Abstract
Glutamate metabolism was studied in co-cultures of mouse cerebellar neurons (predominantly glutamatergic) and astrocytes. One set of cultures was superfused (90 min) in the presence of either [U-13C]glucose (2.5 mM) and lactate (1 mM) or [U-13C]lactate (1 mM) and glucose (2.5 mM). Other sets of cultures were incubated in medium containing [U-13C]lactate (1 mM) and glucose (2.5 mM) for 4 h. Regardless of the experimental conditions cell extracts were analyzed using mass spectrometry and nuclear magnetic resonance spectroscopy. 13C labeling of glutamate was much higher than that of glutamine under all experimental conditions indicating that acetyl-CoA from both lactate and glucose was preferentially metabolized in the neurons. Aspartate labeling was similar to that of glutamate, especially when [U-13C]glucose was the substrate. Labeling of glutamate, aspartate and glutamine was lower in the cells incubated with [U-13C]lactate. The first part of the pyruvate recycling pathway, pyruvate formation, was detected in singlet and doublet labeling of alanine under all experimental conditions. However, full recycling, detectable in singlet labeling of glutamate in the C-4 position was only quantifiable in the superfused cells both from [U-13C]glucose and [U-13C]lactate. Lactate and alanine were mostly uniformly labeled and labeling of alanine was the same regardless of the labeled substrate present and higher than that of lactate when superfused in the presence of [U-13C]glucose. These results show that metabolism of pyruvate, the precursor for lactate, alanine and acetyl-CoA is highly compartmentalized.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
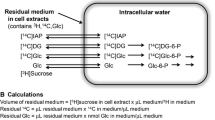
Both glucose and lactate can be converted to pyruvate which is a key metabolite of intermediary metabolism in neurons and astrocytes [1]; it is (1) formed from glucose via glycolysis; (2) the keto acid of alanine; (3) the product of lactate dehydrogenation; (4) the product of oxidative decarboxylation of malate or oxaloacetate, an important step in pyruvate recycling. Hence, once formed, pyruvate can be processed in different ways (see Fig. 1). It can be: carboxylated to oxaloacetate in astrocytes, an important anaplerotic process for the replenishment of TCA cycle intermediates; oxidatively decarboxylated to acetyl-CoA for subsequent further oxidative breakdown in the TCA cycle; transaminated to alanine or reduced to lactate.
Scheme showing the metabolic fates of pyruvate (Pyr) and the formation of metabolites (in bold) discussed in the present work. Glucose is metabolized to pyruvate, a multi-step process known as glycolysis which takes place in the cytosol (not shown). Pyruvate is the central metabolite linking glucose and lactate (Lac) metabolism, as lactate is oxidized to pyruvate. Pyruvate can either be transaminated to alanine (Ala) by alanine aminotransferase (ALAT) or transported into the mitochondria where it is decarboxylated to acetyl-CoA which condenses with oxaloacetate to form citrate (Cit). Citrate is in two steps transformed to α-ketoglutarate (α-KG), which is in equilibrium with glutamate (Glu) catalyzed by aspartate aminotransferase (AAT). Malate (Mal) is formed in four steps from α-ketoglutarate and oxaloacetate (OAA) in one step from malate. Aspartate (Asp) is formed from oxaloacetate by AAT. Pyruvate can be synthesized from either malate or oxaloacetate by malic enzyme (ME) or by the concerted action of phosphoenolpyruvate carboxykinase (PEPCK) and pyruvate kinase (PK), respectively. If pyruvate formed in this way is reintroduced into the tricarboxylic acid (TCA) cycle by decarboxylation to acetyl-CoA (by pyruvate dehydrogenase, PDH), this comprises the pathway known as pyruvate recycling (as indicated by the dashed grey line). In this way, the carbon skeleton is completely oxidized which is a necessary step in oxidative degradation of glucogenic amino acids. AAT aspartate aminotransferase, Ala alanine, ALAT alanine aminotransferase, Asp aspartate, Cit citrate, Glu glutamate, α-KG α-ketoglutarate, Lac lactate, LDH lactate dehydrogenase, Mal malate, ME malic enzyme, OAA oxaloacetate, PDH pyruvate dehydrogenase, PEPCK phosphoenolpyruvate carboxykinase, PK pyruvate kinase, Pyr pyruvate, TCA tricarboxylic acid
Pyruvate recycling is the pathway in which pyruvate derived from malate (via malic enzyme) or oxaloacetate (via phosphoenolpyruvate carboxykinase plus pyruvate kinase) is reintroduced into the TCA cycle as acetyl-CoA subsequent to oxidative decarboxylation by pyruvate dehydrogenase (Fig. 1). This pathway, which is necessary for complete oxidation of the carbon skeleton of glucogenic amino acids was first described in the liver by Freidmann [2]. In the central nervous system, it was initially observed by Cerdan et al. [3] and was at that time proposed to occur primarily in neurons. However, other investigators have presented evidence in favour of this pathway being localized in astrocytes [4–7]. Thus, uncertainty remains with regard to the significance, the cellular localization and physiological role of this pathway.
