Abstract
MicroRNAs (miRNAs) are short non-coding RNAs that regulate gene expression post-transcriptionally by binding to their cognate target mRNAs. Emerging evidence suggests that miRNAs are critical regulators of neuronal functions. The expression pattern of miRNAs in the peripheral nervous system after peripheral nerve injury suggest that miRNAs may have important and yet unknown roles in the mechanisms of pain. Thus, we examined the role of miR-96 in neuropathic pain using a rat model of the condition chronic constriction sciatic nerve injury (CCI). We found that miR-96 alleviated neuropathic pain. The level of miR-96 was decreased within the ipsilateral dorsal root ganglion (DRG) after peripheral nerve injury but the Nav1.3 level was increased. Specifically, Intrathecal administration of miR-96 suppressed the expression of Nav1.3 induced by CCI. Further examination revealed that miR-96 inhibited the Nav1.3 mRNA expression in the embryonic DRG neurons in vitro. Our findings suggest that miR-96 participate in the regulation of neuropathic pain through inhibiting the expression of Nav1.3 in the DRG of CCI rats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNA (miRNA) is a class of single-stranded, endogenous, non-coding RNA 18–24 nucleotides long. They exert direct effects on gene expression by binding to the 3′ untranslated region of target mRNA, which can lead to the degradation of the protein encoded by the target mRNA. It is estimated that more than 30 % of the human genome may be subjected to be regulated by miRNAs [1]. MiRNAs play crucial roles in the regulation of many biological processes, including differentiation, development, apoptosis, metabolism and proliferation [2, 3]. Furthermore, miRNAs have been implicated in viral infection [4], coronary artery diseases [5] and inflammatory lung disease [6], as well as in the development and progression of cancers [7]. Thus, disease-associated miRNAs represent a novel target for therapeutic intervention.
In the nervous system, miRNAs are emerging as important regulators of neuronal survival, neurodegeneration [8], dendritic outgrowth [9], transcriptional regulation and trophic factor regulation [10]. Studies also provide a link between miRNAs and brain diseases. MiR-133b is significantly down-regulated in the midbrain of patients with Parkinson’s disease (PD) [11]. Furthermore, miR-7 reduces alpha-synuclein protein, the major component of Lewy bodies in sporadic PD [12]. Recent data suggest that miRNA are aberrantly expressed in many animal pain models. Ligation of the sciatic nerve induce changes in expression of 201 miRNAs in dorsal root ganglion (DRG) [13]; miRNA expression levels, including those of miR-223, miR-290, miR-137 and miR-219-5P, are altered following traumatic spinal cord injury (SCI) [14]. Spinal nerve ligation (SNL) induced changes in microRNA expression profile in DRG [15, 16]. The miR-183 family is a sensory organ specific cluster of microRNAs that includes miR-96, miR-182 and miR-183. MiR-96, which is abundant in DRG, is down-regulated in DRG following SNL [17]. These data suggest that miR-96 may have an important role in the neuropathic pain.
Nav1.3 plays especially important roles in neuropathic pain. It is predicted that Nav1.3 is one of the miR-96 target genes in rat (TargetScanHuman, release 6.2). In this study, we explored the possibilities that miR-96 would critically regulate neuropathic pain behaviors and the proposed miR-96 effect would be mediated by Nav1.3 in DRG of rats following sciatic nerve ligation. For verification the interaction between miR-96 and Nav1.3, we further investigated the effects of miR-96 on the expression of Nav1.3 in DRG of embryonic rats which is abundant of Nav1.3 [18]. In addition, the expression levels of Nav1.7 and Nav1.8 in DRG of CCI rats were evaluated after intrathecal miR-96. The results indicated that sciatic nerve ligation altered the expressions of miR-96 and Nav1.3 and miR-96 attenuated the expression of Nav1.3. Our results suggest the therapeutic potency of miR-96 on the chronic neuropathic pain.
Materials and Methods
Animals
Male Sprague–Dawley rats weighing 190–220 g and the pregnant rats were used. The rats were fed a standard laboratory diet under controlled temperature and 12 h light/dark cycle. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Medical College of Nanjing University. All efforts were undertaken to minimize the number of animals used and their discomfort. All the DRGs collected from adult rats were ipsilateral to the lesion.
