Abstract
Introduction
Conflicting results have been reported in the association between glioblastoma proximity to the subventricular zone (SVZ) and enrichment of cancer stem cell properties. Here, we examined this hypothesis using magnetic resonance (MR) images derived from 217 The Cancer Imaging Archive (TCIA) glioblastoma subjects.
Methods
Pre-operative MR images were segmented automatically into contrast enhancing (CE) tumor volumes using Iterative Probabilistic Voxel Labeling (IPVL). Distances were calculated from the centroid of CE tumor volumes to the SVZ and correlated with gene expression profiles of the corresponding glioblastomas. Correlative analyses were performed between SVZ distance, gene expression patterns, and clinical survival.
Results
Glioblastoma located in proximity to the SVZ showed increased mRNA expression patterns associated with the cancer stem-cell state, including CD133 (P = 0.006). Consistent with the previous observations suggesting that glioblastoma stem cells exhibit increased DNA repair capacity, glioblastomas in proximity to the SVZ also showed increased expression of DNA repair genes, including MGMT (P = 0.018). Reflecting this enhanced DNA repair capacity, the genomes of glioblastomas in SVZ proximity harbored fewer single nucleotide polymorphisms relative to those located distant to the SVZ (P = 0.003). Concordant with the notion that glioblastoma stem cells are more aggressive and refractory to therapy, patients with glioblastoma in proximity to SVZ exhibited poorer progression free and overall survival (P < 0.01).
Conclusion
An unbiased analysis of TCIA suggests that glioblastomas located in proximity to the SVZ exhibited mRNA expression profiles associated with stem cell properties, increased DNA repair capacity, and is associated with poor clinical survival.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The development of the human cerebrum is a carefully orchestrated process whereby neuronal precursor cells migrate radially from the stem cell niche in the subventricular zone (SVZ) [1]. During neural development, SVZ is the site of extraordinary proliferation where nearly a quarter of a million new neural precursor cells are generated per minute [2]. As the neural precursors migrate centrifugally from the SVZ, they differentiate into distinct cell types and lose the capacity for self-renewal and multi-potency. Waves of radially migrating neural precursor and differentiated cells ultimately populate the cerebrum, forming layers of exquisitely intricate cyto-architecture [3]. After completion of neural development, the SVZ continues to maintain a niche that sustains neural stem cells, which continue to radially migrate and differentiate. In this way, cells located in proximity to the SVZ niche are more likely to harbor stem-like properties relative to those located distant to the SVZ.
The unique SVZ “stem” niche harbors implications related to glioblastoma, the most common form of adult brain cancer [4]. Many investigators have suggested that glioblastomas located in proximity to the SVZ may have arisen from the populations of cells that harbor stem or stem-like properties and retained these properties during neoplastic transformation. Supporting this hypothesis, some studies have reported that glioblastomas contacting the SVZ exhibit cancer stem-cell like properties, including aggressive clinical course [5], increased likelihood of recurrence [6], and poor survival [7,8,9]. However, these results have not been reproduced by others investigators [10, 11]. In general, these studies involved limited patient numbers and are confounded by the subjectivity in the determination of SVZ contact. Here, we utilized Iterative Probabilistic Voxel Labeling (IPVL), an automated tumor segmentation algorithm developed in our laboratory [12] to quantitatively determine glioblastoma location relative to the SVZ in 217 The Cancer Imaging Atlas (TCIA) patients. We then tested whether glioblastoma proximity to the SVZ is associated with survival pattern or gene signatures associated with the cancer stem cell state.
Methods
Data acquisition and Subtype Classification
In total, 217 subjects with artifact-free pre-operative T1 weighted MR images were downloaded from the TCIA (https://cancerimagingarchive.net) in November 2014. Level 3 probe collapsed Messenger RNA (mRNA) expression data: Affymetrix HT-HG-U133A GeneChip and RNAseq, was downloaded for available patients via the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/). Corresponding copy number variation (CNV) and single nucleotide polymorphism (SNPs) were also down-loaded and analyzed as previously described [13]. Boxplot analysis was employed to discard outliers in the SNP and CNV data. Tumors with extreme CNVs in more than 1% of their genes are excluded in this context. Affymetrix expression data were normalized by robust multichip average (RMA) [14]. RNAseq data were RSEM normalized [15]. When not already available in published literature, genomic subtypes were determined for subjects employing single sample Gene Set Enrichment Analysis (ssGSEA) as described previously [13, 16].