A number of metabolic studies have been carried out by incubating neural cells in medium containing different substrates and/or agents acting on enzymes, receptors or transporters. In order to mimic the conditions these cells experience in the brain, a superfusion paradigm has been developed [8–10]. The substrates are often labeled with isotopes which can be radioactive or containing nuclei with magnetic moments. The latter is typically used for nuclear magnetic resonance spectroscopy (MRS) and/or mass spectrometry (MS). Thus, [U-13C]glucose and [U-13C]lactate have been used to analyze metabolism in cultured neurons and astrocytes both by MRS and MS. Extensive labeling of glutamate, aspartate, glutamine (in astrocytes), GABA (in GABAergic neurons) and alanine has been reported [11–16]. Co-cultures in which neurons grow on a pre-cultured layer of astrocytes simulate the close relationship of these cells in the brain and are used as a model for studying interactions between these cell types. In the present study such co-cultures obtained from dissociated cerebellum were incubated for 4 h in medium containing [U-13C]lactate (1 mM) and unlabeled glucose (2.5 mM). Alternatively, in order to imitate the dynamics of the extracellular space in the brain more closely, additionally cultures were superfused (90 min) in Hepes-buffered saline containing either [U-13C]glucose (2.5 mM) and lactate (1 mM) or [U-13C]lactate (1.0 mM) and glucose (2.5 mM). In both experimental paradigms, the incorporation of 13C-label in intracellular metabolites was analyzed by MS as well as 13C MRS.
Experimental procedure
Materials
Seven-day-old mice were obtained from either the animal facility at the Department of Pharmacology, The Danish University of Pharmaceutical Sciences or from Taconic M&B (Ry, Denmark). Plastic tissue culture flasks were purchased from NUNC A/S (Roskilde, Denmark), fetal calf serum from SeraLab Ltd. (Sussex, UK). Culture medium and poly-d-lysine (MW > 300,000) were obtained from Sigma Chemical Co. (St Louis, MO, USA) and penicillin was from Leo Pharma (Ballerup, Denmark). The Phenomenex EZ:faast LC-MS kit (Torrance, CA, USA) was used for amino acid analysis. [U-13C]glucose, [U-13C]lactate and 99.9% w/v D2O were from Cambridge Isotopes Laboratories (Woburn, MA, USA), and ethylene glycol from Merck (Darmstadt, Germany). N-methyl-N-(tert-butyldimethylsilyl)trifluor acetamid (MTBSTFA) in the presence of 1% w/v tert-butyldimethyl-chlorosilane (t-BDMS-Cl) was obtained from Regis Technologies (Morton Grove, IL, USA). All other chemicals used were of the purest grade available from regular commercial sources.
Cell cultures
Co-cultures of cerebellar neurons and astrocytes were prepared in a two-step process as detailed by Westergaard et al. [17]. In step one, cerebellar astrocytes were cultured from dissociated cerebella from 7-day-old mice as detailed by Hertz et al. [18] and Waagepetersen et al. [19]. Briefly, the dissected cerebella were mechanically dissociated by squeezing the tissue through 80 μm nylon sieves into the culture medium and subsequently the cell suspension was seeded in 80-cm2 culture flasks (15 ml/flask corresponding to four animals). Medium was changed twice a week during the culturing period. In step two, a fresh suspension of cerebellar neurons in neuronal medium was prepared, and added on top of a 2-week-old culture of cerebellar astrocytes [17, 19]. In brief, cerebellar neurons were obtained from 7-day-old mice essentially as described by Schousboe et al. [20]. After dissection of the cerebellum, the tissue was exposed to a mild trypsinization (0.25 mg/ml trypsin, 15 min, 37°C) followed by trituration in a DNase solution (75 i.u./ml) containing a trypsin inhibitor (0.53 mg/ml) from soybeans. The cells were suspended (3.5 × 106 cells/ml) in a slightly modified Dulbecco’s medium [21] containing 24.5 mM KCl, 12 mM glucose, 7 μM p-aminobenzoate, 50 μM kainate and 10% v/v fetal calf serum. In order to prevent further proliferation of astrocytes in the co-culture, cytosine arabinoside was added to a final concentration of 20 μM after 48 h in culture. The cultures were maintained for another 7–8 days, at which point the neurons exhibit pronounced glutamatergic characteristics. This is partly due to the treatment with kainate, which inhibits GABAergic cell function [22, 23]. The media of the individual co-cultures were supplemented twice with an aliquot (150 μl) of glucose (1.2 M) to reach a minimum concentration of 12 mM.
