Purpose
This study was conducted to study global gene expression profiles elicited by (−)-epigallocatechin-3-gallate (EGCG) in mouse liver and small intestine, as well as to identify EGCG-regulated Nrf2-dependent genes.
Methods
C57BL/6J and C57BL/6J/Nrf2(−/−) mice were given an oral dose of EGCG at 200 mg/kg or treated with vehicle. Both liver and small intestine were collected 3 h and 12 h after treatment. Total RNA was extracted from the tissues and gene expression profiles were analyzed using Affymetrix mouse genome 430 2.0 array and GeneSpring 6.1 software. Microarray data were validated using quantitative real-time reverse transcription-PCR chain reaction analysis.
Results
Genes that were either induced or suppressed more than two fold by EGCG treatment compared with vehicle treatment in the same genotype group were filtered using the GeneSpring software. Among these well-defined genes, 671 EGCG-regulated Nrf2-dependent genes and 256 EGCG-regulated Nrf2-independent genes were identified in liver, whereas 228 EGCG-regulated Nrf2-dependent genes and 98 EGCG-regulated Nrf2-independent genes were identified in the small intestine. Based on their biological functions, these genes mainly fall into the category of ubiquitination and proteolysis, electron transport, detoxification, transport, cell growth and apoptosis, cell adhesion, kinase and phosphatases, and transcription factors.
Conclusions
Genes expressed in mouse liver are more responsive to oral treatment of EGCG than those expressed in small intestine. EGCG could regulate many genes in both organs in an Nrf2-dependent manner. The identification of genes related to detoxification, transport, cell growth and apoptosis, cell adhesion, kinase, and transcription regulated by EGCG not only provide potential novel insight into the effect of EGCG on global gene expression and chemopreventive effects, but also point to the potential role of Nrf2 in these processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The medicinal benefits of drinking green tea have been known in Asian countries since ancient times. From the in vivo animal cancer model studies, (−)-epigallocatechin-3-gallate (EGCG) or green tea extract has been shown to inhibit tumorigenesis on different organ sites (1). These include 7,12-dimethylbenz[a]anthracene (DMBA)-initiated skin tumor (2) in sensitive to mouse carcinogenesis (SENCAR); UVB radiation-induced photocarcinogenesis in SKH-1 hairless mice (3,4); lung tumorigenesis (5) in A/J mice; prostate tumorigenesis in athymic mice (6) and transgenic murine prostate cancer model (TRAMP) mice (7); breast tumor xenograft in athymic mice (6,8); and DMBA initiation in the Sprague–Dawley (S–D) rat model (9). During the past two decades, many case-control and cohort epidemiological studies have been conducted to investigate the effects of green tea consumption on the incidence of different types of human cancer, including stomach cancer (10,11) pancreatic cancer (12), colorectal cancer (12), lung cancer (13), breast cancer (14,15), prostate cancer (16), and ovarian cancer (17). The epidemiologic studies on tea drinking and stomach cancer, however, are inconclusive (18). In addition to its possible cancer chemoprevention effects, green tea consumption has also been shown to reduce the risk of cardiovascular disease (19) and to protect against coronary atherosclerosis in men (20).
Inspired by these findings, many studies have been carried out to unravel the protective mechanisms of green tea and especially the major polyphenol constituent, EGCG, by using in vitro cell culture and in vivo rodent cancer models. Several molecular mechanisms in the anticarcinogenesis effects of EGCG have been implicated (21). EGCG could cause G1 cell cycle arrest by inducing the expression of cyclin-dependent kinase inhibitors and downregulating hyperphosphorylated pRb protein (22,23), and subsequently induce cancer cell apoptosis through mitochondrial pathway, thereby increasing the ratio of Bax/Bcl-2 (24). EGCG could also inhibit cancer cell invasion and metastasis by downregulating matrix metalloproteinases (MMPs) and increasing the cell adhesion function (25,26). Additionally, the inhibitory effects of EGCG on lipoxygenase (LOX)-dependent arachidonic acid metabolism (27), fatty acid metabolism (28,29), and NOS (30) are not only related to its cancer prevention effect but may also be related to its protective effects against cardiovascular disease. In terms of regulating cellular signaling pathways, EGCG has been shown to inhibit many tumor-associated signaling pathways, including transforming growth factor-beta pathway (TGF-β) (31), vascular endothelial growth factor receptor (VEGFR) pathway (32), epidermal growth factor receptor (EGFR) pathway (33–35), platelet-derived growth factor (PDGF) pathway (36,37), NF-κB (30), AP-1, PI3K/Akt, and MAPK pathways (26,38,39).
Basic leucine zipper family transcription factor nuclear factor E2-related factor 2 (Nrf2) is involved in the regulation of antioxidant response element (ARE)-mediated gene transcription (40). ARE is an cis-acting element [5′-(G/A)TGA(G/C)nnnGC(G/A)-3′] found in the 5′-flanking region of many phase II drug metabolizing/detoxifying enzyme genes such as glutathione S-transferase (GST), UDP-glucuronosyltransferase (UGT), and NAD(P)H: quinone oxidoreductase-1 (NQO1) (41). Under a homeostatic condition, Nrf2 is sequestered in the cytoplasm by Kelch-like ECH-associated protein (Keap1) (42). Exposure of cells to oxidative stress or phase II gene/ARE inducers will trigger the release of Nrf2 from Keap1 and facilitates the nuclear translocation of Nrf2 (43). The nuclear translocation of Nrf2 and subsequent dimerization with small Maf-F/G/K protein and coactivators such as cAMP response element binding protein (CREB)-binding protein (CBP) will drive the transcriptional activation of its target genes (44). Phase II detoxification enzyme and antioxidant enzyme genes are the main targets of Nrf2/ARE-mediated gene transcription, and therefore Nrf2 is believed to play an important role in cancer chemoprevention and regarded as a potential molecular target of cancer chemoprevention (41). To support this role, studies in Nrf2-deficient mice have shown that phase II enzyme expression was dramatically attenuated in the Nrf2 knockout mice, and these mice were also much more susceptible to carcinogen-induced carcinogenesis (45,46). Because EGCG induced ARE-mediated gene expression in our previous study (47), the interaction of Nrf2-mediated signaling pathway, together with other mechanisms described above, may contribute to the overall chemopreventive function of EGCG.
In the current study, the global gene expression profiles elicited by oral administration of EGCG in wild-type and Nrf2 knockout mice were compared by microarray analysis. In addition to genes that were regulated by EGCG, regardless of the Nrf2 status, clusters of Nrf2-dependent genes regulated by EGCG were also identified. The identification of these genes will give us some valuable insights in the potential role of Nrf2 in the EGCG-mediated gene regulation. The current study is also the first to investigate the global gene expression profiles elicited by EGCG in the in vivo mouse model where the role of Nrf2 is examined.
