Abstract
Purpose
Peritumoural brain zone (PT) of glioblastoma (GBM) is the area where tumour recurrence is often observed. We aimed to identify differentially regulated genes between tumour core (TC) and PT to understand the underlying molecular characteristics of infiltrating tumour cells in PT.
Methods
17 each histologically characterised TC and PT tissues of GBM along with eight control tissues were subjected to cDNA Microarray. PT tissues contained 25–30% infiltrating tumour cells. Data was analysed using R Bioconductor software. Shortlisted genes were validated using qRT-PCR. Expression of one selected candidate gene, PDZ Binding Kinase (PBK) was correlated with patient survival, tumour recurrence and functionally characterized in vitro using gene knock-down approach.
Results
Unsupervised hierarchical clustering showed that TC and PT have distinct gene expression profiles compared to controls. Further, comparing TC with PT, we observed a significant overlap in gene expression profile in both, despite PT having fewer infiltrating tumour cells. qRT-PCR for 13 selected genes validated the microarray data. Expression of PBK was higher in PT as compared to TC and recurrent when compared to newly diagnosed GBM tumours. PBK knock-down showed a significant reduction in cell proliferation, migration and invasion with increase in sensitivity to radiation and Temozolomide treatment.
Conclusions
We show that several genes of TC are expressed even in PT contributing to the vulnerability of PT for tumour recurrence. PBK is identified as a novel gene up-regulated in PT of GBM with a strong role in conferring aggressiveness, including radio-chemoresistance, thus contributing to recurrence in GBM tumours.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma (GBM) is the most common primary malignant neoplasm of brain in adults [1]. Despite aggressive treatment regimen, GBM patients have a very poor prognosis and the tumour almost always recurs [2]. During surgical resection of GBM, a variable number of invading tumour cells are invariably left behind in the peritumoural brain zone (PT) which is most often the site of recurrence [3]. Delineating the biology of these subsisting cells could facilitate understanding of tumour recurrence in GBM.
Few attempts have been made to identify the molecular alterations of tumour cells in PT. Studies till date have shown that tumour core (TC) and PT have distinct gene expression profiles [4]. While both TC and PT are heterogeneous across patients, tumour cells in PT are more migratory and invasive compared to TC [5, 6]. Also, the brain adjacent to tumour (BAT) which is histologically normal was found to have a neoplastic gene expression profile [7]. However, there is a need to further characterize PT of GBM in order to identify genes/pathways that have functional relevance in the pathobiology of GBM.
In view of this, in the present study, we identified the alterations in gene expression in PT of GBM tumours in comparison with TC and non-neoplastic brain tissue (control) by employing a whole genome gene expression profiling approach. The analysis revealed several differentially expressed genes between TC and PT. Among these, PDZ-binding kinase (PBK) was found to be significantly up-regulated in PT compared to TC, despite fewer tumour cells in the PT. We correlated PBK expression with patient survival, tumour recurrence and subsequently demonstrated a role for PBK in tumour cell proliferation, invasion, and migration, as well as radio-chemo resistance.
Materials and methods
The study was carried out on a prospective and retrospective cohort of GBM patients. The prospective cohort comprised adult patients with cerebral lobar GBM (n = 25). Prior to surgery, all patients underwent magnetic resonance (MR) imaging of brain, with MR spectroscopy and perfusion imaging and TC areas were demarcated. PT was defined as the area in the neuro-parenchyma immediately adjacent to the enhancing portion of TC, with high choline peaks on MR spectroscopy and increased perfusion. Appropriate areas of PT amenable for safe resection, were biopsied at the beginning of surgical decompression, with the aid of neuronavigation techniques. Around 3–4 PT tissues were collected per patient and histologically assessed for the percentage of tumour cells. Subsequently, TC portion was resected. The respective samples from TC and PT areas were dissected into two parts: one part was fixed in 10% buffered neutral formalin and processed for paraffin sections and second part was placed in RNA later and stored at − 80 °C. Control tissues were collected from lateral temporal cortex from patients undergoing surgery for drug resistant epilepsy due to mesial temporal sclerosis. Sections from formalin fixed paraffin embedded (FFPE) blocks from TC and PT of each case were studied independently by two neuropathologists (VS, TCY). The corresponding tissues collected in RNA later were used for microarray and qRT-PCR experiments. A total of 17 TC and PT tissues each along with eight control tissues were used for cDNA microarray experiments. For validation, in addition to these tissues, another eight TC and PT each and six control tissues were used. The study was approved by the Institutional Ethics Committee and informed consent was obtained from the patient or the closest relative available.
The details of retrospective cohort along with description of the methods used for RNA isolation, Microarray, analysis of microarray data, quantitative real time PCR (qRT-PCR), Immunohistochemistry (IHC), generation of stable knock-down, western blotting and all the in vitro assay protocols are provided in Supplementary File 1. The microarray raw data has been submitted to GEO database with accession number GSE116520.