Image preprocessing and registration
MR images were corrected for gradient nonlinearity using previously described methods [17]. Images were additionally preprocessed and intensity corrected utilizing N4 bias field correction [18], then affine and nonlinear registrations were performed to the Montreal Neurological Institute (MNI) 152 nonlinear 1 mm3 template employing methods from Advanced Normalization Tools (ANTS) [19]. Visual inspection of resulting images was performed by three independent reviewers (T.C.S, J.M.T, & K.S.P) to ensure successful and accurate preprocessing was completed for all subjects.
Tumor segmentation
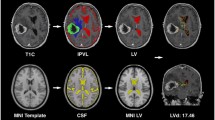
Contrasting-enhancing (CE) regions of tumors were segmented using the iterative probabilistic voxel labeling (IPVL) algorithm developed in the laboratory [12] and the enclosed volume was filled using a 3D morphological open-close operation. Centroid density maps were generated using a 15-mm centroid of each filled CE volume. Visual inspection by two independent operators (T.C.S, J.M.T) was performed to ensure adequacy of region filling and centroid estimation.
Subventricular zone distance
To measure SVZ distance (Fig. 1) with respect to each tumor’s filled CE volume, the MNI template’s lateral ventricle segmentation was used as a basis of comparison. SVZ distances were calculated by taking the mean of the distance from the nearest MNI template ventricular border to the centroid point within a subject’s CE volume. Statistical analyses of SVZ distance with respect to patient survival was performed within the statistics software package SPSS (IBM Corp, New York, United States). When stratification was required, tumors were classified by the median of the SVZ distance: low SVZ (SVZ distance < 19.23 mm) and high SVZ (SVZ distance 19.23 mm).
ssGSEA and statistical analyses
ssGSEA was employed to calculate normalized enrichment scores for gene signatures according to established methods [16], using custom software in MATLAB (Statistics Toolbox Release 2012b, The MathWorks Inc., Natick, MA). Statistical analysis of SVZ distance, gene expression, and boot-strap analysis was performed in MATLAB. All mRNA expression analyses were performed in the Affymetrix HT-HG-U133A RMA normalized data. Gene expression or ssGSEA scores were compared between SVZlow and SVZhigh tumors. To mitigate type I error, 100,000 permutation tests were performed during hypothesis testing in our sample to determine the likelihood of the outcome measured occurring by random chance [20]. When exploring the relationship between SVZ distances, ssGSEA scores and SNP burden, we employed a linear regression and reported statistical correlation and p-value.
Results
SVZ distance and the stem-cell state
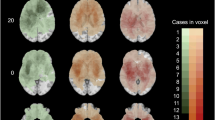
Demographic data of the 217 TCIA patients are as previously described [21] and are shown in Table 1. The cohort consisted of 59.8% (n = 128) male with a median age of 59.3 ± 14.1. Images from these patients were segmented using Iterative Probabilistic Voxel Labeling (IPVL) [12]. The centroid for each CE tumor volume was calculated. Distances from the tumor centroid to the lateral ventricular border were calculated (Fig. 1).
CD133 is a glycoprotein expressed in neuronal and glial stem cells that reside in the SVZ [4, 22]. It is a key surface marker for isolation of normal and glioblastoma stem cells [23]. High expression of CD133 was associated with self-renewal [24], increased DNA repair capacity [25], and other properties associated with the stem-cell state and aggressive clinical course [26]. In this context, we tested whether glioblastomas with centroid in proximity to the SVZ harbored increased CD133 expression. For this analysis, each patient’s glioblastoma is classified as “high” or “low” for SVZ distance based on the median value of SVZ for the entire cohort. We found that glioblastomas located in proximity to the SVZ (low SVZ score) are associated with higher CD133 expression (P = 0.006, Table 2).