Materials and Methods
Animal and Treatment
Nrf2 knockout mice Nrf2 (−/−) (C57BL/SV129) were described previously (48). Nrf2 (−/−) mice were backcrossed with C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME USA). DNA was extracted from the tail of each mouse and genotype of the mouse was confirmed by polymerase chain reaction (PCR) by using primers (3′-primer, 5′-GGA ATG GAA AAT AGC TCC TGC C-3′; 5′-primer, 5′-GCC TGA GAG CTG TAG GCC C-3′; and lacZ primer, 5′-GGG TTT TCC CAG TCA CGA C-3′). Nrf2 (−/−) mice-derived PCR products show only one band of ∼200 bp, Nrf2 (+/+) mice-derived PCR products showed a band of ∼300 bp, whereas both bands were shown in Nrf2 (+/−) mice PCR products. Male C57BL/6J/Nrf2(−/−) mice from third generation of backcrossing were used in this study. Age-matched male C57BL/6J mice were purchased from Jackson Laboratory. Mice between 9 and 12 weeks old were used and housed at Rutgers Animal Facility with free access to water and food under 12 h light/dark cycles. After 1 week of acclimatization, mice were put on AIN-76A diet (Research Diets Inc., New Brunswick, NJ, USA) for another week. Mice were then treated with EGCG (LKT Laboratories Inc., St. Paul, MN, USA) at a dose of 200 mg/kg (dissolved in 50% PEG 400 solution at concentration of 20 mg/mL) by oral gavages. The control groups were given vehicle only (50% PEG 400 solution). Each treatment was administrated to a group of four animals for both C57BL/6J and C57BL/6J/Nrf2(−/−) mice. Mice were sacrificed at 3 and 12 h after EGCG treatments or 3 h after vehicle treatment (control group). Livers and small intestines were removed and stored in RNA Later (Ambion, Austin, TX, USA) solution.
Sample Preparation for Microarray Analyses
Total RNA from liver and small intestine tissues were isolated by using a method of TRIzol (Invitrogen, Carlsbad, CA, USA) extraction coupled with the RNeasy kit from Qiagen (Valencia, CA, USA). Briefly, tissues were homogenized in trizol and then extracted with chloroform by vortexing. A small volume (1.2 mL) of aqueous phase after chloroform extraction and centrifugation was adjusted to 35% ethanol and loaded onto an RNeasy column. The column was washed, and RNA was eluted following the manufacturer's recommendations. RNA qualities were examined by electrophoresis, and concentrations were determined by UV spectrometry.
Microarray Hybridization and Data Analysis
Affymetrix (Affymetrix, Santa Clara, CA, USA) mouse genome 430 2.0 array was used to probe the global gene expression profile in mice following EGCG treatment. The mouse genome 430 2.0 Array is a high-density oligonucleotide array that comprised over 45,101 probe sets representing over 34,000 well-substantiated mouse genes. The library file for the array is available at http://www.affymetrix.com/support/technical/libraryfilesmain.affx. After RNA isolation, all the subsequent technical procedures including quality control and concentration measurement of RNA, cDNA synthesis and biotin-labeling of cRNA, hybridization, and scanning of the arrays were performed at Cancer Institute of New Jersey (CINJ) Core Expression Array Facility of Robert Wood Johnson Medical School (New Brunswick, NJ). Each chip was hybridized with cRNA derived from a pooled total RNA sample from four mice per treatment group, per time point, per organ, and per genotype (a total of 12 chips were used in this study) (Fig. 1). Briefly, double-stranded cDNA was synthesized from 5 μg of total RNA and labeled using the ENZO BioArray RNA transcript labeling kit (Enzo Life Sciences, Inc., Farmingdale, NY, USA) to generate biotinylated cRNA. Biotin-labeled cRNA was purified and fragmented randomly according to Affymetrix's protocol. A total of 200 μL of sample cocktail containing 15 μg of fragmented and biotin-labeled cRNA was loaded onto each chip. Chips were hybridized at 45°C for 16 h and washed with fluidics protocol EukGE-WS2v5 according to Affymetrix's recommendation. At the completion of the fluidics protocol, the chips were placed into the Affymetrix GeneChip Scanner, where the intensity of the fluorescence for each feature was measured. The expression value (average difference) for each gene was determined by calculating the average of differences in intensity (perfect match intensity − mismatch intensity) between its probe pairs. The expression analysis file created from each sample (chip) was imported into GeneSpring 6.1 (Silicon Genetics, Redwood City, CA, USA) for further data characterization. Briefly, a new experiment was generated after importing data from the same organ in which data were normalized by the array to the 50th percentile of all measurements on that array. Data filtration based on flags present in at least one of the samples was first performed, and a corresponding gene list based on those flags was generated. Lists of genes that were either induced or suppressed more that two fold between treated vs. vehicle group of same genotype were created by filtration-on-fold function within the presented flag list. By using color-by-Venn-Diagram function, lists of genes that were regulated more than two fold only in C57BL/6J mice in both liver and small intestine were created. Similarly, lists of gene that were regulated over two fold regardless of genotype were also generated.
Quantitative Real–Time PCR for Microarray Data Validation
To verify the microarray data, several genes (including the housekeeping gene GAPDH) from different categories were chosen for quantitative real-time PCR analyses. The specific primers for these genes were designed by using Primerexpress software (Applied Biosystems, Foster City, CA, USA) and listed in Table I. Instead of using pooled RNA from each group, RNA samples isolated from individual mouse as described above were used in real-time PCR analyses. For real-time PCR, the following procedure was followed: briefly, first-strand cDNA was synthesized using 4 μg of total RNA following the protocol of SuperScript III First-Strand cDNA Synthesis System (Invitrogen) in a 40-μl reaction volume. PCR reactions were carried out using 100 times diluted cDNA product, 60 nM of each primer, and SYBR Green master mix (Applied Biosystems) in 10 μl reactions. The PCR parameters were set using SDS 2.1 software (Applied Biosystems) and involved the following stages: 50°C for 2 min, 1 cycle; 95°C for 10 min, 1 cycle; 95°C for 15 s → 55°C for 30 s → 72°C for 30 s, 40 cycles; and 72°C for 10 min, 1 cycle. Levels of quantitative reverse transcription product were measured using SYBR Green fluorescence collected during real-time PCR on an Applied Biosystems PRISM 7900HT system. A control cDNA dilution series was created for each gene to establish a standard curve. After conclusion of the reaction, dissociation curve analysis was performed using the SDS 2.1 software to ascertain the integrity of the PCR reaction product and the absence of primer dimers. Gene expression was determined by normalization with control gene GAPDH. The correlation between the corresponding microarray data and the real-time PCR data was validated via Spearman rank correlation.
Results
EGCG–Altered Gene Expression Pattern in Mouse Liver and Small Intestine
After data normalization, 58.3% (26,289) of the probes passed the filtration based on flags present in at least one of the six liver sample arrays. Among these probes, about 8.6–10.2% of them were either induced or suppressed over two fold by EGCG regardless of genotype and treatment time. Moreover, there was no large difference in the number of probes being regulated by EGCG between C57BL/6J and C57BL/6J/Nrf2(−/−) groups or between different time points. Expression levels of 671 well-defined genes were either induced (554) or suppressed (117) over two fold by EGCG only in the wild-type mice at both time points, whereas 256 well-defined genes were either induced (205) or suppressed (51) over two fold by EGCG in the liver of both genotype groups (Fig. 2A). Similar changes in gene expression profiles were also observed in small intestine. Overall, the expression levels of 61.7% (27,815) probes were detected at least in one of the small intestine sample arrays. Compared with the results from liver sample arrays, a smaller percentage (2.9–3.9%) of probes were either induced or suppressed over two fold by EGCG in wild-type or Nrf2(−/−) mice at both time points. Further analyses by the software showed that 228 well-defined genes were regulated by more than two fold (162 up and 66 down) only in C57BL/6J mice, but not in Nrf2(−/−) mice at both time points by EGCG; meanwhile, 97 (84 up and 13 down) well-defined genes were regulated over two fold by EGCG regardless of genotype at both time points in the small intestine (Fig. 2B).
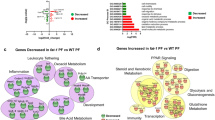
Regulation of Nrf2-dependent and -independent gene expression by EGCG in mouse liver (1A) and small intestine (1B). Gene expression patterns in the liver and the small intestine were analyzed at 3 or 12 h after a single oral dose of 200 mg/kg EGCG; genes that were either induced or suppressed greater than two fold were listed. The positive number on the y-axis refers to the number of genes being induced; the negative number on the y-axis refers to number of genes being suppressed.