Incubation experiments
Following the culture period (7–8 days), the medium was discarded and the cells were rinsed using 2 × 7.5 ml phosphate-buffered saline containing 137 mM NaCl, 2.7 mM KCl, 7.3 mM Na2HPO4, 0.9 mM CaCl2, 0.5 mM MgCl2, pH 7.4 and subsequently incubated for 4 h in serum-free culture medium (12 ml) containing 1 mM [U-13C]lactate in combination with unlabeled glucose (2.5 mM). The incubation was ended by separating the medium from the cells which were rinsed twice (0.9% w/v NaCl) and extracted with 70% v/v ethanol. The cells were scraped off the flask and the cell–ethanol mixture was centrifuged at 15,000g (15 min) to separate the soluble extract from the insoluble cellular proteins. The supernatants were subsequently lyophilized and reconstituted in water for MS and MRS analysis.
Superfusion experiments
The culture medium was replaced with a Hepes-buffered, Mg2+-free saline solution (HBS, 10 mM Hepes at pH 7.4, 5 mM KCl, 135 mM NaCl, 1.5 mM CaCl2, 2.5 mM glucose and 1 mM lactate). The HBS was equilibrated with atmospheric O2 and maintained at 37°C during the experiment. Subsequently, the cultures were placed in a superfusion system [10]. The cell layer was covered with a nylon mesh (80 μm) and the cells were superfused (5 ml/min) with the HBS containing either [U-13C]glucose (2.5 mM) plus lactate (1 mM) or [U-13C]lactate (1 mM) plus glucose (2.5 mM) for 90 min. The cells were subsequently washed twice (0.9% w/v NaCl) and metabolites were extracted using 70% v/v ethanol. The cell extracts were lyophilized and re-dissolved in 99.9% w/v D2O for MRS analysis, thereafter lyophilized again and dissolved in deionized water for MS analysis.
Mass spectrometric analysis
Liquid chromatography-mass spectrometric (LC-MS) analysis was performed employing a Shimadzu LCMS-2010 mass spectrometer coupled to a Shimadzu 10 A VP HPLC system. The Phenomenex EZ:faast amino acid analysis kit for LC-MS was used for derivatization and subsequent analysis of labeling in alanine, glutamate, aspartate and glutamine. For gas chromatography-mass spectrometry (GC-MS), the samples were adjusted to pH 2 and dried under atmospheric air. Lactate and amino acids were derivatized with N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide (MTBSTFA) in the presence of 1% w/v tert-butylchlorodimethylsilane chloride (t-BDMS-Cl) [24]. The derivatized metabolites were analyzed by a Hewlett Packard 5890 Series II gas chromatograph linked to a Hewlett Packard 5971 A mass spectrometer. For both LC-MS and GC-MS analysis, atom percent excess (13C) of the derivatized amino acids and lactate was determined after allowing for naturally abundant 13C, as described by Biemann [25].
13C MRS
Lyophilized samples were dissolved in 99.9% w/v D2O containing 0.1% w/v ethylene glycol as internal standard for quantification and pH was adjusted to 6.8–7.0. Proton decoupled 13C MR spectra were accumulated on a BRUKER DRX500 spectrometer (BRUKER Analytik GmbH, Rheinstetten, Germany). The following acquisition parameters were applied: 30° pulse angle, acquisition time of 1.3 s and a relaxation delay of 0.5 s. The number of scans was typically 100,000. Correction factors for incomplete relaxation and nuclear Overhauser effects were applied to the integrals of the individual peaks.
1H MRS
The content of glutamate in cell extracts of superfused cultures was quantified using 1H MR spectra which were acquired on a BRUKER DRX500 spectrometer with the following acquisition parameters: 90° pulse angle, an acquisition time of 1.36 s and a relaxation delay of 10 s. Four hundred scans were accumulated for each sample. Water suppression was achieved by applying a low-power presaturation pulse at the water frequency.
HPLC
The cellular content of glutamate in cultures incubated in the presence of [U-13C]lactate (1 mM) and glucose (2.5 mM) was determined by HPLC analysis after derivatization with o-phthaldialdehyde essentially as described by Geddes and Wood [26].