To confirm this finding, we tested whether the observed SVZ association can be recapitulated using an independent gene signature associated with glioblastoma “stemness”. Murat et al. reported a stem cell-related signature through analysis of clinical glioblastoma specimens [27]. Supporting our hypothesis, the ssGSEA score for stemness inversely correlated with SVZ distance (R2 = -0.4714, P = 0.0012, Table 3). Meaning, glioblastomas with centroid in proximity of SVZ are more likely to harbor higher expression of the glioblastoma stem cell signature as reported by Murat et al. [27].
In aggregate, these analyses suggest that glioblastomas with centroid located in proximity to the SVZ are more likely to harbor gene expression patterns with a stem-cell state.
SVZ distance and DNA repair capacity
The predominant mechanism of TMZ resistance in glioblastoma involves the expression of the DNA repair protein O6-Methyl-Guanine MethylTransferase (MGMT). MGMT encodes an evolutionarily conserved DNA repair protein whose primary function is to restore TMZ damaged guanine to their native undamaged state. High expression of MGMT has been consistently associated with TMZ resistance in glioblastoma xenografts [28] and in independent clinical trials [7,8,9, 29,30,31]. Importantly, induction or maintenance of glioblastoma stem cell state and MGMT expression appear coupled [32, 33]. That is, glioblastomas in a stem-cell state are more likely to exhibit higher expression of MGMT [32]. In this context, we tested whether glioblastoma with low SVZ score (centroid in proximity to the SVZ) is associated with increased expression of MGMT. Our analysis indicated that glioblastoma with a centroid located in proximity to the SVZ (low SVZ score) exhibited higher expression of MGMT relative to those located farther from the SVZ (high SVZ score, P = 0.01, Table 2). Of note, our previous study demonstrated that MGMT expression level was associated with MGMT promoter methylation status in TCGA glioblastomas [34].
While MGMT plays a central role in TMZ resistance, MGMT-independent DNA repair processes also contribute to TMZ resistance [35]. Using a signature derived from clinical glioblastoma specimens, Gaspar et al. showed that an MGMT independent repair process contributed to clinical temozolomide response [35]. We tested whether mRNA expression of genes in this signature also correlated with the SVZ distance of glioblastomas. Like MGMT, our analysis showed that the ssGSEA score for this gene signature inversely correlated with SVZ distance. Specifically, lower signature expression (indicative of TMZ resistance) was found in glioblastoma in closer proximity to SVZ (R2 = 0.4073, P = 0.0061, Table 3).
In aggregate, these analyses support the notion that glioblastomas located in proximity to the SVZ are more likely to be associated with increased DNA repair capacity and TMZ resistance.
SVZ distance and single nucleotide polymorphism
Glioblastoma cells intrinsically harbor high levels of oxidative stress and endogenous DNA damages [36]. Failure to repair these processes lead to genomic instability, resulting in the accumulation of single nucleotide polymorphisms (SNPs) in the genome. Our results indicate that glioblastomas in proximity of SVZ harbored higher capacity for DNA repair against these damages. Since faithful repair of these damages prevents mutation formation, a corollary of the increased repair capacity for SVZ proximal glioblastomas is that they should harbor fewer SNPs relative to those distant to the SVZ. We tested these hypotheses.
We normalized glioblastoma SNPs into a range of + 2 or − 2 values and tested the linear dependence between SVZ distances for patients age older than 65 versus normalized SNP (Fig. 2). This cut-off was selected because age > 65 is a commonly accepted threshold for age related research (https://www.who.int/healthinfo/survey/ageingdefnolder/en/). Moreover, the survival patterns for glioblastoma patients age > 65 significantly differ from those age ≤ 65 [37, 38]. The Pearson’s Correlation scores for this analysis was 0.38 (P = 0.003). This analysis points to direct correlations between SNP frequency and SVZ distance. Such correlation was not seen in patients age < 65, supporting the literature that the clinical behavior of glioblastoma in younger patients is distinct from that of the elderly [37].