Quantitative Real–Time PCR Validation of Microarray Data
To verify the data generated from the microarray, 10 genes from different categories (Table I) were chosen to confirm the EGCG regulative effects by using quantitative real-time PCR analyses as described in Materials and Methods. Values for each gene were normalized by the values of corresponding GAPDH gene and the ratios of treated/vehicle were calculated. Spearman correlation was calculated and it showed that the data generated from microarray analyses are well correlated with the results obtained from quantitative real-time PCR (Fig. 3) with a correlation coefficient R2 of 0.751, with the exception of two high-value points that drove the correlation down quite a bit.
Correlation of microarray data and quantitative real-time PCR data. Fold of changes in gene expression measured by real-time PCR was plotted against the corresponding fold of changes in microarray data. The Spearman correlation was calculated as R2 = 0.751, which indicated the data from the two methods were in good correlation.
EGCG-Regulated Nrf2-Dependent Genes in Liver and Small Intestine
Genes that were altered only in wild-type mice, but not in Nrf2(−/−) mice, by EGCG were considered EGCG-regulated Nrf2-dependent genes. A selected group of these types of genes were categorized based on their biological functions, such as ubiquitination and proteolysis, electron transport, detoxification, transport, cell growth and apoptosis, cell adhesion, kinase and phosphatase, and transcription (Table II).
In the category of ubiquitination and proteolysis, liver gene expression is much more sensitive to EGCG treatment than in small intestine. In liver, EGCG induced several ubiquitination-related genes including ubiquitin fusion degradation 1-like (UFD1L), ubiquitin-specific protease 14 (USP14), and ubiquitin-conjugating enzyme E2I (UBE2I). Interestingly, a previous study showed that these genes were also similarly regulated in an Nrf2-dependent manner by dithiolethione (49). Another big category of genes identified were xenobiotic metabolism enzyme genes including phase I, phase II, and transporter genes. EGCG induced Nrf2-dependent genes including CYP4A10, catalytic subunit of glutamate-cysteine ligase (GCLC), gamma-glutamyltransferase 1 (GGT1), aldehyde reductase-like 6 (ALDRL6), sialyltransferase 10 (ST3GAL6) in liver, and heme oxygenase 1 (HMOX1, HO-1) in the small intestine. Interestingly, arachidonate 12-lipoxygenase (ALOX12), nitric oxide synthase1 (NOS1), and endothelial cell nitric oxide synthase 3 (NOS3) genes were all strongly suppressed. In the liver, EGCG induced several ATP-binding cassette family genes (MDR1 and TAP2) and transporter genes involved in the H+ (ATP5G2), Cu2+ (ATP7A, ATP7B), Cl− (MCLCA1), and fatty acid (FABP4) transport. Many solute family member genes (SLC4A4, SLC9A8, SLC12A4, SLC12A6, SLC13A2, SLC16A1, SLC18A2, and SLC37A3) involved in transporting cellular products such as organic cation, sodium-dependent dicarboxylate, monocarboxylic acid, sodium/hydrogen, and glycerol-3-phosphate were all induced in the liver by EGCG in an Nrf2-dependent manner. As for the transporter genes in the small intestine, hemopexin (HPXN) and major urinary protein 3 (MUP3) genes, which are early response genes, their expression levels were dramatically induced by EGCG. Solute family transporter genes such as SLC4A11, SLC12A9, SLC17A1, SLC35A2, and FPN1 were also induced by EGCG. Interestingly, the same solute carrier family gene SLC6A14 (ATB0,+) was the most highly induced solute carrier family gene in both liver and small intestine, suggesting a possible dominant role for Nrf2 in EGCG-elicited regulation of this gene.
Groups of genes related to apoptosis, cell adhesion, and signaling pathways were also regulated by EGCG in both liver and small intestine. These include the induction of apoptotic protease activating factor 1 (APAF1) and BCL2-associated transcription factor 1 genes in the liver and inhibition of cell cycle control related p21-activated kinase 2 and 3 (PAK2 and PAK3) genes in the small intestine. The cell adhesion-related gene, cadherin 4 (CDH4), was the most highly induced gene in this category both in liver and small intestine by EGCG. Although EGCG has been reported to regulate many signaling pathways by disturbing the phosphorylation status of kinase or phosphatase, the microarray results clearly indicated that EGCG treatment could also regulate the gene expression of many kinases and phosphatases depending on the status of Nrf2. For example, insulin-like growth factor 1 receptor (IGFIR) gene expression was strongly suppressed both in liver and small intestine, and a member of RAS oncogene family Rab38 (RAB38) was inhibited in the small intestine.
A wide variety of transcription-related genes were regulated by EGCG in an Nrf2-dependent manner. The microarray data indicated that more of these genes were regulated in the liver than in the small intestine by EGCG. Genes that were induced including inhibitor of kappaB kinase gamma (IKBKG, IKKγ), CREB binding protein (CREBBP, CBP), retinoblastoma-like 2 (RBL2), retinoid X receptor gamma (RXRG, RXRγ), and histone deacetylase 8 (HDAC8). Several interesting transcription factors, such as paired-like homeobox 2a (PHOX2A), runt related transcription factor 1 (RUNX1), and peroxisome proliferators activated receptor alpha (PPARA, PPARα) genes were suppressed by EGCG in the liver.
EGCG-Regulated Nrf2-Independent Genes in Liver and Small Intestine
With the exception of those genes that were regulated only in C57BL/6J mice described above, a list of genes that were upregulated or downregulated more than two fold in both C57/BL/6J and Nrf2(−/−) mice by EGCG were also identified and classified into similar functional categories (Table III). For genes related to proteolysis, carboxypeptidases (CPA1 and CPB1), elastases (ELA2 and ELA3B), and protease serine 2 (PRSS2) genes were the most sensitive genes to EGCG treatments in the small intestine as they were induced by more than 100-fold in all the time points in both groups. It is interesting that hydroxyacid oxidase 3 (HAO3) and UGT2B5 were induced considerably more in C57BL/6J mice than in Nrf2(−/−) mice, suggesting that their regulation by EGCG may be also Nrf2 genotype-dependent.
EGCG also induced several cell adhesion related genes, such as integrin alpha 8 (ITGA8) and procollagen type IV alpha 5 (COL4A5) in the liver, in an Nrf2-independent manner. Several EGCG-regulated kinases and phosphatases related to the phosphorylation of receptor-couple tyrosine kinase were identified in the liver. These include G protein-coupled receptor kinase 6 (GRK6), receptor-like tyrosine kinase, and protein tyrosine phosphatase receptor type G that were all induced by EGCG treatment. The double cortin and calcium/calmodulin-dependent protein kinase-like 1 gene (DCAMKL1) was the only gene that was suppressed by EGCG in both genotypes.
Discussion
EGCG is a promising cancer chemopreventive agent and its anticancer effects have been investigated in numerous rodent carcinogenesis and tumor models. Because nuclear transcription factor Nrf2 regulates the expression of genes related to cellular defense and detoxification function, and the loss of Nrf2 function in mice results in increased susceptibility to carcinogenesis (45), it is of interest to investigate the role of Nrf2 in EGCG-elicited global gene expression profiles in vivo. One distinct expression pattern found in our current study is that more genes were regulated by EGCG in the liver than in the small intestine in both genotypes. Although the oral administration of EGCG could generate very high concentrations of EGCG in the intestinal tissue (50), differences in the gene expression patterns between the liver and the small intestine could be related to differences in the abundance of nuclear transcription factors and/or other signaling molecules in response to EGCG between the cells of these two tissues. For example, the Nrf2 expression level and nuclear coactivators available to interact with Nrf2 may determine in part how large the pool of its target genes could be regulated. Interestingly, Nrf2 expression level has been found to be much higher in the liver than in the small intestine in humans (51).
