foster, B. M. Chauhan, F. Shirazi, I. Seguin, E. J. Phillips, and E. Mills. Effect of high-dose vitamin C on hepatic cytochrome P450 3A4 activity. Pharmacotherapy. 25:1725–1728 (2005).
C. D. Chiang, E. J. Song, V. C. Yang, and C. C. Chao. Ascorbic acid increases drug accumulation and reverses vincristine resistance of human non-small-cell lung-cancer cells. Biochem. J. 301(Pt 3):759–764 (1994).
M. Hollmann, E. Brode, G. Greger, H. Muller-Peltzer, and N. Wetzelsberger. Biperiden effects and plasma levels in volunteers. Eur. J. Clin. Pharmacol. 27:619–621 (1984).
S. Kudo, and M. Odomi. Involvement of human cytochrome P450 3A4 in reduced haloperidol oxidation. Eur. J. Clin. Pharmacol. 54:253–259 (1998).
R. Grimaldi, E. Perucca, G. Ruberto, C. Gelmi, F. Trimarchi, M. Hollmann, and A. Crema. Pharmacokinetic and pharmacodynamic studies following the intravenous and oral administration of the antiparkinsonian drug biperiden to normal subjects. Eur. J. Clin. Pharmacol. 29:735–737 (1986).
S. A. Factor, W. J. Weiner, and F. Hefti. Acetaminophen metabolism by cytochrome P450 monooxygenases in Parkinson’s disease. Ann. Neurol. 26:286–288 (1989).
S. Vickers, E. K. Stuart, J. R. Bianchine, H. B. Hucker, M. E. Jaffe, R. E. Rhodes, and W. J. Vandenheuvel. Metabolism of carbidopa (1-(−)-alpha-hydrazino-3,4-dihydroxy-alpha-methylhydrocinnamic acid monohydrate), an aromatic amino acid decarboxylase inhibitor, in the rat, rhesus monkey, and man. Drug Metab. Dispos. 2:9–22 (1974).
J. Barre. [Pharmacokinetic properties of cefixime]. Presse. Med. 18:1578–1582 (1989).
T. Niwa, T. Shiraga, T. Hashimoto, and A. Kagayama. Effect of cefixime and cefdinir, oral cephalosporins, on cytochrome P450 activities in human hepatic microsomes. Biol. Pharm. Bull. 27:97–99 (2004).
W. F. Marshall, and J. E. Blair. The cephalosporins. Mayo Clin. Proc. 74:187–195 (1999).
U. Loos, E. Musch, M. Malek, and E. Riedel. Comparative pharmacokinetics of chlorambucil and prednimustine after oral administration. Oncology. 48:334–342 (1991).
M. Baumhakel, D. Kasel, R. A. Rao-Schymanski, R. Bocker, K. T. Beckurts, M. Zaigler, D. Barthold, and U. Fuhr. Screening for inhibitory effects of antineoplastic agents on CYP3A4 in human liver microsomes. Int. J. Clin. Pharmacol. Ther. 39:517–528 (2001).
C. S. Morrow, P. K. Smitherman, S. K. Diah, E. Schneider, and A. J. Townsend. Coordinated action of glutathione S-transferases (GSTs) and multidrug resistance protein 1 (MRP1) in antineoplastic drug detoxification. Mechanism of GST A1-1- and MRP1-associated resistance to chlorambucil in MCF7 breast carcinoma cells. J. Biol. Chem. 273:20114–20120 (1998).
G. C. Bolton, G. D. Allen, B. E. Davies, C. W. Filer, and D. J. Jeffery. The disposition of clavulanic acid in man. Xenobiotica. 16:853–863 (1986).
M. A. Wynalda, J. M. Hutzler, M. D. Koets, T. Podoll, and L. C. Wienkers. in vitro metabolism of clindamycin in human liver and intestinal microsomes. Drug Metab. Dispos. 31:878–887 (2003).
D. Mazur, B. S. Schug, G. Evers, V. Larsimont, H. Fieger-Buschges, W. Gimbel, A. Keilbach-Bermann, and H. H. Blume. Bioavailability and selected pharmacokinetic parameters of clindamycin hydrochloride after administration of a new 600 mg tablet formulation. Int. J. Clin. Pharmacol. Ther. 37:386–392 (1999).