Biochemical analysis
The content of protein was determined according to Lowry et al. [27] using bovine serum albumin as the standard. The typical protein content of a cerebellar co-culture was 4 mg.
Calculation of percent molecular carbon labeling (MCL)
To obtain a measure of total incorporation of 13C label, the average percent of labeled carbon atoms for each metabolite was calculated based on the data obtained from either LC-MS or GC-MS, a concept introduced by Bak et al. [9]. For example, glutamate may contain anywhere between one and five 13C atoms (designated M + 1 to M + 5, see Results for details). To provide a measure of the total labeling of the glutamate pool the percent of the individual isotopomers (i.e. M + 1, M + 2, M + 3, etc.) are first multiplied by the number of carbons labeled (i.e. 1 for M + 1, 2 for M + 2, etc.), subsequently summed up, and expressed as a percent of the total number of carbon atoms. To illustrate this, the following example of the calculation for glutamate in a fictive experiment is provided: If glutamate is labeled as follows M + 1, 20%; M + 2, 10%; M + 3, 5%; M + 4, 3%; M + 5, 2%, the calculation will be ([(M + 1, 20% × 1) + (M + 2, 10% × 2) + (M + 3, 5% × 3) + (M + 4, 3% × 4) + (M + 5, 2% × 5)]/500)100% = 15.4%. Hence, percent MCL is a measure of the total labeling of glutamate (or any other metabolite), and is therefore the most precise measure of total incorporation of label.
Data analysis
The mass spectrometric data were corrected for natural abundance of 13C and the isotopic enrichment was calculated according to Biemann [25]. Data analysis was performed using Microsoft Excel 2002 and GraphPad Prism v4.01. Relevant peaks in the 13C and 1H MR spectra were identified and integrated using XWINNMR software. The amounts of 13C were quantified from the integrals of the peak areas, using ethylene glycol as internal standard. The protons on the C-4 carbon of glutamate were quantified in the 1H spectra using ethylene glycol as an internal standard. Results are given as mean ± SEM. The data were analyzed using one-way ANOVA and significant differences between means were determined by Tukey–Kramer post hoc test. The level of statistical significance was 95%.
Results
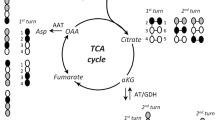
The results are divided into two sections based on the method of analysis, as all cell extracts have been analyzed by both MRS and MS. A schematic presentation of some of the isotopomers of glutamate generated from [U-13C]lactate and [U-13C]glucose is shown in Fig. 2.
A simplified scheme of TCA cycle metabolism of [1,2-13C]acetyl-CoA derived from [U-13C]glucose or [U-13C]lactate. In the first turn, unlabeled oxaloacetate condenses with labeled acetyl-CoA. (a) The scheme shows all possible combinations of labeled glutamate and glutamine derived from three turns of the TCA cycle. (b),(c) Glutamate and alanine labeling from pyruvate recycling is shown using unlabeled oxaloacetate and first turn only. Many additional isotopomers are formed both from unlabeled acetyl-CoA and [1-13C] -or [2-13C] acetyl-CoA or/and labeled oxaloacetate in the TCA cycle. Labeled carbon atoms are represented by black circles. TCA tricarboxylic acid
Mass spectrometry
The labeling of intracellular glutamate, glutamine and aspartate after incubation in medium containing [U-13C]lactate (1 mM) and glucose (2.5 mM) or after superfusion in a physiological HBS containing either [U-13C]glucose (2.5 mM) and lactate (1 mM) or [U-13C]lactate (1 mM) and glucose (2.5 mM) is presented in Table 1, as MCL (for details see Experimental procedures) and percent double labeling (M + 2). Labeling of glutamine (MCL) was lower than for aspartate and glutamate (P < 0.05, not indicated in Table 1). MCL of aspartate or glutamate was similar regardless of the experimental conditions. Incubating cells in medium containing [U-13C]lactate led to similar labeling (MCL) as that observed in superfusion experiments with [U-13C]glucose, except for glutamine labeling which was lower from [U-13C]glucose than from [U-13C]lactate. The percent double labeling in glutamate and glutamine was higher in the cells incubated in the presence of [U-13C]lactate compared to cells superfused in the presence of [U-13C]glucose (P < 0.05). The cellular content of glutamate was lower (P < 0.001) after superfusion (15.3 ± 1.9 nmol/mg protein) compared to that observed after incubation (38.1 ± 2.4 nmol/mg protein (value from [19]).