Experimental Design
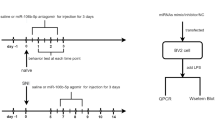
Three experiments were performed in this study. In the first experiment, to analyze the expression pattern of mature rat miR-96 and Nav1.3 after CCI, rats were randomly divided into two groups: sham group (sham, n = 8) and chronic constriction injury group (CCI, n = 24). The expression levels of miR-96 and Nav1.3 in L4–L6 DRG were detected at days 1, 7, 14 and 21 after CCI. The sham operations were performed 7 days before harvesting of DRGs.
In the second experiment, the rats at day 10 after CCI were randomly divided into three groups with 8 rats in each group: a vehicle group (CCI + Vehicle), a scramble group (CCI + scramble miRNA) and a miR-96 group (CCI + miR-96 mimics). In the miR-96 and scramble groups, 50 μl of the scrambled miRNA or miR-96 mimics (60 μg/25 μl; Invitrogen, Carlsbad, CA, USA) and atelocollagen (Koken, Tokyo, Japan) complexes were prepared according to the procedure of Shoji and Takeshita [19, 20].
In the third experiment, the embryonic DRG neurons were cultured and transfected with miR-96 mimics or scramble miRNA. Then the mRNA expression levels of Nav1.3, Nav1.7 and Nav1.8 were detected.
Intrathecal Catheter Implantation
Intrathecal catheters were implanted according to the method described [21]. In briefly, rats were intraperitoneally anesthetized with 10 % chloral hydrate (300 mg/kg). Polyethylene catheters (PE-10) were inserted via an incision in the cisterna magna, and advanced 7.0–7.5 cm caudally to the level of the lumbar enlargement. Twenty-four hours later, rats exhibiting neurological deficits were excluded from the experiments. Proper location of the catheter was confirmed by hind limb paralysis after the injection of 10 μl of 2 % lidocaine.
Chronic Constriction Injury (CCI) Model
The CCI model was established as previously described [22]. Briefly, rats were anesthetized with sodium pentobarbital (40 mg/kg, i.p.). The sciatic nerve was exposed and ligated loosely with 4–0 catgut thread at 4 sites with an interval of 1 mm. Meanwhile, a sham surgery was performed with the sciatic nerve exposed but not ligated.
Intrathecal Injections
Intrathecal drug administration was accomplished using a microinjection syringe connected to the intrathecal catheter. The complexes containing miR-96 or scambled miRNA were administered intrathecally in 50 μl volume once daily for 3 days beginning 10 days after CCI.
Evaluation of the Pain Threshold
Animals were habituated to the test environment daily (a 60 min session) for 4 days before baseline testing. The testing procedure was performed according to a previous method [23]. Mechanical withdrawal threshold (MWT) was determined to evaluate mechanical hyperalgesia by calibrated von Frey filaments (BME-403, Tianjing, china). Thermal hyperalgesia was measured using thermal paw stimulation system (BME-410C, Tianjin, China) and expressed as thermal withdrawal latency (PWL). Each rat was measured three times and the mean value was taken as the threshold value.
Preparation of DRG Cultures
Embryonic rats at embryonic day 18 (E18) were used for the DRG culture preparations. The bilateral dorsal root ganglia (DRGs) were rapidly removed under the microscope and enzymatically digested at 37 °C for 15 min in DMEM containing 2 mg/ml collagenase and 1 mg/ml trypsin (Sigma, St. Louis, USA). After washing three times with culture medium single neurons were obtained by gentle agitation through a Pasteur pipette. The dissociated DRG neurons were plated into 6-well plates at 5 × 105 cells/well and incubated at 37 °C with 5 % CO2 for 24 h. Then the cells were maintained in culture medium containing cytarabine (ara-C) (5 μg/ml) for another 24 h to inhibit growth of non-neuronal cells. The composition of the culture medium was D-MEM/F-12 (1:1, Gibco, Grand Island, NY) supplemented with 10 % fetal bovine serum (Gibco, Grand Island, USA), 10 ng/ml NGF (Invitrogen Corporation, Grand Island, USA), 0.1 mg/ml l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma, St. Louis, USA).