Genes that were mostly affected by the Nrf2 status were drug/xenobiotic metabolism enzymes, kinase, and tran scription factors encoding genes. In the current study, several genes belonging to CYP2C, CYP2D, CYP2J, and CYP4A families were identified as EGCG-regulated Nrf2-dependent genes for the first time. The regulation of these cytochrome P450 genes, especially CYP2J and CYP4A genes, implicated that EGCG may be involved in vascular homeostasis, such as the metabolism of fatty acid and epoxyeicosatrienoic acids (52). In agreement with the putative role of Nrf2 in regulating phase II and antioxidant enzyme genes and with the previous report (47) in which EGCG could induce ARE-mediated gene transcription, gamma-glutamyltransferase 1, GCLC, and heme oxygenase 1 (HMOX1, HO-1) were identified as EGCG-induced Nrf2-dependent genes. The regulation of UGT2B5 is also considered Nrf2 genotype-dependent because it was more inducible by EGCG in C57BL/6J mice than in Nrf2(−/−) mice. Interestingly, this gene was also found to be Nrf2-dependently induced by dithiolethiones in a previous mouse microarray study (49). Another putative Nrf2 target gene cytochrome c oxidase, subunit VIIa (COX7A2) (53), was also induced by EGCG only in wild-type mouse liver. The identification of these genes strongly supported the role of Nrf2 in exerting EGCG's chemopreventive effects and validated the microarray data through biological or functional aspects. Transport-function related genes are one of the biggest gene categories regulated by EGCG in both genotypes of mice. Among these transporter genes, several ABC family transporters (such as MDR1) and many solute carrier family members (such as organic anion/cation transporters) were induced in the liver and/or small intestine in an Nrf2-dependent manner. Interestingly, EGCG regulates more Nrf2-dependent transporter genes than Nrf2-independent transporter genes, suggesting that Nrf2 plays a critical role in mediating EGCG-induced expression of transporter genes. It is also interesting that several transporter genes' expressions (such as ABCA5, SLC7A4, and SLCO1A6) were suppressed by EGCG in both types of mice. Although nuclear receptor pregnane X receptor (PXR) and constitutive androstane receptor (CAR) have been implicated in regulating the expression of numerous transporters (such as MDR, MRP and OATP) (54), the role of Nrf2 has not been fully investigated. In a recent study, Nrf2 activators butylated hydroxyanisole, oltipraz, and ethoxyquin were found to induce Mrp2-6 in C57BL/6J mouse liver, and Mrp3 induction was suggested to be mediated by Nrf2 and AhR (55). Therefore, the current study clearly suggests that Nrf2 not only mediated the transcription of phase II drug metabolism enzyme genes, but could also regulate the expression of phase III transporters. Because EGCG could regulate a wide variety of drug metabolism enzyme genes including these phase II detoxification and phase III transporter genes as indicated by the microarray data, and Nrf2 was also found to be involved in these processes, one of the potential molecular mechanisms underlying the anticarcinogenesis effects of EGCG could be the enhancement of the cellular defense system as well as the excretion or efflux of the carcinogen/metabolites by regulating Nrf2-mediated gene transcription.
Previous studies have shown that EGCG could cause cell cycle arrest and induce apoptosis in many cancer cells (23,24). From the array data, EGCG could induce APAF1 gene by more than eight fold in the liver. The induction of Apaf-1 gene by EGCG is consistent with the recent studies (56,57) in which EGCG treatment induced the expression of Apaf-1 in breast cancer cells. P21-activated kinase 2 and 3 (PAK2 and PAK3) were suggested to be essential for Ras-induced upregulation of cyclin D1 during G1 to S transition (58); therefore, suppression of PAK2 and PAK3 by EGCG may relate to its ability to cause G1 cell cycle arrest. EGCG has been shown to inhibit cancer cell invasion and metastasis by increasing cell adhesive ability through upregulation of the beta 1 integrin subunit (26,57,59). In the microarray study, EGCG was found to induce integrin alpha 6 in the liver, and the induction of Rho-associated coiled-coil forming kinase ROCK1 and ROCK2 by EGCG may lead to enhanced integrin-mediated cell adhesion (60). As the impaired expression of cadherin genes was associated with cancer invasion and metastasis (61), the induction of cadherin genes cadherin 4 (CDH4) and cadherin 22 (CDH22) both in the liver and the small intestine by EGCG could also contribute to its cancer chemopreventive effects.
EGCG could block the activation of many signaling pathways such as VEGF, EGF, PDGF, NF-κB, ERK, and PI3K/Akt pathways as indicated by previous studies (4,32,33,36,39). Blocking these pathways is believed to play a central role in exerting the cancer chemopreventive effects of EGCG. The microarray data indicated that EGCG could regulate the expression of many kinase components related to these pathways. TNF receptor-associated factor 3 (CRAF1, TRAF3), which was induced by EGCG more than four fold, could heterodimerize with TRAF2 and inhibit the activation of NF-κB induced by TRAF2 (62), and therefore the induction of TRAF3 and IKKγ genes seems to be consistent with EGCG's inhibitory effect on NF-κB signaling pathways reported in many previous studies (38), which may be critical in EGCG's chemopreventive effects. The inhibition of insulin-like growth factor 1 receptor gene (IGF1R) by EGCG in both liver and small intestine is interesting because IGF-1R signaling is involved in the proliferation, invasion, and metastasis of many tumors including colorectal cancer and hepatocarcinoma by subsequently activation of ERK and PI3K/Akt pathways (63). Therefore, this is the first identification of IGF-1R as a target of EGCG in vivo, and the suppression of IGF-IR by EGCG in our study is consistent with a recent study showing that green tea polyphenols inhibited insulin-like growth factor I (IGF-I) signaling pathway in a prostate cancer mice model (7). The inhibition of IGF-1R gene expression was also accompanied with the suppression of Rab38 gene (RAB38), which is a member of RAS oncogene family in the small intestine. PKC mu (PKD1) was recently shown to phosphorylate E-cadherin, and PKD1 was downregulated in advanced human prostate cancer (64). Therefore, inducing of PKD1 in the liver at 3 h and in the small intestine by EGCG may result in stabilization of the cadherin/catenin complex. This may lead to increased cell aggregation and decreased cellular motility, contributing to inhibition of tumor metastasis. Arachidonate 12-lipoxygenase (ALOX12) converts arachidonic acid to 12(S)-hydroxyeicosatetraenoic acid (HETE), which is a signaling molecule implicated in tumor angiogenesis, growth, metastasis, and inhibition of apoptosis through the activation of NF-κB pathway (65). This enzyme and its product may also be involved in atherosclerosis and inflammation. Inhibition of this enzyme has been shown to induce apoptosis in gastric cancer cells (66). The inhibition of MAP/microtubule affinity-regulating kinase 1 (MARKL1) by EGCG is also interesting because overexpression of this kinase was found in many hepatic carcinoma cells and accompanied by accumulation of β-catenin (67).