The mass spectrometry data for alanine and lactate in Table 2 are expressed as mono (M + 1), double (M + 2) and triple (M + 3) labeling of carbon atoms. Triple labeling was the predominant form of alanine and lactate, but double and especially mono labeling were also present with the exception of cells superfused in the presence of [U-13C]glucose. Incubation in medium containing [U-13C]lactate led to higher mono and triple labeling of alanine and lactate compared to superfusion, except for the triple labeling of lactate in cells superfused in HBS containing [U-13C]lactate. The triple labeling of alanine was similar to that of lactate using [U-13C]lactate as the labeled precursor both after superfusion and incubation; however, employing [U-13C]glucose, the enrichment of alanine was almost three-fold higher (P < 0.01).
Magnetic resonance spectroscopy
Table 3 summarizes data obtained from 13C MRS. In the present study, the glutamate C-3 resonance consists of a singlet ([3-13C]glutamate) and a doublet ([2,3-13C]glutamate). Likewise, as seen in Fig. 3, the glutamate C-4 resonance consists of a singlet ([4-13C]glutamate) and a doublet ([4,5-13C]glutamate). The latter is synthesized from α-ketoglutarate generated in the first turn of the TCA cycle and is part of the double labeling measured by MS. Percent enrichments were calculated from the cellular content of glutamate and the amounts of 13C labeling. 13C Enrichment in glutamate C-3 and amount of [4,5-13C]glutamate was not dependent on the experimental conditions. It should be noted that [4,5-13C] glutamate is uniformly labeled and contribution from the naturally occurring isotopomer is negligible. The amount of 13C present in glutamate C-3 and [4,5-13C]glutamate was highest in the cells incubated with [U-13C]lactate, and in the cells superfused in the presence of [U-13C]glucose and [U-13C]lactate the amounts were comparable. [4-13C]/[5-13C]glutamate is formed if α-[4,5-13C]ketoglutarate is metabolized further to [1,2-13C]/[3,4-13C]malate or [1,2-13C]/[3,4-13C]oxaloacetate which then leaves the TCA cycle and the 13C label from the resulting pyruvate is reintroduced into the TCA cycle as [2-13C]/[1-13C]acetyl-CoA (Figs. 1, 2b,c). In the present study only [4-13C]glutamate was quantifiable. This so-called “pyruvate recycling” was extensive in the superfused cells but not quantifiable in the cells incubated in the presence of [U-13C]lactate (Table 3).
13C NMR spectrum of cell extract of co-cultures of cerebellar neurons and astrocytes after exposure to [U-13C]glucose (2.5 mM) and lactate (1 mM) in a superfusion paradigm (5 ml/min for 90 min). For details see Experimental procedures. The glutamate C-4 and C-3 peaks are shown
Discussion
Catabolism of glucose and lactate involves pyruvate which is a key metabolite in both energy homeostasis and amino acid synthesis. Extensive compartmentation of pyruvate metabolism in cerebellar co-cultures of astrocytes and neurons was observed, when [U-13C]glucose or [U-13C]lactate were the substrates, in agreement with a previous study [10].
In the neuronal-astrocytic co-cultures, used in the present study, alanine labeling was modest regardless of the experimental conditions. Bak et al. [9] showed that superfused cultures of cerebellar neurons contained highly labeled alanine (∼38%; MCL) when lactate was the labeled substrate, whereas labeling from [U-13C]glucose was only modest (∼7%; MCL) [9]. Incubating cultured astrocytes under the same conditions as in the present study, the labeling of alanine was only 8% [28]. These data indicates that in the co-cultures the labeling of alanine is diluted by the astrocytic pool of alanine. Interestingly, labeling of alanine was higher than that of lactate when [U-13C]glucose was present in the superfusion medium, indicating that the pyruvate pool from which alanine was synthesized was different from that used for lactate synthesis. In fact the same was observed in neurons superfused in the presence of [U-13C]glucose [9] indicating that synthesis of alanine from glucose-derived pyruvate in the co-cultures mostly occurs in the neuronal compartment. That alanine synthesis preferentially takes place in neurons has been suggested previously [19, 29]. When labeled lactate was present in the superfusion or incubation medium, intracellular lactate was more highly labeled than when [U-13C]glucose was present. However, alanine labeling was not higher during superfusion with a medium containing [U-13C]lactate than when [U-13C]glucose was present in the superfusion medium, again pointing towards either intra- or inter-cellular compartmentation of pyruvate metabolism. Intracellular compartmentation of pyruvate metabolism has been suggested from studies of both cultured neurons and astrocytes [16, 30, 31].