Transfection of miR-96
The miR-96 mimics and scramble miRNA were transfected into the DRG neurons using Lipofectamine 2000 (Invitrogen Corporation, Grand Island, USA) according to the manufacturer’s instructions. The final concentration of small RNAs was 60 nM. Total RNA was harvested 72 h after transfection.
RNA Extraction, Reverse Transcription, and Real-Time PCR Quantification
Total RNA, including miRNAs, of rat DRG was isolated with the mirVana™ miRNA Isolation Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. 1,000 ng total RNA was used as a template for a 20 μl reverse transcription reaction using TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). By using ABI7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA), we performed quantitative PCR for miR-96, Nav1.3, Nav1.7 and Nav1.8. The expression levels of miR-96 were normalized to U6, while levels of Nav1.3, Nav1.7 and Nav1.8 were normalized to GAPDH. The microRNA specific primers and Nav1.3, Nav1.7 and Nav1.8 probes were purchased from Applied Biosystems. All assays were performed in triplicate.
Western Blot
Samples of 30 μg of total protein were separated by 6.5 % SDS–polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. After incubation with Nav1.3 primary antibody (LifeSpan BioSciences, Seattle, WA, USA), the membrane was incubated with peroxidase conjugated secondary antibodies (Cell SignalingTechnology, Danvers, MA, USA). Immunodetection was completed using Pierce-enhanced chemiluminescence substrate (Thermo Scientific, Waltham, MA, USA). GAPDH (Santa Cruz Biotecnology, Santa Cruz, CA, USA) was used as a loading control.
Immunohistochemistry
Six DRGs from 6 rats were used in each group. The Formalin-fixed, paraffin-embedded tissues were cut, and every fifth section (5 μm thickness) was used for Immunohistochemistry (IHC) detection. At least 40 sections were detected in each group. IHC was carried out as previously described [24]. Primary antibody for Nav1.3 (LifeSpan BioSciences, Seattle, WA, USA) was used. Protein was localized by incubation with diaminobenzidine (DAB) and H2O2 for 2 min. Finally, the sections were dehydrated in ethanol and mounted with neutral balsam.
Statistical Analysis
Data reflect the mean ± S.E.M. Comparisons of means between two groups were done with t test and those between multiple groups with one way analysis of variance (ANOVA). A value of P < 0.05 was considered statistically significant.
Results
Nav1.3 mRNA Up-Regulated in L4–L6 DRG of CCI Rats
To assess changes in Nav1.3 mRNA expression in the DRG of CCI rat, Real-time PCR techniques were used to quantify mRNA prepared from L4–L6 DRG. Nav1.3 mRNA levels were detected at days 1, 7, 14 and 21 after CCI and sham group. The results indicated that Nav1.3 increased in DRG of CCI rats (Fig. 1a). At day 1 after CCI, Nav1.3 mRNA level was 1.7 folds compared to sham group (P > 0.05). Then it constantly increased until day 14 after CCI which mRNA level was 4.6 folds than that of sham group (P < 0.01). At day 21 after CCI, Nav1.3 mRNA levels begin to decrease. At this point, it was 3.0 folds compared to sham group (P < 0.01).
Expression changes of Nav1.3 mRNA and miR-96 in the L4–L6 DRG of CCI rats. a Nav1.3 mRNA was up-regulated in the DRG of CCI rats. Nav1.3 mRNA levels were detected by Real-time qPCR at days of 1, 7, 14 and 21 after CCI. b MiR-96 was down-regulated in the DRG of CCI rats. MiR-96 levels were detected by Real-time qPCR at days of 1, 7, 14 and 21 after CCI. Data are shown as mean ± S.E.M, n = 6. **P < 0.01 and ***P < 0.001 versus sham group
MiR-96 Down-Regulated in L4–L6 DRG of CCI Rats
The time course expression of miR-96 was detected following CCI using real-time PCR. The results indicated that CCI decreased miR-96 expression in DRG (Fig. 1b). At day 1 after CCI, miR-96Nav1.3 was about 0.78 fold compared to sham group (P > 0.05). MiR-96 levels decreased following day 7 after CCI (P < 0.001) and remained at low levels. At day 21 after CCI, MiR-96 mRNA levels were still lower compared to that of the sham group (P < 0.001).