Among the transcription factors that were Nrf2-dependently regulated by EGCG, retinoid X receptor gamma (RXRγ), which was induced by EGCG in our study, has been reported to induce terminal differentiation in squamous cell carcinoma lines, suggesting a potential tumor suppressor function for this transcription factor (68). The suppression of oncogenic transcription factor runt related transcription factor (RUNX1) gene is important because a recent study showed that RUNX1 could interplay with DNA methyltransferases (DNMT) by forming a complex (69), and EGCG has been reported to inhibit DNMT and reactivate methylation silenced genes by demethylating the hypermethylated promoter region (70). Krupple-like factor 4 (KLF4) gene is highly expressed in epithelial tissues, such as the gut. Decreased or loss of KLF4 expression has been observed in many gastric cancers (71), therefore, induction of KLF4 by EGCG in small intestine (as shown in our study) suggested another potential mechanism of EGCG for colon cancer prevention. Peroxisome proliferator-activated receptor alpha (PPARα) is involved in fatty acid metabolism, because previous studies have shown that EGCG induced cancer cell growth inhibition and apoptosis may be associated with its ability to block fatty acid synthesis (28). The downregulation of this gene by EGCG in the small intestine is in accord with these previous findings.
For Nrf2-dependent genes that were induced by EGCG treatment, the molecular mechanisms have been well studied. It is believed that exposure of phase II detoxification enzyme inducers, including some chemopreventive agents in the cells, could result in the nuclear accumulation or phosphorylation of Nrf2 as well as its coactivators by many putative mechanisms that have been discussed previously (40). Although considerably more Nrf2-dependent genes were upregulated by EGCG in this study, many interesting genes were also suppressed by EGCG treatment in an Nrf2-dependent manner. The Nrf2-mediated downregulation of gene transcription mechanism has not been well studied. However, in a recent study, Dhakshinamoorthy et al. (72) discovered that overexpression of transcription factor Bach1 in HepG2 cellscould negatively regulate the expression of NAD(P)H:quinone oxidoreductase 1 (NQO1) and ARE luciferase by binding to ARE as a heterodimer with small Maf proteins. Therefore, the Nrf2-dependent induction or suppression of gene expression by EGCG may not only depend on the direct effects of Nrf2, but may also depend on interactions with other transcription factors, coactivators, or corepressors of Nrf2 transcriptional activation complex in the nucleus.
Although this study focused on genes regulated by EGCG in an Nrf2-dependent manner, many EGCG-regulated genes, which are Nrf2-independent, were also identified and classified into similar categories except that the number of genes was smaller. For genes related to ubiquitination and proteolysis, clusters of carboxypeptidases, elastase 3B (ELA3B), and protein serine 2 genes (PRSS2) were the most highly induced genes by EGCG in this study. A chymotrypsin-like unnamed protein product gene was also induced more than 100-fold in the small intestine, and it is interesting to note that a previous study suggested that EGCG could selectively inhibit the chymotrypsin-like activity of proteasome (73). Therefore, further investigation will be needed to address the role of EGCG on proteasome gene expression and activity. Genes such as transporters in ABC and SLC families, cell adhesion protein, G-protein coupled receptor kinases, tyrosine kinases, and transcription factors were also interesting and their role in EGCG chemoprevention deserves further investigation.
In summary, our microarray analysis provides some novel insights into the global gene expression profiles elicited in the mouse liver and small intestine by EGCG. Among these EGCG-regulated genes, clusters of Nrf2-dependent gene were identified by comparing gene expression profiles between C57BL/6J and C57BL/6J/Nrf2(−/−) mice. Many of these genes were identified as EGCG-regulated Nrf2-dependent genes for the first time, such as many transport-related genes. The ability to regulate a wide variety of Nrf2-dependent genes related to ubiquitination, drug metabolism, cell growth and adhesion, phosphorylation, and transcription by EGCG may contribute to the overall anticarcinogenesis and/or the beneficial effects of green tea consumption. Results from this study also provide important and novel insights into the molecular mechanisms underlying EGCG's cancer chemoprevention effects as well as the role of Nrf2 in its biological functions. Future studies on other naturally occurring cancer chemopreventive agents focusing on specific molecular targets or signaling pathways identified in this study would greatly extend our current knowledge on cancer chemoprevention (34).
Abbreviations
- ABC:
-
ATP-binding cassette
- ALOX:
-
arachidonate 12-lipoxygenase
- ARE:
-
antioxidant response element
- DNMT:
-
DNA methyltransferases
- EGCG:
-
(−)-epigallocatechin-3-gallate
- IGF-1R:
-
insulin-like growth factor 1 receptor gene
- MAPK:
-
mitogen-activated protein kinase
- MMP:
-
matrix metalloprotease
- NF-κB:
-
nuclear factor kappa B
- Nrf2:
-
nuclear factor E2-related factor 2
- NOS:
-
nitric oxide synthase
References
C. S. Yang P. Maliakal X. Meng (2002) ArticleTitleInhibition of carcinogenesis by tea Annu. Rev. Pharmacol. Toxicol. 42 25–54 Occurrence Handle10.1146/annurev.pharmtox.42.082101.154309 Occurrence Handle11807163
S. K. Katiyar R. Agarwal Z. Y. Wang A. K. Bhatia H. Mukhtar (1992) ArticleTitle(−)-Epigallocatechin-3-gallate in Camellia sinensis leaves from Himalayan region of Sikkim: inhibitory effects against biochemical events and tumor initiation in Sencar mouse skin Nutr. Cancer 18 73–83 Occurrence Handle1408948
Y. P. Lu Y. R. Lou J. G. Xie Q. Y. Peng J. Liao C. S. Yang M. T. Huang A. H. Conney (2002) ArticleTitleTopical applications of caffeine or (−)-epigallocatechin gallate (EGCG) inhibit carcinogenesis and selectively increase apoptosis in UVB-induced skin tumors in mice Proc. Natl. Acad. Sci. USA 99 12455–12460 Occurrence Handle10.1073/pnas.182429899 Occurrence Handle12205293
F. Afaq N. Ahmad H. Mukhtar (2003) ArticleTitleSuppression of UVB-induced phosphorylation of mitogen-activated protein kinases and nuclear factor kappa B by green tea polyphenol in SKH-1 hairless mice Oncogene 22 9254–9264 Occurrence Handle10.1038/sj.onc.1207035 Occurrence Handle14681684
S. T. Shi Z. Y. Wang T. J. Smith J. Y. Hong W. F. Chen C. T. Ho C. S. Yang (1994) ArticleTitleEffects of green tea and black tea on 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone bioactivation, DNA methylation, and lung tumorigenesis in A/J mice Cancer Res. 54 4641–4647 Occurrence Handle8062257