U. S. Rao, R. L. Fine, and G. A. Scarborough. Antiestrogens and steroid hormones: substrates of the human P-glycoprotein. Biochem. Pharmacol. 48:287–292 (1994).
T. J. Mikkelson, P. D. Kroboth, W. J. Cameron, L. W. Dittert, V. Chungi, and P. J. Manberg. Single-dose pharmacokinetics of clomiphene citrate in normal volunteers. Fertil. Steril. 46:392–396 (1986).
D. Ardid, and G. Guilbaud. Antinociceptive effects of acute and ‘chronic’ injections of tricyclic antidepressant drugs in a new model of mononeuropathy in rats. Pain. 49:279–287 (1992).
A. Nagy, and R. Johansson. The demethylation of imipramine and clomipramine as apparent from their plasma kinetics. Psychopharmacology (Berl). 54:125–131 (1977).
L. E. Evans, J. H. Bett, J. R. Cox, J. P. Dubois, and T. Van Hees. The bioavailability of oral and parenteral chlorimipramine (Anafranil). Prog. Neuropsychopharmacol. 4:293–302 (1980).
A. E. Balant-Gorgia, M. Gex-Fabry, and L. P. Balant. Clinical pharmacokinetics of clomipramine. Clin. Pharmacokinet. 20:447–462 (1991).
C. A. Knupp, W. C. Shyu, R. Dolin, F. T. Valentine, C. McLaren, R. R. Martin, K. A. Pittman, and R. H. Barbhaiya. Pharmacokinetics of didanosine in patients with acquired immunodeficiency syndrome or acquired immunodeficiency syndrome-related complex. Clin. Pharmacol. Ther. 49:523–535 (1991).
S. Kaul, W. C. Shyu, U. A. Shukla, K. A. Dandekar, and R. H. Barbhaiya. Absorption, disposition, and metabolism of [14C]didanosine in the beagle dog. Drug Metab. Dispos. 21:447–453 (1993).
N. Shiraki, A. Hamada, K. Yasuda, J. Fujii, K. Arimori, and M. Nakano. Inhibitory effect of human immunodeficiency virus protease inhibitors on multidrug resistance transporter P-glycoproteins. Biol. Pharm. Bull. 23:1528–1531 (2000).
C. A. Joseph, and P. A. Dixon. A possible cytochrome P-450-mediated N-oxidation of diethylcarbamazine. J. Pharm. Pharmacol. 36:711–712 (1984).
K. L. Mealey, R. Barhoumi, R. C. Burghardt, S. Safe, and D. T. Kochevar. Doxycycline induces expression of P glycoprotein in MCF-7 breast carcinoma cells. Antimicrob. Agents Chemother. 46:755–761 (2002).
G. R. Bailie, and C. A. Johnson. Comparative review of the pharmacokinetics of vitamin D analogues. Semin. Dial. 15:352–357 (2002).
S. A. a. D. E. Rangel-Castro I.J. The ergocalciferol content of dried pigmented and albino Cantharellus cibarius fruit bodies. Mycological Research. 106:70–73 (2002).
K. E. Thummel, C. Brimer, K. Yasuda, J. Thottassery, T. Senn, Y. Lin, H. Ishizuka, E. Kharasch, J. Schuetz, and E. Schuetz. Transcriptional control of intestinal cytochrome P-4503A by 1alpha,25-dihydroxy vitamin D3. Mol. Pharmacol. 60:1399–1406 (2001).
S. W. Park, N. Lomri, L. A. Simeoni, J. P. Fruehauf, and E. Mechetner. Analysis of P-glycoprotein-mediated membrane transport in human peripheral blood lymphocytes using the UIC2 shift assay. Cytometry. A. 53:67–78 (2003).
J. L. Sommerfeldt, J. L. Napoli, E. T. Littledike, D. C. Beitz, and R. L. Horst. Metabolism of orally administered [3H]ergocalciferol and [3H]cholecalciferol by dairy calves. J. Nutr. 113:2595–2600 (1983).
C. S. Lee, J. G. Gambertoglio, D. C. Brater, and L. Z. Benet. Kinetics of oral ethambutol in the normal subject. Clin. Pharmacol. Ther. 22:615–621 (1977).