Intrathecal miR-96 Alleviates the Neuropathic Pain
The TWL and MWT were measured to evaluate the effects of miR-96 on pain thresholds of the CCI rats. At day 10 after CCI when the neuropathic pain was established, the drugs were intrathecally administered once daily for 3 days. Since day 1 after drugs administration, both thermal hyperalgesia and mechanical allodynia induced by CCI were attenuated by intrathecal administration with miR-96 (P < 0.001), but not scrambled miRNA (Fig. 2).
miR-96 alleviates the neuropathic pain. The TWL (a) and MWT (b) were measured. Vehicle, scrambled miRNA or miR-96 mimics were intrathecally delivered once daily for 3 days, as indicated by arrows. Data are shown as mean ± S.E.M, n = 5. ### P < 0.001 versus scramble group; ***P < 0.001 versus vehicle group
MiR-96 Inhibits the Expression of Nav1.3 in L4–L6 DRG of CCI Rats
In order to identify the relationship between miR-96 and sodium channel subtypes in the DRG after CCI, miR-96 was delivered into the rat following day 10 after CCI for 3 days through intrathecal injection. The expression of Nav1.3 protein and mRNA was detected by Western blotting and real-time PCR. The results showed miR-96 mimics inhibited both Nav1.3 protein and mRNA expression (Figs. 3a, 4a, b). However, miR-96 did not affect the mRNA expressions of Nav1.7 and Nav1.8 (Fig. 3a).
MiR-96 inhibited the mRNA expression of Nav1.3 in DRG. a Nav1.3 mRNA levels were decreased in L4–L6 DRG of CCI rats after intrathecal injection of miR-96 mimics. Vehicle, scrabled miRNA or miR-96 mimics were intrathecally delivered once daily for 3 days. Nav1.3, Nav1.7 and Nav1.8 mRNA levels were detected by Real-time qPCR. b MiR-96 decreased the expression of Nav1.3 mRNA in the embryonic DRGs in vitro. The embryonic DRGs were transfected with miR-96. Nav1.3, Nav1.7 and Nav1.8 mRNA levels were detected by Real-time qPCR. Data are shown as mean ± S.E.M, n = 6. ## P < 0.01 versus scramble group; **P < 0.01 versus vehicle group
MiR-96 decreased the expression of Nav1.3 protein in L4–L6 DRG of CCI rats. Nav1.3 protein levels were decreased in L4–L6 DRG CCI rats after intrathecal injection of miR-96 mimics. a Nav1.3 protein levels were evaluated by Western Blotting. b Quantitative data indicating the expression of Nav1.3 protein. c MiR-96 decreased the immunoreactivities of Nav1.3 in L4–L6 DRG of CCI rats. Nav1.3 immunoreactivity (brown) was detected in DRG neurons of CCI + vehicle group (b), CCI + scramble miRNA group (c) and CCI + miR-96 mimics group (d). Omission of the primary antibodies abolished staining in DRG neurons (a). Arrows indicate Nav1.3-positive neurons; solid arrowheads indicate Nav1.3-negative neurons. Scale bar is 100 μm. d Percentage of Nav1.3-positive neurons in the vehicle, scramble miRNA and miR-96 mimics groups. Data are shown as mean ± S.E.M. n = 3 in a, b; n = 4 in c, d. ## P < 0.01 versus scramble group; *P < 0.05 and **P < 0.01 versus vehicle group
MiR-96 Decreases the Immunoreactivities of Nav1.3 in L4–L6 DRG of CCI Rats
The glia and neurons can be distinguished clearly under the light microscope. Immunoreactivity was detected by immunohistochemistry. Nav1.3 immunohistochemical staining was located in the cytoplasm of the DRG neurons. The percentage of Nav1.3+ neurons from the total DRG neuronal number was analyzed. The results showed miR-96 mimics inhibited Nav1.3 immunoreactivities (Fig. 4c, d). In addition, we found that about 30 % of NaV1.3 positive neurons also expressed activated transcription factor-3 (ATF3), a marker of injured neurons (data not shown).
MiR-96 Down-Regulates the Expression of Nav1.3 in the Embryonic DRG In Vitro
To further test the regulation of Nav1.3 by miR-96, we overexpressed miR-96 by infecting DRG of E18 rats with either a miR-96 mimic expression vector or a control vector containing a scrambled sequence. 72 h after infection, Nav1.3 mRNA expression level in DRG neurons infected by the miR-96 vector was down-regulated. However, the mRNA expression levels of Nav1.7 and Nav1.8 did not change (Fig. 3b).