[U-13C]Pyruvate can be converted to [1,2-13C]acetyl-CoA which can enter the TCA cycle and labeled glutamate, aspartate and glutamine can be formed from their respective precursors. Glutamine is exclusively synthesized in the astrocytic compartment of the co-cultures due to the astrocytic localization of glutamine synthetase [32]. However, glutamate and aspartate are preferentially localized in the neuronal compartment estimated from results obtained in cultures of only neurons or astrocytes [19]. Glutamate and aspartate were the most highly labeled amino acids and glutamine labeling was less than half of that observed in glutamate regardless of the experimental conditions. This indicates that pyruvate and thus acetyl-CoA originating from lactate as well as glucose is metabolized more extensively in neurons than in astrocytes [33]. Interestingly, aspartate labeling was very similar to that of glutamate when glucose was the labeled precursor but double labeling was different comparing these metabolites when cells were superfused in the presence of [U-13C]lactate. This is another indication of intracellular compartmentalized pyruvate metabolism since aspartate is localized mainly in neurons (see above).
Using MRS analysis [4,5-13C]glutamate (1.turn) and label in C-3 glutamate (subsequent turns) were detected in the co-cultures incubated in the presence of [U-13C]lactate, whereas significantly less labeled glutamate was observed in superfused cells. Surprisingly, the amount of [4,5-13C]glutamate was similar under all experimental conditions. This, taken together with the fact that the amount of glutamate was greatly reduced in the superfused cells, shows that labeling was similar in the different experimental paradigms and that superfusion with HBS caused release of glutamate from the neurons. However, the content of alanine, which is also present mostly in neurons in cerebellum [19] was unchanged. This indicates that most of the alanine aminotransferase is operating in a compartment not affected by a reduced glutamate content. In line with this, in cultured neurons superfused in the presence of [2-15N]glutamine an increased labeling of glutamate and aspartate with no concomitant increase in alanine labeling was observed following induced neuronal activity [8]. Interestingly, the amount of double labeling was similar to that of [4,5-13C]glutamate (one turn see Fig. 2a) after superfusion (90 min), but this double labeling was higher than the amount of [4,5-13C]glutamate in cells incubated for 4 h. This indicates that double labeling can be used as an estimate of one turn labeling after superfusion for 90 min but not after 4 h in an incubation paradigm.
Pyruvate recycling
Pyruvate recycling is a pathway for complete oxidation of glutamate. When cells were incubated or superfused in medium containing [U-13C]lactate in the present study, alanine and lactate were not only triple labeled but also mono and double labeled, demonstrating that the first part of the pyruvate recycling pathway was operating (Fig. 2b,c). A similar labeling pattern was detected in alanine but not in lactate when [U-13C]glucose was present in the superfusion medium. The reason for the latter may be the dilution of lactate by the unlabeled lactate present in the superfusion medium.
Complete pyruvate recycling from [U-13C]pyruvate can be detected in the form of [4-13C]glutamate [7]. Recycling was not quantifiable when co-cultures were incubated in a medium containing [U-13C]lactate. In previous studies part of the pyruvate recycling pathway could be demonstrated in astrocytes incubated in medium containing [U-13C]glutamate or [U-13C]aspartate, whereas no recycling was detectable in neurons [34]. Another study using [3-13C]glutamate confirmed that recycling takes place in astrocytes but not in neurons or cerebral cortical co-cultures [7]. Using MS analysis, which is more sensitive than MRS, recent experiments have shown that pyruvate recycling from [U-13C]glutamate is taking place in cerebellar neurons [35]. Interestingly, when cerebellar co-cultures were superfused with [U-13C]glucose or [U-13C]lactate considerable recycling was detected. The extent of recycling was indicated by the presence of [4-13C]glutamate (3–5% of total glutamate). Since 10% of all glutamate is [4,5-13C]glutamate, which is the major isotopomer, pyruvate recycling may be considered to be of quantitative significance in the superfused cultures. Interestingly, the lack of pyruvate recycling in co-cultures of cerebral cortical astrocytes and neurons [7] could indicate that recycling is more important in the cerebellum or in glutamatergic than in GABAergic structures.
Conclusion
The main findings of the present study can be summarized by the following: (1) pyruvate metabolism is compartmentalized as reflected by differential labeling of lactate and alanine; (2) pyruvate recycling was considerable in cerebellar co-cultures superfused with labeled glucose or lactate as revealed by the presence of [4-13C]glutamate.