Y. Nishimura, N. Kurata, E. Sakurai, and H. Yasuhara. Inhibitory effect of antituberculosis drugs on human cytochrome P450-mediated activities. J. Pharmacol. Sci. 96:293–300 (2004).
M. J. Ruse, and R. H. Waring. The effect of methimazole on thioamide bioactivation and toxicity. Toxicol. Lett. 58:37–41 (1991).
M. Giaccone, A. Bartoli, G. Gatti, R. Marchiselli, F. Pisani, M. A. Latella, and E. Perucca. Effect of enzyme inducing anticonvulsants on ethosuximide pharmacokinetics in epileptic patients. Br. J. Clin. Pharmacol. 41:575–579 (1996).
K. Bachmann, C. A. Chu, and V. Greear. In vivo evidence that ethosuximide is a substrate for cytochrome P450IIIA. Pharmacology. 45:121–128 (1992).
K. Bachmann, Y. He, J. G. Sarver, and N. Peng. Characterization of the cytochrome P450 enzymes involved in the in vitro metabolism of ethosuximide by human hepatic microsomal enzymes. Xenobiotica. 33:265–276 (2003).
A. Crowe, and Y. K. Teoh. Limited P-glycoprotein mediated efflux for anti-epileptic drugs. J. Drug Target. 14:291–300 (2006).
R. A. Tokola, and P. J. Neuvonen. Pharmacokinetics of antiepileptic drugs. Acta Neurol. Scand. Suppl. 97:17–27 (1983).
R. D. Smyth, M. Pfeffer, A. Scalzo, and R. L. Comis. Bioavailability and pharmacokinetics of etoposide (VP-16). Semin. Oncol. 12:48–51 (1985).
D. J. Stewart, D. Nundy, J. A. Maroun, L. Tetreault, and J. Prior. Bioavailability, pharmacokinetics, and clinical effects of an oral preparation of etoposide. Cancer Treat. Rep. 69:269–273 (1985).
J. M. van Maanen, J. de Vries, D. Pappie, E. van den Akker, V. M. Lafleur, J. Retel, J. van der Greef, and H. M. Pinedo. Cytochrome P-450-mediated O-demethylation: a route in the metabolic activation of etoposide (VP-16-213). Cancer Res. 47:4658–4662 (1987).
B. L. Leu, and J. D. Huang. Inhibition of intestinal P-glycoprotein and effects on etoposide absorption. Cancer Chemother. Pharmacol. 35:432–436 (1995).
G. Inselmann, U. Holzlohner, and H. T. Heidemann. Effect of 5-fluorocytosine and 5-fluorouracil on human and rat hepatic cytochrome P 450. Mycoses. 32:638–643 (1989).
D. de Graaf, R. C. Sharma, E. B. Mechetner, R. T. Schimke, and I. B. Roninson. P-glycoprotein confers methotrexate resistance in 3T6 cells with deficient carrier-mediated methotrexate uptake. Proc. Natl. Acad. Sci. U. S. A. 93:1238–1242 (1996).
A. J. Clifford, A. Arjomand, S. R. Dueker, P. D. Schneider, B. A. Buchholz, and J. S. Vogel. The dynamics of folic acid metabolism in an adult given a small tracer dose of 14C-folic acid. Adv. Exp. Med. Biol. 445:239–251 (1998).
L. Balant, J. Fabre, and G. R. Zahnd. Comparison of the pharmacokinetics of glipizide and glibenclamide in man. Eur. J. Clin. Pharmacol. 8:63–69 (1975).
M. Develoux. [Griseofulvin]. Ann. Dermatol. Venereol. 128:1317–1325 (2001).
E. O. Farombi, O. Akinloye, C. O. Akinmoladun, and G. O. Emerole. Hepatic drug metabolizing enzyme induction and serum triacylglycerol elevation in rats treated with chlordiazepoxide, griseofulvin, rifampicin and phenytoin. Clin. Chim. Acta. 289:1–10 (1999).
S. Kives, P. M. Hahn, E. White, F. Z. Stanczyk, and R. L. Reid. Bioavailability of the Yuzpe and levonorgestrel regimens of emergency contraception: vaginal vs. oral administration. Contraception. 71:197–201 (2005).