SVZ distance and survival
Previous studies have demonstrated that patients afflicted with glioblastomas that contact SVZ exhibit poor survival relative to those with tumor without such contacts [5]. These results are consistent with our finding that glioblastoma in proximity to the SVZ tend to harbor properties associated with the cancer stem cell state, since these properties tend to render tumor progression or recurrence more likely. We wish to determine whether the previously reported survival associations [7,8,9] can be recapitulated in our dataset. For this analysis, each patient’s glioblastoma is again classified as “high” or “low” for SVZ distance based on the median value of SVZ distance for the entire cohort. Our analysis showed that patients with SVZ proximal (or lower than median SVZ distance) glioblastomas were associated with poor overall survival and progression free survival in comparison to patients with SVZ distal glioblastomas (Fig. 3a, b). We next determined whether the association between SVZ distance and survival remained robust after controlling for patient age. In a multi-variate model, age and SVZ distance independently associated with overall survival (Table 4).
a Association of SVZ distance with overall survival. Kaplan Meier Survival curve demonstrating association of lower SVZ distance and reduced overall survival for all glioblastomas (p = 0.02, log rank, n = 217). SVZ stratified as high or low based on median value. b Association of SVZ distance with progression-free survival. Kaplan Meier Survival curve demonstrating association of lower SVZ distance and reduced progression-free survival for all glioblastomas (p = 0.03, log rank, n = 196). SVZ stratified as high or low based on median value. c SVZ distance and LVd independently associate with overall survival. SVZ and LVd stratified as high or low based on median value. p = 0.02 (log rank, n = 217)
Finally, we examined the interaction between SVZ and, LVd, another MR variable that we previously published [39]. LVd is defined as the magnitude of displacement from the center of mass of the lateral ventricle volume in glioblastoma patients relative to that of a normal, reference brain. It is a measure of the mass effect associated with the glioblastoma tumor mass. We had previously demonstrated that higher LVd was associated with poor patient survival. In a bivariate model that incorporated LVd and SVZ, both variables independently associate with survival (Fig. 3c). In a multi-variate model that included age, LVd, and SVZ, all three variables independently contribute to survival (Table 5). We believe that this analysis further demonstrates the robust association between SVZ and overall survival.
Discussion
Making sense of the conflicting literature on glioblastoma phenotypes based on SVZ contact [7,8,9,10,11] is challenging. The published studies determined SVZ contact based on expert readers, and MR interpretation by the human eye is necessarily subjective [40]. For instance, would a one millimeter distance between the glioblastoma and SVZ be sufficient to disqualify contact? What image slice and in what orientation should be used for this determination? Should questionable contact on a single slice be sufficient to qualify determination of “contact”? These issues are further confounded by variability between readers in terms of the defining of the glioblastoma tumor border [41] as well as the limited number of subjects studied. Here, we address the subject matter by employing an automated segmentation algorithm [12] to quantitatively determine glioblastoma distance to the SVZ in 217 TCIA subjects and subsequently correlating these distances to the gene expression patterns and clinical survival. Our results suggest that glioblastomas in proximity to the SVZ exhibited increased expression of gene signatures associated with the cancer stem cell phenotype, including signatures associated with increased DNA repair capacity. Consistent with the aggressive behavior of glioblastoma stem cells, patients afflicted with glioblastomas in proximity to the SVZ exhibited from poor overall survival. Overall, our study supports the notion that glioblastoma in proximity to the SVZ are molecularly and phenotypically distinct from those in proximity to the cortex.
In the context of the current glioma literature, we propose two potential explanations to account for the differences between SVZ proximal and cortex proximal glioblastomas. First, the finding supports the hypothesis that glioblastomas located in proximity to the SVZ may have arisen from the populations of cells that harbor stem or stem-like properties and retained these properties during neoplastic transformation. In experimental models, gliomas formed from SVZ proximal cells are more invasive than those forming from non-SVZ cells [42]. Alternatively, the microenvironment of the SVZ may induce glioblastoma de-differentiation into a more stem-like state [43]. This notion is supported by the phenomenon of induced pluripotent state [44] and observation that the molecular physiology of gliomas may be modified by its microenvironment [45]. These observations, coupled with the capacity of the SVZ microenvironment to sustain neural stem cells [46] suggest the plausibility of the second hypothesis. Of note, these two hypotheses are not mutually exclusive and elements of both hypotheses may contribute to our observations.