S. Liao Y. Umekita J. Guo J. M. Kokontis R. A. Hiipakka (1995) ArticleTitleGrowth inhibition and regression of human prostate and breast tumors in athymic mice by tea epigallocatechin gallate Cancer Lett. 96 239–243 Occurrence Handle10.1016/0304-3835(95)03948-V Occurrence Handle7585463
V. M. Adhami I. A. Siddiqui N. Ahmad S. Gupta H. Mukhtar (2004) ArticleTitleOral consumption of green tea polyphenols inhibits insulin-like growth factor-I-induced signaling in an autochthonous mouse model of prostate cancer Cancer Res. 64 8715–8722 Occurrence Handle10.1158/0008-5472.CAN-04-2840 Occurrence Handle15574782
M. R. Sartippour D. Heber J. Ma Q. Lu V. L. Go M. Nguyen (2001) ArticleTitleGreen tea and its catechins inhibit breast cancer xenografts Nutr. Cancer 40 149–156 Occurrence Handle10.1207/S15327914NC402_11 Occurrence Handle11962250
K. T. Kavanagh L. J. Hafer D. W. Kim K. K. Mann D. H. Sherr A. E. Rogers G. E. Sonenshein (2001) ArticleTitleGreen tea extracts decrease carcinogen-induced mammary tumor burden in rats and rate of breast cancer cell proliferation in culture J. Cell. Biochem. 82 387–398 Occurrence Handle10.1002/jcb.1164 Occurrence Handle11500915
Y. Hoshiyama T. Kawaguchi Y. Miura T. Mizoue N. Tokui H. Yatsuya K. Sakata T. Kondo S. Kikuchi H. Toyoshima N. Hayakawa A. Tamakoshi Y. Ohno T. Yoshimura (2004) ArticleTitleA nested case-control study of stomach cancer in relation to greentea consumption in Japan Br. J. Cancer 90 135–138 Occurrence Handle10.1038/sj.bjc.6601512 Occurrence Handle14710220
V. W. Setiawan Z. F. Zhang G. P. Yu Q. Y. Lu Y. L. Li M. L. Lu M. R. Wang C. H. Guo S. Z. Yu R. C. Kurtz C. C. Hsieh (2001) ArticleTitleProtective effect of green tea on the risks of chronic gastritis and stomach cancer Int. J. Cancer 92 600–604 Occurrence Handle10.1002/ijc.1231 Occurrence Handle11304697
B. T. Ji W. H. Chow A. W. Hsing J. K. McLaughlin Q. Dai Y. T. Gao W. J. Blot J. F. Fraumeni SuffixJr (1997) ArticleTitleGreen tea consumption and the risk of pancreatic and colorectal cancers Int. J. Cancer 70 255–258 Occurrence Handle10.1002/(SICI)1097-0215(19970127)70:3<255::AID-IJC1>3.0.CO;2-W Occurrence Handle9033623
L. Zhong M. S. Goldberg Y. T. Gao J. A. Hanley M. E. Parent F. Jin (2001) ArticleTitleA population-based case-control study of lung cancer and green tea consumption among women living in Shanghai, China Epidemiology 12 695–700 Occurrence Handle10.1097/00001648-200111000-00019 Occurrence Handle11679799
A. H. Wu M. C. Yu C. C. Tseng J. Hankin M. C. Pike (2003) ArticleTitleGreen tea and risk of breast cancer in Asian Americans Int. J. Cancer 106 574–579 Occurrence Handle10.1002/ijc.11259 Occurrence Handle12845655
A. H. Wu C. C. Tseng D. Berg ParticleVan Den M. C. Yu (2003) ArticleTitleTea intake, COMT genotype, and breast cancer in Asian-American women Cancer Res. 63 7526–7529 Occurrence Handle14612555
L. Jian L. P. Xie A. H. Lee C. W. Binns (2004) ArticleTitleProtective effect of green tea against prostate cancer: a case-control study in southeast China Int. J. Cancer 108 130–135 Occurrence Handle14618627
M. Zhang A. H. Lee C. W. Binns X. Xie (2004) ArticleTitleGreen tea consumption enhances survival of epithelial ovarian cancer Int. J. Cancer 112 465–469 Occurrence Handle10.1002/ijc.20456 Occurrence Handle15382073
Y. Tsubono Y. Nishino S. Komatsu C. C. Hsieh S. Kanemura I. Tsuji H. Nakatsuka A. Fukao H. Satoh S. Hisamichi (2001) ArticleTitleGreen tea and the risk of gastric cancer in Japan N. Engl. J. Med. 344 632–636 Occurrence Handle10.1056/NEJM200103013440903 Occurrence Handle11228277
K. Nakachi S. Matsuyama S. Miyake M. Suganuma K. Imai (2000) ArticleTitlePreventive effects of drinking green tea on cancer and cardiovascular disease: epidemiological evidence for multiple targeting prevention BioFactors 13 49–54 Occurrence Handle11237198
S. Sasazuki H. Kodama K. Yoshimasu Y. Liu M. Washio K. Tanaka S. Tokunaga S. Kono H. Arai Y. Doi T. Kawano O. Nakagaki K. Takada S. Koyanagi K. Hiyamuta T. Nii K. Shirai M. Ideishi K. Arakawa M. Mohri A. Takeshita (2000) ArticleTitleRelation between green tea consumption and the severity of coronary atherosclerosis among Japanese men and women Ann. Epidemiol. 10 401–408 Occurrence Handle10.1016/S1047-2797(00)00066-1 Occurrence Handle10964006
Z. Hou J. D. Lambert K. V. Chin C. S. Yang (2004) ArticleTitleEffects of tea polyphenols on signal transduction pathways related to cancer chemoprevention Mutat. Res. 555 3–19 Occurrence Handle15476848
L. Y. Chung T. C. Cheung S. K. Kong K. P. Fung Y. M. Choy Z. Y. Chan T. T. Kwok (2001) ArticleTitleInduction of apoptosis by green tea catechins in human prostate cancer DU145 cells Life Sci. 68 1207–1214 Occurrence Handle10.1016/S0024-3205(00)01020-1 Occurrence Handle11228105
N. Ahmad V. M. Adhami S. Gupta P. Cheng H. Mukhtar (2002) ArticleTitleRole of the retinoblastoma (pRb)-E2F/DP pathway in cancer chemopreventive effects of green tea polyphenol epigallocatechin-3-gallate Arch. Biochem. Biophys. 398 125–131 Occurrence Handle10.1006/abbi.2001.2704 Occurrence Handle11811957
M. Masuda M. Suzui I. B. Weinstein (2001) ArticleTitleEffects of epigallocatechin-3-gallate on growth, epidermal growth factor receptor signaling pathways, gene expression, and chemosensitivity in human head and neck squamous cell carcinoma cell lines Clin. Cancer Res. 7 4220–4229 Occurrence Handle11751523
Y. D. Jung L. M. Ellis (2001) ArticleTitleInhibition of tumour invasion and angiogenesis by epigallocatechin gallate (EGCG), a major component of green tea Int. J. Exp. Pathol. 82 309–316 Occurrence Handle10.1046/j.1365-2613.2001.00205.x Occurrence Handle11846837
H. S. Kim M. H. Kim M. Jeong Y. S. Hwang S. H. Lim B. A. Shin B. W. Ahn Y. D. Jung (2004) ArticleTitleEGCG blocks tumor promoter-induced MMP-9 expression via suppression of MAPK and AP-1 activation in human gastric AGS cells Anticancer Res. 24 747–753 Occurrence Handle15161022
J. Hong T. J. Smith C. T. Ho D. A. August C. S. Yang (2001) ArticleTitleEffects of purified green and black tea polyphenols on cyclooxygenase- and lipoxygenase-dependent metabolism of arachidonic acid in human colon mucosa and colon tumor tissues Biochem. Pharmacol. 62 1175–1183 Occurrence Handle10.1016/S0006-2952(01)00767-5 Occurrence Handle11705450
K. Brusselmans E. Schrijver ParticleDe W. Heyns G. Verhoeven J. V. Swinnen (2003) ArticleTitleEpigallocatechin-3-gallate is a potent natural inhibitor of fatty acid synthase in intact cells and selectively induces apoptosis in prostate cancer cells Int. J. Cancer 106 856–862 Occurrence Handle10.1002/ijc.11317 Occurrence Handle12918062
C. W. Yeh W. J. Chen C. T. Chiang S. Y. Lin-Shiau J. K. Lin (2003) ArticleTitleSuppression of fatty acid synthase in MCF-7 breast cancercells by tea and tea polyphenols: a possible mechanism for their hypolipidemic effects Pharmacogenomics J. 3 267–276 Occurrence Handle12931129