V. Hatorp, K. T. Hansen, and M. S. Thomsen. Influence of drugs interacting with CYP3A4 on the pharmacokinetics, pharmacodynamics, and safety of the prandial glucose regulator repaglinide. J. Clin. Pharmacol. 43:649–660 (2003).
M. Frohlich, N. Albermann, A. Sauer, I. Walter-Sack, W. E. Haefeli, and J. Weiss. In vitro and ex vivo evidence for modulation of P-glycoprotein activity by progestins. Biochem. Pharmacol. 68:2409–2416 (2004).
F. Z. Stanczyk, and S. Roy. Metabolism of levonorgestrel, norethindrone, and structurally related contraceptive steroids. Contraception. 42:67–96 (1990).
H. R. Maxon, W. A. Ritschel, C. P. Volle, M. A. Eldon, I. W. Chen, M. F. Fernandez, J. Cline, and G. Mayfield. Pilot study on the absolute and relative bioavailability of Synthroid and Levothroid, two brands of sodium levothyroxine. Int. J. Clin. Pharmacol. Ther. Toxicol. 21:379–382 (1983).
C. Liddle, B. J. Goodwin, J. George, M. Tapner, and G. C. Farrell. Separate and interactive regulation of cytochrome P450 3A4 by triiodothyronine, dexamethasone, and growth hormone in cultured hepatocytes. J. Clin. Endocrinol. Metab. 83:2411–2416 (1998).
T. Mitin, L. L. Von Moltke, M. H. Court, and D. J. Greenblatt. Levothyroxine up-regulates P-glycoprotein independent of the pregnane X receptor. Drug Metab. Dispos. 32:779–782 (2004).
P. T. Giao, and P. J. de Vries. Pharmacokinetic interactions of antimalarial agents. Clin. Pharmacokinet. 40:343–373 (2001).
F. Ezzet, R. Mull, and J. Karbwang. Population pharmacokinetics and therapeutic response of CGP 56697 (artemether + benflumetol) in malaria patients. Br. J. Clin. Pharmacol. 46:553–561 (1998).
J. M. Wright, M. Orozco-Gonzalez, G. Polak, and C. T. Dollery. Duration of effect of single daily dose methyldopa therapy. Br. J. Clin. Pharmacol. 13:847–854 (1982).
E. Myhre, H. E. Rugstad, and T. Hansen. Clinical pharmacokinetics of methyldopa. Clin. Pharmacokinet. 7:221–233 (1982).
G. R. Haenen, F. P. Jansen, N. P. Vermeulen, and A. Bast. Activation of the microsomal glutathione S-transferase by metabolites of alpha-methyldopa. Arch. Biochem. Biophys. 287:48–52 (1991).
O. Heikinheimo. Clinical pharmacokinetics of mifepristone. Clin. Pharmacokinet. 33:7–17 (1997).
G. R. Jang, S. A. Wrighton, and L. Z. Benet. Identification of CYP3A4 as the principal enzyme catalyzing mifepristone (RU 486) oxidation in human liver microsomes. Biochem. Pharmacol. 52:753–761 (1996).
O. Fardel, A. Courtois, B. Drenou, T. Lamy, V. Lecureur, P. Y. le Prise, and R. Fauchet. Inhibition of P-glycoprotein activity in human leukemic cells by mifepristone. Anticancer Drugs. 7:671–677 (1996).
P. J. Hoskin, G. W. Hanks, G. W. Aherne, D. Chapman, P. Littleton, and J. Filshie. The bioavailability and pharmacokinetics of morphine after intravenous, oral and buccal administration in healthy volunteers. Br. J. Clin. Pharmacol. 27:499–505 (1989).
D. Projean, P. E. Morin, T. M. Tu, and J. Ducharme. Identification of CYP3A4 and CYP2C8 as the major cytochrome P450 s responsible for morphine N-demethylation in human liver microsomes. Xenobiotica. 33:841–854 (2003).
S. P. Letrent, J. W. Polli, J. E. Humphreys, G. M. Pollack, K. R. Brouwer, and K. L. Brouwer. P-glycoprotein-mediated transport of morphine in brain capillary endothelial cells. Biochem. Pharmacol. 58:951–957 (1999).
E. D. Kharasch, C. Hoffer, D. Whittington, and P. Sheffels. Role of P-glycoprotein in the intestinal absorption and clinical effects of morphine. Clin. Pharmacol. Ther. 74:543–554 (2003).