Discussion
Neuropathic pain, arising from lesions to peripheral nerves, is present in many neurological diseases and frequently occurs in patients with diabetes [25], cancer or AIDS [26].
DRG is anatomically discrete peripheral nervous system (PNS) structure located between the dorsal root and the spinal nerve. It contains cell bodies of the primary neurons of the somatosensory pathway. These neurons receive stimuli from the peripheral tissue via their peripheral axon, and transmit this information to the spinal dorsal horn. DRG is recognized as one of the organs which are damaged in peripheral sensory neuropathic pain states [27]. It has been well recognized that nerve lesion induces change of gene and protein expression in DRG which correspond to induced neuropathic pain [28, 29]. It has been shown that Knockdown of Nav1.3 in rat DRG attenuates nerve injury-induced neuropathic pain [30]. DRG was thought to be a potential new therapeutic target for neuropathic pain [31].
Intrathecal injection is an efficient technique to investigate characteristics of the peripheral nervous system (PNS) in the rat model of neuropathic pain. Intrathecal administration of lappaconitine inhibits nociceptive behaviors by decreasing the expression of the P2X3 receptors in the DRG neurons of the CCI rats [23]. Down-regulation of the sodium channels in DRG neurons contributes to the therapeutic effect of IL-10 on neuropathic pain [32].
Voltage-gated sodium channels play a key role in the initiation and generation of action potentials in neurons. These sodium channels also contribute to the dynamic regulation of neuronal excitability and are good candidates for modulation by second messenger pathways. There are several voltage-gated sodium channels, including Nav1.2, Nav1.3, Nav1.7, Nav1.8 and Nav1.9 these are linked to neuropathic pain. Among these, only Nav1.3 is up-regulated in DRG neurons following peripheral nerve axotomy [33, 34]. Nav1.3 is correlated with ectopic firing and development of neuropathic pain. It is significantly up-regulated in the adult DRG in a range of neuropathic pain models and conditions [18, 33, 35, 36]. Mechanical allodynia and thermal hyperalgesia were reversed by intrathecal injection of Nav1.3 specific antisense oligodeoxynucleotides in CCI rats [36]. By contrast, neither mechanical allodynia nor thermal hyperalgesia was reversed by NaV1.3 antisense treatment in SNI rats [37]. Intrathecal lidocaine pretreatment alleviates neuropathic pain by attenuating Nav1.3 up-regulation and decreasing spinal microglial activation [33]. The behavioral hypersensitivity is partially alleviated by oral administration of two sodium channel blockers, mexiletine and lamotrigine [37]. In this study, we detected the time course expression of Nav1.3 in rat DRG after CCI. Nav1.3 was up-regulated since day 7 after CCI. This data verified that Nav1.3 involved in chronic neuropathic pain.
Aberrant expression of miRNAs is a common feature of several human diseases. Recently, the expression profiles of miRNAs are discovered in neuropathic pain [14, 15, 38]. In addition, miR-96 was decreased in the DRG after SNI [17]. However, the expression change of miR-96 in the DRG of CCI rats is unknown. Our results show that miR-96 was down-regulated in DRG of CCI rats. This suggests that miR-96 may be involved in the pathologic regulation in CCI rats. The role of individual miRNAs in primary sensory neurons has not been extensively investigated. In understanding the role of miRNAs in the control of nociceptive neuron function, it is necessary to determine the mRNAs targeted by miRNAs and their roles on neuron function. To investigate the role of miR-96 on chronic neuropathic pain in CCI rats, miR-96 mimic was injected intrathecally with atelocollagen.
Atelocollagen, a highly purified and biocompatible typeIcollagen, can complex with small RNAs through electrostatic interactions, forming nanoparticles with high delivery efficiency [19, 20]. Using this technique, miR-96 was intrathecally delivered. The results showed that thermal hyperalgesia and mechanical allodynia induced by CCI were attenuated by intrathecal administration with miR-96. It is predicted that Nav1.3 is one of the miR-96 target genes. We further investigated the effects of miR-96 mimics on Nav1.3 expression in DRG after CCI. The results indicated that miR-96 efficiently inhibited Nav1.3 up-regulation caused by CCI. Furthermore, miR-96 down-regulated the expression of Nav1.3 but not Nav1.7 or Nav1.8 in DRG of E18 rats in vitro. These data provided the further evidence that miR-96 can inhibit specifically the Nav1.3 expression in DRG.