Y. L. Lin J. K. Lin (1997) ArticleTitle(−)-Epigallocatechin-3-gallate blocks the induction of nitric oxide synthase by down-regulating lipopolysaccharide-induced activity of transcription factor nuclear factor-kappaB Mol. Pharmacol. 52 465–472 Occurrence Handle9281609
P. C. Chen D. S. Wheeler V. Malhotra K. Odoms A. G. Denenberg H. R. Wong (2002) ArticleTitleA green tea-derived polyphenol, epigallocatechin-3-gallate, inhibits IkappaB kinase activation and IL-8 gene expression in respiratory epithelium Inflammation 26 233–241 Occurrence Handle10.1023/A:1019718718977 Occurrence Handle12238566
Y. D. Jung M. S. Kim B. A. Shin K. O. Chay B. W. Ahn W. Liu C. D. Bucana G. E. Gallick L. M. Ellis (2001) ArticleTitleEGCG, a major component of green tea, inhibits tumour growth by inhibiting VEGF induction in human colon carcinoma cells Br. J. Cancer 84 844–850 Occurrence Handle10.1054/bjoc.2000.1691 Occurrence Handle11259102
N. Ahmad D. K. Feyes A. L. Nieminen R. Agarwal H. Mukhtar (1997) ArticleTitleGreen tea constituent epigallocatechin-3-gallate and induction of apoptosis and cell cycle arrest in human carcinoma cells J. Natl. Cancer Inst. 89 1881–1886 Occurrence Handle10.1093/jnci/89.24.1881 Occurrence Handle9414176
Y. C. Liang S. Y. Lin-Shiau C. F. Chen J. K. Lin (1999) ArticleTitleInhibition of cyclin-dependent kinases 2 and 4 activities as well as induction of Cdk inhibitors p21 and p27 during growth arrest of human breast carcinoma cells by (−)-epigallocatechin-3-gallate J. Cell. Biochem. 75 1–12 Occurrence Handle10.1002/(SICI)1097-4644(19991001)75:1<1::AID-JCB1>3.0.CO;2-N Occurrence Handle10462699
N. Ahmad P. Cheng H. Mukhtar (2000) ArticleTitleCell cycle dysregulation by green tea polyphenol epigallocatechin-3-gallate Biochem. Biophys. Res. Commun. 275 328–334 Occurrence Handle10.1006/bbrc.2000.3297 Occurrence Handle10964666
A. Chen L. Zhang J. Xu J. Tang (2002) ArticleTitleThe antioxidant (−)-epigallocatechin-3-gallate inhibits activated hepatic stellate cell growth and suppresses acetaldehyde-induced gene expression Biochem. J. 368 695–704 Occurrence Handle10.1042/BJ20020894 Occurrence Handle12223099
A. A. Weber T. Neuhaus R. A. Skach J. Hescheler H. Y. Ahn K. Schror Y. Ko A. Sachinidis (2004) ArticleTitleMechanisms of the inhibitory effects of epigallocatechin-3 gallate on platelet-derived growth factor-BB-induced cell signaling and mitogenesis FASEB J. 18 128–130 Occurrence Handle14630705
F. Yang H. S. Oz S. Barve W. J. Villiers Particlede C. J. McClain G. W. Varilek (2001) ArticleTitleThe green tea polyphenol (−)-epigallocatechin-3-gallate blocks nuclear factor-kappa B activation by inhibiting Ikappa B kinase activity in the intestinal epithelial cell line IEC-6 Mol. Pharmacol. 60 528–533 Occurrence Handle11502884
M. Nomura A. Kaji Z. He W. Y. Ma K. Miyamoto C. S. Yang Z. Dong (2001) ArticleTitleInhibitory mechanisms of tea polyphenols on the ultraviolet B-activated phosphatidylinositol 3-kinase-dependent pathway J. Biol. Chem. 276 46624–46631 Occurrence Handle10.1074/jbc.M107897200 Occurrence Handle11591714
C. Chen A. N. Kong (2004) ArticleTitleDietary chemopreventive compounds and ARE/EpRE signaling Free Radic. Biol. Med. 36 1505–1516 Occurrence Handle10.1016/j.freeradbiomed.2004.03.015 Occurrence Handle15182853
J. S. Lee Y. J. Surh (2005) ArticleTitleNrf2 as a novel molecular target for chemoprevention Cancer Lett. 224 171–184 Occurrence Handle10.1016/j.canlet.2004.09.042 Occurrence Handle15914268
K. Itoh N. Wakabayashi Y. Katoh T. Ishii K. Igarashi J. D. Engel M. Yamamoto (1999) ArticleTitleKeap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain Genes Dev. 13 76–86 Occurrence Handle10.1101/gad.13.17.2328 Occurrence Handle9887101
A. T. Dinkova-Kostova W. D. Holtzclaw R. N. Cole K. Itoh N. Wakabayashi Y. Katoh M. Yamamoto P. Talalay (2002) ArticleTitleDirect evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants Proc. Natl. Acad. Sci. USA 99 11908–11913 Occurrence Handle10.1073/pnas.172398899 Occurrence Handle12193649
G. Shen V. Hebbar S. Nair C. Xu W. Li W. Lin Y. S. Keum J. Han M. A. Gallo A. N. Kong (2004) ArticleTitleRegulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein J. Biol. Chem. 279 23052–23060 Occurrence Handle10.1074/jbc.M401368200 Occurrence Handle15020583
A. Enomoto K. Itoh E. Nagayoshi J. Haruta T. Kimura T. O'Connor T. Harada M. Yamamoto (2001) ArticleTitleHigh sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes Toxicol. Sci. 59 169–177 Occurrence Handle10.1093/toxsci/59.1.169 Occurrence Handle11134556
M. Ramos-Gomez M. K. Kwak P. M. Dolan K. Itoh M. Yamamoto P. Talalay T. W. Kensler (2001) ArticleTitleSensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in Nrf2 transcription factor-deficient mice Proc. Natl. Acad. Sci. USA 98 3410–3415 Occurrence Handle10.1073/pnas.051618798 Occurrence Handle11248092
C. Chen R. Yu E. D. Owuor A. N. Kong (2000) ArticleTitleActivation of antioxidant-response element (ARE), mitogen-activated protein kinases (MAPKs) and caspases by major green tea polyphenol components during cell survival and death Arch. Pharm. Res. 23 605–612 Occurrence Handle11156183
K. Chan R. Lu J. C. Chang Y. W. Kan (1996) ArticleTitleNrf2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development Proc. Natl. Acad. Sci. USA 93 13943–13948 Occurrence Handle10.1073/pnas.93.24.13943 Occurrence Handle8943040
M. K. Kwak N. Wakabayashi K. Itoh H. Motohashi M. Yamamoto T. W. Kensler (2003) ArticleTitleModulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival J. Biol. Chem. 278 8135–8145 Occurrence Handle10.1074/jbc.M211898200 Occurrence Handle12506115
J. D. Lambert M. J. Lee H. Lu X. Meng J. J. Hong D. N. Seril M. G. Sturgill C. S. Yang (2003) ArticleTitleEpigallocatechin-3-gallate is absorbed but extensively glucuronidated following oral administration to mice J. Nutr. 133 4172–4177 Occurrence Handle14652367
J. Y. Chan X. L. Han Y. W. Kan (1993) ArticleTitleIsolation of cDNA encoding the human NF-E2 protein Proc. Natl. Acad. Sci. USA 90 11366–11370 Occurrence Handle8248255
E. Grasso V. Longo F. Coceani P. Giovanni Gervasi (2005) ArticleTitleCytochrome P450 expression and catalytic activity in coronary arteries and liver of cattle Biochim. Biophys. Acta 1722 116–123 Occurrence Handle15716024
K. Chantrel-Groussard L. Delpy M. H. Ratinaud M. Cogne (2001) ArticleTitleCharacterization of the murine gene for subunit VIIaL of cytochrome c oxidase C. R. Acad. Sci. III 324 1117–11123 Occurrence Handle11803812
C. Xu C. Y. Li A. N. Kong (2005) ArticleTitleInduction of phase I, II and III drug metabolism/transport by xenobiotics Arch. Pharm. Res. 28 249–268 Occurrence Handle15832810
J. M. Maher, X. Cheng, A. L. Slitt, M. Z. Dieter, and C. D. Klaassen. Induction of the Mrp family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab. Dispos. (2005).