T. E. Bapiro, A. C. Egnell, J. A. Hasler, and C. M. Masimirembwa. Application of higher throughput screening (HTS) inhibition assays to evaluate the interaction of antiparasitic drugs with cytochrome P450s. Drug Metab. Dispos. 29:30–35 (2001).
A. Rojas, R. J. Hodgkiss, M. R. Stratford, M. F. Dennis, and H. Johns. Pharmacokinetics of varying doses of nicotinamide and tumour radiosensitisation with carbogen and nicotinamide: clinical considerations. Br. J. Cancer. 68:1115–1121 (1993).
A. Petley, B. Macklin, A. G. Renwick, and T. J. Wilkin. The pharmacokinetics of nicotinamide in humans and rodents. Diabetes. 44:152–155 (1995).
G. La Piana, D. Marzulli, M. I. Consalvo, and N. E. Lofrumento. Cytochrome c-induced cytosolic nicotinamide adenine dinucleotide oxidation, mitochondrial permeability transition, and apoptosis. Arch. Biochem. Biophys. 410:201–211 (2003).
S. Zhou, S. Yung Chan, B. Cher Goh, E. Chan, W. Duan, M. Huang, and H. L. McLeod. Mechanism-based inhibition of cytochrome P450 3A4 by therapeutic drugs. Clin. Pharmacokinet. 44:279–304 (2005).
M. E. Letelier, P. Izquierdo, L. Godoy, A. M. Lepe, and M. Faundez. Liver microsomal biotransformation of nitro-aryl drugs: mechanism for potential oxidative stress induction. J. Appl. Toxicol. 24:519–525 (2004).
S. M. Murta, W. G. dos Santos, C. Anacleto, P. Nirde, E. S. Moreira, and A. J. Romanha. Drug resistance in Trypanosoma cruzi is not associated with amplification or overexpression of P-glycoprotein (PGP) genes. Mol. Biochem. Parasitol. 117:223–228 (2001).
C. Paulos, J. Paredes, I. Vasquez, S. Thambo, A. Arancibia, and G. Gonzalez-Martin. Pharmacokinetics of a nitrofuran compound, nifurtimox, in healthy volunteers. Int. J. Clin. Pharmacol. Ther. Toxicol. 27:454–457 (1989).
R. K. Liedtke, S. Ebel, B. Missler, W. Haase, and L. Stein. Single-dose pharmacokinetics of macrocrystalline nitrofurantoin formulations. Arzneimittelforschung. 30:833–836 (1980).
V. Leskovac, and M. Popovic. Mechanism of reduction of nitrofurantoin on liver microsomes. Pharmacol. Res. Commun. 12:13–27 (1980).
J. D. Conklin. The pharmacokinetics of nitrofurantoin and its related bioavailability. Antibiot. Chemother. 25:233–252 (1978).
H. R. Kwon, P. Green, and S. H. Curry. Pharmacokinetics of nitroglycerin and its metabolites after administration of sustained-release tablets. Biopharm. Drug Dispos. 13:141–152 (1992).
N. H. Lee, and F. M. Belpaire. Biotransformation of nitroglycerin by organic nitrate reductase. Arch. Int. Pharmacodyn. Ther. 196 Suppl 196:165 (1972).
R. A. Okerholm, F. E. Peterson, F. J. Keeley, T. C. Smith, and A. J. Glazko. Bioavailability of norethindrone in human subjects. Eur. J. Clin. Pharmacol. 13:35–39 (1978).
C. H. Verhoeven, T. T. van Munster, G. M. Groothuis, R. M. Vos, and I. M. Rietjens. Identification of the human P450 enzymes involved in the in vitro metabolism of the synthetic steroidal hormones Org 4060 and Org 30659. Xenobiotica. 32:109–118 (2002).
W. Y. Kim, and L. Z. Benet. P-glycoprotein (P-gp/MDR1)-mediated efflux of sex-steroid hormones and modulation of P-gp expression in vitro. Pharm. Res. 21:1284–1293 (2004).
W. G. Schoonen, G. H. Deckers, M. E. de Gooijer, R. de Ries, and H. J. Kloosterboer. Hormonal properties of norethisterone, 7alpha-methyl-norethisterone and their derivatives. J. Steroid. Biochem. Mol. Biol. 74:213–222 (2000).