The finding that SVZ proximal and cortex proximal glioblastomas differ in molecular physiology harbor significant therapeutic implications. The enriched stem-cell like properties of the SVZ proximal glioblastomas and associated resistance to DNA damage suggest that intensification of DNA damaging regimen for these tumors may improve clinical outcome. Several published studies have provided data in support of this notion [47,48,49,50]. Employment of such intensified regimens should be weighed in the context of increased risk for treatment related morbidities, including neuro-cognitive decline [51]. The distinct molecular physiology of SVZ-proximal and cortex-proximal glioblastomas further suggest that these tumors may exhibit differential response to targeted agents. For instance, SVZ-proximal glioblastomas may be more sensitive to agents targeting pathways required for maintaining the cancer stem cell state. In this context, SVZ distance may serve as a biomarker for response to select targeted agents.
A fundamental assumption of our study is that the bulk of the glioblastoma cells did not migrate from its native site to another region during the process of tumor formation. We believe that this assumption is reasonable for the following reasons. First, studies in murine glioblastoma models where lineage tracing afford definition of the cell of origin reveal no evidence of such mass tumor bulk migration [52]. While it is possible that a few tumor initiating cells migrated away from the SVZ, this hypothesis remains unsubstantiated [6]. Second, gliomas can be formed by the introduction of oncogenes into cortical astrocytes as well as astrocytes in proximity to the SVZ [53]. Thus, tumor formation in distinct regions of the cerebrum can occur in the absence of significant cell migration. Finally, mathematical modeling of glioblastoma growth based on serial MRIs also reveals no evidence of en bulk tumor migration during tumor growth [10, 54].
While a strength of our study is that we bypass reader subjectivity through application of an automated segmentation algorithm, no algorithm is perfect. Technical caveats of our IPVL segmentation algorithm have been reviewed in our previous articles [12, 21]. In addition, TCGA provides no information regarding the location within the tumor in which specimens were retrieved. As such, a potential spatial sampling bias exists which may limit the interpretation of our results. Finally, quantification of the extent of resection was not collected in most TCGA glioblastomas. As such, we were unable to tease out the contribution of surgical resection to the survival association reported here. Within the limit of these technical caveats, our results indicate distinct molecular physiology between SVZ-proximal and cortex-proximal glioblastomas. These findings bear significant implication in terms of clinical prognosis and therapeutic development.
In future works, analysis using an external dataset would serve to validate our results. While our work examined specific genes and gene signatures, future analyses may interrogate the expression landscape in an unbiased manner. In addition, trials targeting SVZ proximal glioblastomas through dose-dense TMZ or escalation of radiation therapy warrant consideration, since these measures may counteract the enhanced DNA-repair capacity.
Conclusions
Quantitative MR image analysis of 217 TCIA glioblastoma subjects in the context of gene expression profiles revealed that tumor proximity to the SVZ is associated with increased expression of gene signatures suggestive of enrichment in cancer stem cell state. Patients with SVZ-proximal glioblastomas exhibited poor clinical survival.