In conclusion, our results have shown the expression pattern of Nav1.3 and miR-96 in DRG after CCI. MiR-96 suppressed Nav1.3 expression in DRG after CCI and alleviated the chronic neuropathic pain. These findings suggest that miR-96 involves in the regulation of neuropathic pain by targeting Nav1.3 in CCI rats. To our knowledge, this is the first time to investigate the roles of miR-96 on the neuropathic pain. However, the effect of miR-96 on the spinal cord of CCI rats is unknown. Thus, further investigation will be necessary to clarify this issue.
References
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Ambros V (2006) The functions of animal microRNAs. Nature 431(2004):350–355
Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11:441–450
Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnić VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, Babic M, Zimmer R, Trgovcich J, Koszinowski UH, Jonjic S, Pfeffer S, Dölken L (2012) Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog 8:e1002510
Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z (2007) The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med 13:486–491
Oglesby IK, McElvaney NG, Greene CM (2010) MicroRNAs in inflammatory lung disease—master regulators or target practice? Respir Res 11:148
Ma X, Becker Buscaglia LE, Barker JR, Li Y (2011) MicroRNAs in NF-kappaB signaling. J Mol Cell Biol 3:159–166
Eacker SM, Dawson TM, Dawson VL (2009) Understanding microRNAs in neurodegeneration. Nat Rev Neurosci 10:837–841
Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, Mandel G, Goodman RH (2010) microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci USA 107:20382–20387
Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S (2005) A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA 102:16426–16431
Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A (2007) A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317:1220–1224
Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM (2009) Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci USA 106:13052–13057
Yu B, Zhou S, Wang Y, Ding G, Ding F, Gu X (2011) Profile of microRNAs following rat sciatic nerve injury by deep sequencing: implication for mechanisms of nerve regeneration. PLoS ONE 6:e24612
Liu NK, Wang XF, Lu QB, Xu XM (2009) Altered microRNA expression following traumatic spinal cord injury. Exp Neurol 219:424–429
Von Schack D, Agostino MJ, Murray BS, Li Y, Reddy PS, Chen J, Choe SE, Strassle BW, Li C, Bates B, Zhang L, Hu H, Kotnis S, Bingham B, Liu W, Whiteside GT, Samad TA, Kennedy JD, Ajit SK (2011) Dynamic changes in the microRNA expression profile reveal multiple regulatory mechanisms in the spinal nerve ligation model of neuropathic pain. PLoS ONE 6:e17670
Sakai A, Suzuki H (2013) Nerve injury-induced upregulation of miR-21 in the primary sensory neurons contributes to neuropathic pain in rats. Biochem Biophys Res Commun 435:176–181
Aldrich BT, Frakes EP, Kasuya J, Hammond DL, Kitamoto T (2009) Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience 164:711–723
Waxman SG, Kocsis JD, Black JA (1994) Type III sodium channel mRNA is expressed in embryonic but not adult spinal sensory neurons, and is re-expressed following axotomy. J Neurophysiol 72:466–471
Shoji T, Nakasa T, Yamasaki K, Kodama A, Miyaki S, Niimoto T, Okuhara A, Kamei N, Adachi N, Ochi M (2012) The effect of intra-articular injection of microRNA-210 on ligament healing in a rat model. Am J Sports Med 40:2470–2478
Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, Kosaka N, Kawamata M, Kelnar K, Bader AG, Brown D, Ochiya T (2010) Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther 18:181–187
Yaksh TL, Rudy TA (1976) Chronic catheterization of the spinal subarachnoid space. Physiol Behav 17:1031–1036
Bennett GJ, Xie YK (1988) A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain 33:87–107