G. O. Kokwaro, and G. Taylor. Oxamniquine pharmacokinetics in healthy Kenyan African volunteers. East Afr. Med. J. 68:359–364 (1991).
C. M. Masimirembwa, J. A. Hasler, and I. Johansson. Inhibitory effects of antiparasitic drugs on cytochrome P450 2D6. Eur. J. Clin. Pharmacol. 48:35–38 (1995).
L. F. Prescott. Kinetics and metabolism of paracetamol and phenacetin. Br. J. Clin. Pharmacol. 10 Suppl 2:291S–298S (1980).
J. Mohandas, G. G. Duggin, J. S. Horvath, and D. J. Tiller. Metabolic oxidation of acetaminophen (paracetamol) mediated by cytochrome P-450 mixed-function oxidase and prostaglandin endoperoxide synthetase in rabbit kidney. Toxicol. Appl. Pharmacol. 61:252–259 (1981).
I. Manov, H. Motanis, I. Frumin, and T. C. Iancu. Hepatotoxicity of anti-inflammatory and analgesic drugs: ultrastructural aspects. Acta Pharmacol. Sin. 27:259–272 (2006).
R. F. Bergstrom, D. R. Kay, T. M. Harkcom, and J. G. Wagner. Penicillamine kinetics in normal subjects. Clin. Pharmacol. Ther. 30:404–413 (1981).
P. Netter, B. Bannwarth, P. Pere, and A. Nicolas. Clinical pharmacokinetics of D-penicillamine. Clin. Pharmacokinet. 13:317–333 (1987).
A. E. Pilkington, and R. H. Waring. D-penicillamine metabolism: in vitro studies of S-oxidation mechanisms. Drug Metabol. Drug Interact. 6:85–93 (1988).
C. Chen, and G. M. Pollack. Enhanced antinociception of the model opioid peptide [D-penicillamine] enkephalin by P-glycoprotein modulation. Pharm. Res. 16:296–301 (1999).
J. Huwyler, M. B. Wright, H. Gutmann, and J. Drewe. Induction of cytochrome P450 3A4 and P-glycoprotein by the isoxazolyl-penicillin antibiotic flucloxacillin. Curr. Drug Metab. 7:119–126 (2006).
N. Z. Nyazema, F. F. Mutamiri, I. Mudiwa, A. Chimuka, and J. Ndamba. Immunopharmacological aspects of praziquantel. Cent. Afr. J. Med. 41:284–288 (1995).
S. Kaojarern, S. Nathakarnkikool, and U. Suvanakoot. Comparative bioavailability of praziquantel tablets. DICP. 23:29–32 (1989).
W. Ridtitid, M. Wongnawa, W. Mahatthanatrakul, J. Punyo, and M. Sunbhanich. Rifampin markedly decreases plasma concentrations of praziquantel in healthy volunteers. Clin. Pharmacol. Ther. 72:505–513 (2002).
R. Hayeshi, C. Masimirembwa, S. Mukanganyama, and A. L. Ungell. The potential inhibitory effect of antiparasitic drugs and natural products on P-glycoprotein mediated efflux. Eur. J. Pharm. Sci. 29:70–81 (2006).
R. Preiss, F. Baumann, R. Regenthal, and M. Matthias. Plasma kinetics of procarbazine and azo-procarbazine in humans. Anticancer Drugs. 17:75–80 (2006).
M. W. Coomes, and R. A. Prough. The mitochondrial metabolism of 1,2-disubstituted hydrazines, procarbazine and 1,2-dimethylhydrazine. Drug Metab. Dispos. 11:550–555 (1983).
I. Bygbjerg, P. Ravn, A. Ronn, H. Flachs, and E. F. Hvidberg. Human pharmacokinetics of proguanil and its metabolites. Trop. Med. Parasitol. 38:77–80 (1987).
E. M. Chiluba, K. A. Fletcher, and A. H. Price. The pharmacokinetics of proguanil in human subjects following a single oral dose. Afr. J. Med. Sci. 16:43–46 (1987).
N. A. Helsby, G. Edwards, A. M. Breckenridge, and S. A. Ward. The multiple dose pharmacokinetics of proguanil. Br. J. Clin. Pharmacol. 35:653–656 (1993).