A. M. Roy M. S. Baliga S. K. Katiyar (2005) ArticleTitleEpigallocatechin-3-gallate induces apoptosis in estrogen receptor-negative human breast carcinoma cells via modulation in protein expression of p53 and Bax and caspase-3 activation Mol. Cancer Ther. 4 81–90 Occurrence Handle15657356
M. S. Baliga S. Meleth S. K. Katiyar (2005) ArticleTitleGrowth inhibitory and antimetastatic effect of green tea polyphenols on metastasis-specific mouse mammary carcinoma 4T1 cells in vitro and in vivo systems Clin. Cancer Res. 11 1918–1927 Occurrence Handle10.1158/1078-0432.CCR-04-1976 Occurrence Handle15756018
T. Nheu H. He Y. Hirokawa F. Walker J. Wood H. Maruta (2004) ArticleTitlePAK is essential for RAS-induced upregulation of cyclin D1 during the G1 to S transition Cell Cycle 3 71–74 Occurrence Handle14657670
A. Pilorget V. Berthet J. Luis A. Moghrabi B. Annabi R. Beliveau (2003) ArticleTitleMedulloblastoma cell invasion is inhibited by green tea (−)epigallocatechin-3-gallate J. Cell. Biochem. 90 745–755 Occurrence Handle10.1002/jcb.10667 Occurrence Handle14587030
F. Ikeda H. Terajima Y. Shimahara T. Kondo Y. Yamaoka (2003) ArticleTitleReduction of hepatic ischemia/reperfusion-induced injury by a specific ROCK/Rho kinase inhibitor Y-27632 J. Surg. Res. 109 155–160 Occurrence Handle10.1016/S0022-4804(02)00081-1 Occurrence Handle12643858
K. Matsuura J. Kawanishi S. Fujii M. Imamura S. Hirano M. Takeichi Y. Niitsu (1992) ArticleTitleAltered expression of E-cadherin in gastric cancer tissues and carcinomatous fluid Br. J. Cancer 66 1122–1130 Occurrence Handle1333788
L. He A. C. Grammer X. Wu P. E. Lipsky (2004) ArticleTitleTRAF3 forms heterotrimers with TRAF2 and modulates its ability to mediate NF-{kappa}B activation J. Biol. Chem. 279 55855–55865 Occurrence Handle10.1074/jbc.M407284200 Occurrence Handle15383523
Y. Min Y. Adachi H. Yamamoto A. Imsumran Y. Arimura T. Endo Y. Hinoda C. T. Lee S. Nadaf D. P. Carbone K. Imai (2005) ArticleTitleInsulin-like growth factor I receptor blockade enhances chemotherapy and radiation responses and inhibits tumour growth in human gastric cancer xenografts Gut 54 591–600 Occurrence Handle10.1136/gut.2004.048926 Occurrence Handle15831900
M. Jaggi P. S. Rao D. J. Smith M. J. Wheelock K. R. Johnson G. P. Hemstreet K. C. Balaji (2005) ArticleTitleE-cadherin phosphorylation by protein kinase D1/protein kinase C{mu} is associated with altered cellular aggregation and motility in prostate cancer Cancer Res. 65 483–492 Occurrence Handle15695390
M. Kandouz D. Nie G. P. Pidgeon S. Krishnamoorthy K. R. Maddipati K. V. Honn (2003) ArticleTitlePlatelet-type 12-lipoxygenase activates NF-kappaB in prostate cancer cells Prostaglandins Other Lipid Mediat. 71 189–204 Occurrence Handle10.1016/S1098-8823(03)00042-X Occurrence Handle14518561
B. C. Wong W. P. Wang C. H. Cho X. M. Fan M. C. Lin H. F. Kung S. K. Lam (2001) ArticleTitle12-Lipoxygenase inhibition induced apoptosis in human gastric cancer cells Carcinogenesis 22 1349–1354 Occurrence Handle10.1093/carcin/22.9.1349 Occurrence Handle11532854
T. Kato S. Satoh H. Okabe O. Kitahara K. Ono C. Kihara T. Tanaka T. Tsunoda Y. Yamaoka Y. Nakamura Y. Furukawa (2001) ArticleTitleIsolation of a novel human gene, MARKL1, homologous to MARK3 and its involvement in hepatocellular carcinogenesis Neoplasia 3 4–9 Occurrence Handle10.1038/sj.neo.7900132 Occurrence Handle11326310
D. L. Crowe C. F. Shuler (1998) ArticleTitleIncreased cdc2 and cdk2 kinase activity by retinoid X receptor gamma-mediated transcriptional down-regulation of the cyclin-dependent kinase inhibitor p21Cip1/WAF1 correlates with terminal differentiation of squamous cell carcinoma lines Cell Growth Differ. 9 619–627 Occurrence Handle9716179
S. Liu T. Shen L. Huynh M. I. Klisovic L. J. Rush J. L. Ford J. Yu B. Becknell Y. Li C. Liu T. Vukosavljevic S. P. Whitman K. S. Chang J. C. Byrd D. Perrotti C. Plass G. Marcucci (2005) ArticleTitleInterplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia Cancer Res. 65 1277–1284 Occurrence Handle10.1158/0008-5472.CAN-04-4532 Occurrence Handle15735013
M. Z. Fang Y. Wang N. Ai Z. Hou Y. Sun H. Lu W. Welsh C. S. Yang (2003) ArticleTitleTea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines Cancer Res. 63 7563–7570 Occurrence Handle14633667
D. Wei W. Gong M. Kanai C. Schlunk L. Wang J. C. Yao T. T. Wu S. Huang K. Xie (2005) ArticleTitleDrastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression Cancer Res. 65 2746–2754 Occurrence Handle10.1158/0008-5472.CAN-04-3619 Occurrence Handle15805274
S. Dhakshinamoorthy A. K. Jain D. A. Bloom A. K. Jaiswal (2005) ArticleTitleBach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants J. Biol. Chem. 280 16891–16900 Occurrence Handle10.1074/jbc.M500166200 Occurrence Handle15734732
D. J. Kuhn A. C. Burns A. Kazi Q. P. Dou (2004) ArticleTitleDirect inhibition of the ubiquitin-proteasome pathway by ester bond-containing green tea polyphenols is associated with increased expression of sterol regulatory element-binding protein 2 and LDL receptor Biochim. Biophys. Acta 1682 1–10 Occurrence Handle15158750
Acknowledgments
This work was supported in part by NIH Grants R01 CA094828 and R01-CA092515. We thank Mr. Curtis Krier at the Cancer Institute of New Jersey (CINJ) Core Expression Array Facility for assistance with the microarray analyses.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guoxiang Shen and Changjiang Xu contributed equally to the present study.
Rights and permissions
About this article
Cite this article
Shen, G., Xu, C., Hu, R. et al. Comparison of (−)-Epigallocatechin-3-Gallate Elicited Liver and Small Intestine Gene Expression Profiles Between C57BL/6J Mice and C57BL/6J/Nrf2 (−/−) Mice. Pharm Res 22, 1805–1820 (2005). https://doi.org/10.1007/s11095-005-7546-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-005-7546-8