Ou S, Zhao YD, Xiao Z, Wen HZ, Cui J, Ruan HZ (2011) Effect of lappaconitine on neuropathic pain mediated by P2X3 receptor in rat dorsal root ganglion. Neurochem Int 58:564–573
Chen HP, Fan J, Cui S (2006) Detection and estrogen regulation of leptin receptor expression in rat dorsal root ganglion. Histochem Cell Biol 126:363–369
Ren YS, Qian NS, Tang Y, Liao YH, Yang YL, Dou KF, Toi M (2012) Sodium channel Nav1.6 is up-regulated in the dorsal root ganglia in a mouse model of type 2 diabetes. Brain Res Bull 87:244–249
Katz N (2000) Neuropathic pain in cancer and AIDS. Clin J Pain 16:S41–S48
Shimoshige Y, Enomoto R, Aoki T, Matsuoka N, Kaneko S (2010) The involvement of aldose reductase in alterations to neurotrophin receptors and neuronal cytoskeletal protein mRNA levels in the dorsal root ganglion of streptozotocin-induced diabetic rats. Biol Pharm Bull 33:67–71
Zhou XF, Deng YS, Xian CJ, Zhong JH (2000) Neurotrophins from dorsal root ganglia trigger allodynia after spinal nerve injury in rats. Eur J Neurosci 12:100–105
Sakurai E, Kurihara T, Kouchi K, Saegusa H, Zong S, Tanabe T (2009) Upregulation of casein kinase 1epsilon in dorsal root ganglia and spinal cord after mouse spinal nerve injury contributes to neuropathic pain. Mol Pain 5:74
Samad OA, Tan AM, Cheng X, Foster E, Dib-Hajj SD, Waxman SG (2013) Virus-mediated shRNA knockdown of Na(v)1.3 in rat dorsal root ganglion attenuates nerve injury-induced neuropathic pain. Mol Ther 21:49–56
Sapunar D, Kostic S, Banozic A, Puljak L (2012) Dorsal root ganglion—a potential new therapeutic target for neuropathic pain. J Pain Res 5:31–38
Shen KF, Zhu HQ, Wei XH, Wang J, Li YY, Pang RP, Liu XG (2013) Interleukin-10 down-regulates voltage gated sodium channels in rat dorsal root ganglion neurons. Exp Neurol. doi:10.1016/j.expneurol.2013.01.018
Cheng KI, Lai CS, Wang FY, Wang HC, Chang LL, Ho ST, Tsai HP, Kwan AL (2011) Intrathecal lidocaine pretreatment attenuates immediate neuropathic pain by modulating Nav1.3 expression and decreasing spinal microglial activation. BMC Neurol 11:71
Kim CH, Oh Y, Chung JM, Chung K (2001) The changes in expression of three subtypes of TTX sensitive sodium channels in sensory neurons after spinal nerve ligation. Brain Res Mol Brain Res 95:153–161
Black JA, Cummins TR, Plumpton C, Chen YH, Hormuzdiar W, Clare JJ, Waxman SG (1999) Upregulation of a silent sodium channel after peripheral, but not central, nerve injury in DRG neurons. J Neurophysiol 82:2776–2785
Hains BC, Saab CY, Klein JP, Craner MJ, Waxman SG (2004) Altered sodium channel expression in second-order spinal sensory neurons contributes to pain after peripheral nerve injury. J Neurosci 24:4832–4839
Lindia JA, Kohler MG, Martin WJ, Abbadie C (2005) Relationship between sodium channel NaV1.3 expression and neuropathic pain behavior in rats. Pain 117:145–153
Kusuda R, Cadetti F, Ravanelli MI, Sousa TA, Zanon S, De Lucca FL, Lucas G (2011) Differential expression of microRNAs in mouse pain models. Mol Pain 17:17
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Nos. 30973030, 30800424, 81260318 and 81371967), Natural Science Foundation (No. 20122BAB205061) and Science and Technology Support Program (No. 2009JX01268) of Jiangxi Province, China. Educational Department Foundation (No. GJJ13155) of Jiangxi Province, China. We are grateful for critical reading of the manuscript by Michael Sun, USA.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Hong-Ping Chen, Wei Zhou and Lu-Mei Kang have contributed equally to this study.
Rights and permissions
About this article
Cite this article
Chen, HP., Zhou, W., Kang, LM. et al. Intrathecal miR-96 Inhibits Nav1.3 Expression and Alleviates Neuropathic Pain in Rat Following Chronic Construction Injury. Neurochem Res 39, 76–83 (2014). https://doi.org/10.1007/s11064-013-1192-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-013-1192-z