W. H. Hoffman, and J. N. Miceli. Pharmacokinetics of propylthiouracil in children and adolescents with Graves’ disease in the hyperthyroid and euthyroid states. Dev. Pharmacol. Ther. 11:73–81 (1988).
H. Bjorn, D. R. Hennessy, and C. Friis. The kinetic disposition of pyrantel citrate and pamoate and their efficacy against pyrantel-resistant Oesophagostomum dentatum in pigs. Int. J. Parasitol. 26:1375–1380 (1996).
G. E. Goodman, D. S. Alberts, Y. M. Peng, J. Beaudry, S. A. Leigh, and T. E. Moon. Plasma kinetics of oral retinol in cancer patients. Cancer Treat. Rep. 68:1125–1133 (1984).
L. A. Hansen, C. C. Sigman, F. Andreola, S. A. Ross, G. J. Kelloff, and L. M. De Luca. Retinoids in chemoprevention and differentiation therapy. Carcinogenesis. 21:1271–1279 (2000).
G. Acocella. Clinical pharmacokinetics of rifampicin. Clin. Pharmacokinet. 3:108–127 (1978).
E. G. Schuetz, A. H. Schinkel, M. V. Relling, and J. D. Schuetz. P-glycoprotein: a major determinant of rifampicin-inducible expression of cytochrome P4503A in mice and humans. Proc. Natl. Acad. Sci. U. S. A. 93:4001–4005 (1996).
D. A. Goldstein, Y. K. Tan, and S. J. Soldin. Pharmacokinetics and absolute bioavailability of salbutamol in healthy adult volunteers. Eur. J. Clin. Pharmacol. 32:631–634 (1987).
D. J. Morgan, J. D. Paull, B. H. Richmond, E. Wilson-Evered, and S. P. Ziccone. Pharmacokinetics of intravenous and oral salbutamol and its sulphate conjugate. Br. J. Clin. Pharmacol. 22:587–593 (1986).
S. Kaul, K. A. Dandekar, B. E. Schilling, and R. H. Barbhaiya. Toxicokinetics of 2′,3′-didehydro-3′-deoxythymidine, stavudine (D4T). Drug Metab. Dispos. 27:1–12 (1999).
K. Z. Rana, and M. N. Dudley. Clinical pharmacokinetics of stavudine. Clin. Pharmacokinet. 33:276–284 (1997).
S. L. Glynn, and M. Yazdanian. In vitro blood–brain barrier permeability of nevirapine compared to other HIV antiretroviral agents. J. Pharm. Sci. 87:306–310 (1998).
E. M. Cretton, Z. Zhou, L. B. Kidd, H. M. McClure, S. Kaul, M. J. Hitchcock, and J. P. Sommadossi. In vitro and in vivo disposition and metabolism of 3′-deoxy-2′,3′-didehydrothymidine. Antimicrob. Agents Chemother. 37:1816–1825 (1993).
H. R. Winter, and J. D. Unadkat. Identification of cytochrome P450 and arylamine N-acetyltransferase isoforms involved in sulfadiazine metabolism. Drug Metab. Dispos. 33:969–976 (2005).
U. Klotz. Clinical pharmacokinetics of sulphasalazine, its metabolites and other prodrugs of 5-aminosalicylic acid. Clin. Pharmacokinet. 10:285–302 (1985).
C. Beyeler, B. M. Frey, and H. A. Bird. Urinary 6 beta-hydroxycortisol excretion in rheumatoid arthritis. Br. J. Rheumatol. 36:54–58 (1997).
C. M. Tallaksen, A. Sande, T. Bohmer, H. Bell, and J. Karlsen. Kinetics of thiamin and thiamin phosphate esters in human blood, plasma and urine after 50 mg intravenously or orally. Eur. J. Clin. Pharmacol. 44:73–78 (1993).
Acknowledgement
Akash Khandelwal and Praveen M. Bahadduri contributed equally to this work and should be considered co-first authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khandelwal, A., Bahadduri, P.M., Chang, C. et al. Computational Models to Assign Biopharmaceutics Drug Disposition Classification from Molecular Structure. Pharm Res 24, 2249–2262 (2007). https://doi.org/10.1007/s11095-007-9435-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-007-9435-9