Abbreviations
- GABA:
-
γ-Aminobutyric acid
- GC-MS:
-
Gas chromatography-mass spectrometry
- Hepes:
-
N-2-Hydroxyethyl-piperazine-N′-2-ethanesulfonic acid
- HBS:
-
Hepes-buffered saline
- LC-MS:
-
Liquid chromatography-mass spectrometry
- ME:
-
Malic enzyme
- MRS:
-
Nuclear magnetic resonance spectroscopy
- TCA:
-
Tricarboxylic acid
References
McKenna MC, Gruetter R, Sonnewald U, Waagepetersen HS, Schousboe A (2006) Energy metabolism of the brain. In: Siegel GJ, Albers RW, Brady ST, Price DL (eds) Basic neurochemistry. Elsevier, Amsterdam, pp 531–557
Freidmann B, Goodman EH Jr, Saunders HL, Kostos V, Weinhouse S (1971) An estimation of pyruvate recycling during gluconeogenesis in the perfused rat liver. Arch Biochem Biophys 143:566–578
Cerdan S, Kunnecke B, Seelig J (1990) Cerebral metabolism of [1,2-13C2]acetate as detected by in vivo and in vitro 13C. J Biol Chem 265:12916–12926
Alves PM, Nunes R, Zhang C, Maycock CD, Sonnewald U, Carrondo MJ, Santos H (2000) Metabolism of 3-13C-malate in primary cultures of mouse astrocytes. Dev Neurosci 22:456–462
Bakken IJ, White LR, Aasly J, Unsgard G, Sonnewald U (1997) Lactate formation from [U-13C]aspartate in cultured astrocytes: compartmentation of pyruvate metabolism. Neurosci Lett 237:117–120
Sonnewald U, Westergaard N, Jones P, Taylor A, Bachelard HS, Schousboe A (1996) Metabolism of [U-13C5] glutamine in cultured astrocytes studied by NMR spectroscopy: first evidence of astrocytic pyruvate recycling. J Neurochem 67:2566–2572
Waagepetersen HS, Qu H, Hertz L, Sonnewald U, Schousboe A (2002) Demonstration of pyruvate recycling in primary cultures of neocortical astrocytes but not in neurons. Neurochem Res 27:1431–1437
Bak LK, Sickmann HM, Schousboe A, Waagepetersen HS (2005) Activity of the lactate-alanine shuttle is independent of glutamate-glutamine cycle activity in cerebellar neuronal-astrocytic cultures. J Neurosci Res 79:88–96
Bak LK, Schousboe A, Sonnewald U, Waagepetersen HS (2006) Glucose is necessary to maintain neurotransmitter homeostasis during synaptic activity in cultured glutamatergic neurons. J Cereb Blood Flow Metab (in press). DOI 10.1038/sj.jcbfm.9600281
Drejer J, Honore T, Schousboe A (1987) Excitatory amino acid-induced release of 3H-GABA from cultured mouse cerebral cortex interneurons. J Neurosci 7:2910–2916
Bouzier-Sore AK, Voisin P, Canioni P, Magistretti PJ, Pellerin L (2003) Lactate is a preferential oxidative energy substrate over glucose for neurons in culture. J Cereb Blood Flow Metab 23:1298–1306
Martin M, Portais JC, Labouesse J, Canioni P, Merle M (1993) [1–13C]glucose metabolism in rat cerebellar granule cells and astrocytes in primary culture. Evaluation of flux parameters by 13C- and 1H-NMR spectroscopy. Eur J Biochem 217:617–625
Martin M, Portais JC, Voisin P, Rousse N, Canioni P, Merle M (1995) Comparative analysis of 13C-enriched metabolites released in the medium of cerebellar and cortical astrocytes incubated with [1-13C]glucose. Eur J Biochem 231:697–703
Waagepetersen HS, Bakken IJ, Larsson OM, Sonnewald U, Schousboe A (1998) Comparison of lactate and glucose metabolism in cultured neocortical neurons and astrocytes using 13C-NMR spectroscopy. Dev Neurosci 20:310–320
Waagepetersen HS, Bakken IJ, Larsson OM, Sonnewald U, Schousboe A (1998) Metabolism of lactate in cultured GABAergic neurons studied by 13C nuclear magnetic resonance spectroscopy. J Cereb Blood Flow Metab 18:109–117
Waagepetersen HS, Sonnewald U, Larsson OM, Schousboe A (2001) Multiple compartments with different metabolic characteristics are involved in biosynthesis of intracellular and released glutamine and citrate in astrocytes. Glia 35:246–252
Westergaard N, Fosmark H, Schousboe A (1991) Metabolism and release of glutamate in cerebellar granule cells cocultured with astrocytes from cerebellum or cerebral cortex. J Neurochem 56:59–66
Hertz L, Juurlink BHJ, Hertz E, Fosmark H, Schousboe A (1989) Preparation of primary cultures of mouse (rat) astrocytes. In: Shahar A, de Vellis J, Vernadakis A, Haber B (eds) A dissection and tissue culture manual for the nervous system. Liss, New York, pp 105–108