References
Ming G-L, Song H (2011) Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70:687–702
Cowan WM (1979) The development of the brain. Sci Am 241:112–133
Tan X, Shi S-H (2013) Neocortical neurogenesis and neuronal migration. Wiley Interdiscip Rev Dev Biol 2:443–459
Sanai N, Alvarez-Buylla A, Berger MS (2005) Neural stem cells and the origin of gliomas. N Engl J Med 353:811–822
Berendsen S, van Bodegraven E, Seute T et al (2019) Adverse prognosis of glioblastoma contacting the subventricular zone: biological correlates. PLoS ONE 14:e0222717
Lim DA, Cha S, Mayo MC et al (2007) Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neuro Oncol 9:424–429
Young GS, Macklin EA, Setayesh K et al (2011) Longitudinal MRI evidence for decreased survival among periventricular glioblastoma. J Neurooncol 104:261–269
Chaichana KL, McGirt MJ, Frazier J et al (2008) Relationship of glioblastoma multiforme to the lateral ventricles predicts survival following tumor resection. J Neurooncol 89:219–224
Jafri NF, Clarke JL, Weinberg V et al (2013) Relationship of glioblastoma multiforme to the subventricular zone is associated with survival. Neuro Oncol 15:91–96
Bohman L-E, Swanson KR, Moore JL et al (2010) Magnetic resonance imaging characteristics of glioblastoma multiforme: implications for understanding glioma ontogeny. Neurosurgery 67:1319–1327
Kappadakunnel M, Eskin A, Dong J et al (2010) Stem cell associated gene expression in glioblastoma multiforme: relationship to survival and the subventricular zone. J Neurooncol 96:359–367
Steed TC, Treiber JM, Patel KS et al (2015) Iterative probabilistic voxel labeling: automated segmentation for analysis of The Cancer Imaging Archive glioblastoma images. AJNR Am J Neuroradiol 36:678–685
Brennan CW, Verhaak RGW, McKenna A et al (2013) The somatic genomic landscape of glioblastoma. Cell 155:462–477
Irizarry RA, Hobbs B, Collin F et al (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323
Barbie DA, Tamayo P, Boehm JS et al (2009) Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462:108–112
Gooya A, Pohl KM, Bilello M et al (2012) GLISTR: glioma image segmentation and registration. IEEE Trans Med Imag 31:1941–1954
Tustison NJ, Avants BB, Cook PA et al (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imag 29:1310–1320
Avants BB, Tustison NJ, Stauffer M et al (2014) The Insight ToolKit image registration framework. Front Neuroinform 8:44
Li J, Taich ZJ, Goyal A et al (2014) Epigenetic suppression of EGFR signaling in G-CIMP+ glioblastomas. Oncotarget 5:7342–7356
Steed TC, Treiber JM, Patel K et al (2016) Differential localization of glioblastoma subtype: implications on glioblastoma pathogenesis. Oncotarget 7:24899–24907
Singh SK, Clarke ID, Terasaki M et al (2003) Identification of a cancer stem cell in human brain tumors. Cancer Res 63:5821–5828
Li Z (2013) CD133: a stem cell biomarker and beyond. Exp Hematol Oncol 2:17
Glumac PM, LeBeau AM (2018) The role of CD133 in cancer: a concise review. Clin Transl Med 7:18
Chalmers AJ (2007) Radioresistant glioma stem cells—therapeutic obstacle or promising target? DNA Repair 6:1391–1394
Wang Y, Zhao G, Yu T (2016) CD133 expression may be useful as a prognostic indicator in glioblastoma multiforme: a meta-analysis. Int J Clin Exp Pathol 9:12407–12414
Murat A, Migliavacca E, Gorlia T et al (2008) Stem cell-related“ self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol 26:3015–3024
Kitange GJ, Mladek AC, Carlson BL et al (2012) Inhibition of histone deacetylation potentiates the evolution of acquired temozolomide resistance linked to MGMT upregulation in glioblastoma xenografts. Clin Cancer Res 18:4070–4079
Hegi ME, Diserens A-C, Gorlia T et al (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352:997–1003
Zhang W, Zhang J, Hoadley K et al (2012) miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol 14:712–719
Kushwaha D, Ramakrishnan V, Ng K et al (2014) A genome-wide miRNA screen revealed miR-603 as a MGMT-regulating miRNA in glioblastomas. Oncotarget 5:4026–4039
Chumakova A, Lathia JD (2018) Outlining involvement of stem cell program in regulation of O6-methylguanine DNA methyltransferase and development of temozolomide resistance in glioblastoma: An Editorial Highlight for “Transcriptional control of O6-methylguanine DNA methyltransferase expression and temozolomide resistance in glioblastoma”on page 780. J Neurochem 144:688–690