Waagepetersen HS, Sonnewald U, Larsson OM, Schousboe A (2000) A possible role of alanine for ammonia transfer between astrocytes and glutamatergic neurons. J Neurochem 75:471–479
Schousboe A, Meier E, Drejer J, Hertz L (1989) Preparation of primary cultures of mouse (rat) cerebellar granule cells. In: Shahar A, de Vellis J, Vernadakis A, Haber B (eds) A dissection and tissue culture manual for the nervous system. Liss, New York, pp 183–186
Hertz L, Juurlink BHJ, Fosmark H, Schousboe A (1982) Astrocytes in primary cultures. In: Pfeiffer SE (ed) Neuroscience approached through cell culture. CRC, Boca Raton, pp 175–186
Drejer J, Schousboe A (1989) Selection of a pure cerebellar granule cell culture by kainate treatment. Neurochem Res 14:751–754
Sonnewald U, Olstad E, Qu H, Babot Z, Cristofol R, Sunol C, Schousboe A, Waagepetersen H (2004) First direct demonstration of extensive GABA synthesis in mouse cerebellar neuronal cultures. J Neurochem 91:796–803
Mawhinney TP, Robinett RS, Atalay A, Madson MA (1986). Analysis of amino acids as their tert.-butyldimethylsilyl derivatives by gas-liquid chromatography and mass spectrometry. J Chromatogr 358:231–242
Biemann K (1962) The mass spectra of isotopically labeled molecules. In: Mass spectrometry; Organic chemical applications. McGraw-Hill, New York, pp 223–227
Geddes JW, Wood JD (1984) Changes in the amino acid content of nerve endings (synaptosomes) induced by drugs that alter the metabolism of glutamate and gamma-aminobutyric acid. J Neurochem 42:16–24
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Schousboe A, Sonnewald U, Waagepetersen HS (2003) Differential roles of alanine in GABAergic and glutamatergic neurons. Neurochem Int 43:311–315
Zwingmann C, Richter-Landsberg C, Brand A, Leibfritz D (2000) NMR spectroscopic study on the metabolic fate of [3-13C]alanine in astrocytes, neurons, and cocultures: implications for glia–neuron interactions in neurotransmitter metabolism. Glia 32:286–303
Bouzier AK, Goodwin R, de Gannes FM, Valeins H, Voisin P, Canioni P, Merle M (1998) Compartmentation of lactate and glucose metabolism in C6 glioma cells. A 13C and 1H NMR study. J Biol Chem 273:27162–27169
Cruz F, Villalba M, Garcia-Espinosa MA, Ballesteros P, Bogonez E, Satrustegui J, Cerdan S (2001) Intracellular compartmentation of pyruvate in primary cultures of cortical neurons as detected by 13C NMR spectroscopy with multiple 13C labels. J Neurosci Res 66:771–781
Norenberg MD, Martinez-Hernandez A (1979) Fine structural localization of glutamine synthetase in astrocytes of rat brain. Brain Res 161:303–310
Qu H, Haberg A, Haraldseth O, Unsgard G, Sonnewald U (2000) 13C MR spectroscopy study of lactate as substrate for rat brain. Dev Neurosci 22:429–436
Bakken IJ, White LR, Unsgard G, Aasly J, Sonnewald U (1998) [U-13C]glutamate metabolism in astrocytes during hypoglycemia and hypoxia. J Neurosci Res 51:636–645
Olstad E, Olsen GM, Qu Q, Sonnewald U (2007) Pyruvate recycling in cultured neurons from cerebellum. J Neurosci Res (in press)
Acknowledgments
The expert technical assistance by Mrs. Kirsten Thuesen (Danish University of Pharmaceutical Sciences, Copenhagen, Denmark) as well as Dr. Turid Nilsen and Mr. Lars Evje, M.Sc. (both Norwegian University of Science and Technology), is hereby cordially acknowledged. The Danish Medical Research Council (22-03-0250; 22-4-0314), the Alfred Benzon, Hørslev, Lundbeck, Norwegian Epilepsy and Novo Nordisk Foundations are acknowledged for generous financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special issue dedicated to John P. Blass.
Rights and permissions
About this article
Cite this article
Bak, L.K., Waagepetersen, H.S., Melø, T.M. et al. Complex Glutamate Labeling from [U-13C]glucose or [U-13C]lactate in Co-cultures of Cerebellar Neurons and Astrocytes. Neurochem Res 32, 671–680 (2007). https://doi.org/10.1007/s11064-006-9161-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-006-9161-4