Ramakrishnan V, Xu B, Akers J, Nguyen T, Ma J, Dhawan S, Ning J, Mao Y, Hua W, Kokkoli E, Furnari F, Carter BS, Chen CC (2020) Radiation-induced extracellular vesicle (EV) release of miR-603 promotes an insulin-like growth factor (IGF1) signaling induced stem cell state in glioblastomas. EBioMedicine 55:102736
Ramakrishnan V, Kushwaha D, Koay DC et al (2012) Post-transcriptional regulation of O 6-methylguanine-DNA methyltransferase MGMT in glioblastomas. Cancer Biomark 10:185–193
Gaspar N, Marshall L, Perryman L et al (2010) MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase–mediated HOX/stem cell gene signature. Cancer Res 70(22):9243–9252
Bartkova J, Hamerlik P, Stockhausen M-T et al (2010) Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene 29:5095–5102
Morgan ER, Norman A, Laing K, Seal MD (2017) Treatment and outcomes for glioblastoma in elderly compared with non-elderly patients: a population-based study. Curr Oncol 24:e92–e98
Noorbakhsh A, Tang JA, Marcus LP et al (2014) Gross-total resection outcomes in an elderly population with glioblastoma: a SEER-based analysis. J Neurosurg 120:31–39
Steed TC, Treiber JM, Brandel MG et al (2018) Quantification of glioblastoma mass effect by lateral ventricle displacement. Sci Rep 8:2827
Kerkhof M, Hagenbeek RE, van der Kallen BFW et al (2016) Interobserver variability in the radiological assessment of magnetic resonance imaging (MRI) including perfusion MRI in glioblastoma multiforme. Eur J Neurol 23:1528–1533
Provenzale JM, Ison C, Delong D (2009) Bidimensional measurements in brain tumors: assessment of interobserver variability. AJR Am J Roentgenol 193:W515–W522
Aguirre A, Gallo V (2004) Postnatal neurogenesis and gliogenesis in the olfactory bulb from NG2-expressing progenitors of the subventricular zone. J Neurosci 24:10530–10541
Matarredona ER, Pastor AM (2019) Neural stem cells of the subventricular zone as the origin of human glioblastoma stem cells. Ther Implic Front Oncol 9:779
Heddleston JM, Hitomi M, Venere M et al (2011) Glioma stem cell maintenance: the role of the microenvironment. Curr Pharm Des 17:2386–2401
Hambardzumyan D, Bergers G (2015) Glioblastoma: Defining Tumor Niches. Trends Cancer Res 1:252–265
Doetsch F, Caillé I, Lim DA et al (1999) Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97:703–716
Evers P, Lee PP, DeMarco J et al (2010) Irradiation of the potential cancer stem cell niches in the adult brain improves progression-free survival of patients with malignant glioma. BMC Cancer 10:384
Gupta T, Nair V, Paul SN et al (2012) Can irradiation of potential cancer stem-cell niche in the subventricular zone influence survival in patients with newly diagnosed glioblastoma? J Neurooncol 109:195–203
Lee P, Eppinga W, Lagerwaard F et al (2013) Evaluation of high ipsilateral subventricular zone radiation therapy dose in glioblastoma: a pooled analysis. Int J Radiat Oncol Biol Phys 86:609–615
Chen L, Guerrero-Cazares H, Ye X et al (2013) Increased subventricular zone radiation dose correlates with survival in glioblastoma patients after gross total resection. Int J Radiat Oncol Biol Phys 86:616–622
McDuff SGR, Taich ZJ, Lawson JD et al (2013) Neurocognitive assessment following whole brain radiation therapy and radiosurgery for patients with cerebral metastases. J Neurol Neurosurg Psychiatry 84:1384–1391
Zhu Y, Guignard F, Zhao D et al (2005) Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell 8:119–130
Hambardzumyan D, Amankulor NM, Helmy KY et al (2009) Modeling adult gliomas using RCAS/t-va technology. Transl Oncol 2:89–95
Swanson KR, Bridge C, Murray JD, Alvord EC Jr (2003) Virtual and real brain tumors: using mathematical modeling to quantify glioma growth and invasion. J Neurol Sci 216:1–10
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declare that they is no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Steed, T.C., Treiber, J.M., Taha, B. et al. Glioblastomas located in proximity to the subventricular zone (SVZ) exhibited enrichment of gene expression profiles associated with the cancer stem cell state. J Neurooncol 148, 455–462 (2020). https://doi.org/10.1007/s11060-020-03550-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-020-03550-4