Results
Histopathological analysis reveals a variable number of tumour cells in the PT samples
Histopathological evaluation of tissue sections from TC showed typical GBM features (Fig. S1b1 and b2). The PT sections showed variable percentage of tumour cell infiltration into adjacent grey matter as well as white matter. These cells were identified on histology, supplemented by the MIB-1 staining that helped identify the proliferating cells (Fig. S1c1 and c2). Percentage of infiltrating tumour cells in PT samples ranged from 5 to 70%. In order to maintain uniformity in the sample set, only those PT samples containing approximately 25–30% tumour cells were used for subsequent experiments. The MR image depicting TC and PT and the respective histopathology of a representative case of GBM is depicted in Figure S1a and b.
Comparison of transcriptome profiles of TC, PT and controls
Transcriptome analysis reveals a shared gene expression profile between TC and PT as compared to control
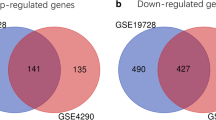
TC and PT samples were compared with control samples to identify the differentially expressed genes. Data analysis showed 7124 genes to be significantly differentially expressed in TC versus control and 5188 genes in PT versus control. The lists of top 25 differentially expressed genes are given in Supplementary Table 1. On unsupervised hierarchical clustering, TC and control as well as PT and control formed two distinct clusters respectively with minimal overlap (Fig. 1a, b). Interestingly, using Venn, we noted that 4176 genes had a similar expression pattern in TC and PT when compared to control (Fig. 1c). This suggested that PT, with fewer infiltrating tumour cells, had a significant overlap with TC in the expression of several genes. There were 436 and 2062 genes specifically differentially expressed in PT and TC, respectively, compared to control tissues. The lists of differentially expressed 5176, 436 and 2062 genes are given in Supplementary Table 2. Among the 4176 genes, top up-regulated genes included TOP2A, UBE2C, IGFBP2, PBK, MELK, NRP1, NTN1 and CD68and the top down-regulated genes included SLC17A7, CHD5, PACSIN1 and DLGAP2.
Gene expression analysis of TC, PT and control. Representative cluster from unsupervised hierarchical clustering shows differentially regulated genes between a TC versus control, b PT versus control and d PT versus TC. Several samples show overlapping gene expression profile between PT and TC. c Venn diagram shows 4176 genes commonly differentially expressed between TC versus control and PT versus control. In the hierarchical cluster red and blue color represents up and down-regulated genes respectively. All the rows represent genes and columns represent samples. PT Peritumoural brain zone, TC tumour core
Transcriptome analysis of PT as compared to TC reveals differentially expressed genes between the two groups
We found 991 genes to be differentially expressed in PT as compared to TC with 391 genes being up-regulated and 600 genes down-regulated. The lists of top 25 differentially expressed genes are given in Supplementary Table 1. On unsupervised hierarchical clustering, whilst a subset of TC and PT samples formed distinct cluster, other samples showed a shared gene expression profile (Fig. 1d). Some of the top upregulated genes in PT included MOG, MAL, MAG, MOBP, KLK6, ENPP2, SH3GL3 and others, known to be associated with neurogenesis and stem cell markers. Some genes upregulated in TC include IL8, NNMT, TGFBI, SOD2, LOX amongst others, which are known to be pro-tumorigenic in GBM.
Gene set enrichment analysis (GSEA) of differentially regulated genes in TC and PT as compared to control reveals commonly enriched pathways
Pathway analysis was carried out using Web-based Gene Set Analysis toolkit and gene set enrichment analysis (GSEA). Both the TC and PT, as compared to control, showed enrichment of pathways related to cell cycle, MAPK signalling, ECM receptor interaction, Focal adhesion, along with immune system related pathways like complement and coagulation cascades and antigen processing and presentation. However, tight junction, calcium signalling and cell adhesion molecules pathways were specifically enriched in the PT as compared to control (Supplementary Table 3) with most of the genes in these pathways down-regulated in PT versus control. Angiogenesis and p53 pathways were specifically enriched in TC.
Gene expression analysis of selected genes by qRT-PCR corroborates with microarray data
A set of 13 genes were selected for qRT-PCR validation based on their fold change (seven highly up-regulated, two highly down-regulated and four moderately regulated). δδCt method was followed for calculation of the fold change of each gene in TC and PT as compared to control. The expression pattern of these genes in qRT-PCR matched with that of the expression pattern in Microarray experiment (Fig. 2a). The expression of all 13 genes in GBM tissues of TCGA and Rembrandt cohort also followed the same pattern as TC in our cohort (Fig. 2b).
Validation of selected differentially expressed genes using qRT-PCR. a Scatter plots depict genes significantly up or down-regulated in TC and PT compared to control (p ≤ 0.05). b Bar diagram depicting differential expression of selected genes in GBM versus control in TCGA and Rembrandt datasets. 1-PBK, 2-MELK, 3-MYL9, 4-MMP9, 5-UBE2C, 6-HOXB2, 7-RCBTB2, 8-SAA2, 9-GBP2, 10-FLNC, 11-TCBA1, 12-PKNOX1, 13-SNCG. TCGA sample number—affymetrix: n = 528, Agilent: n = 572, Rembrandt-n = 227. Y-axis represents the fold changes for TC, PT and control samples
IHC validation showed higher expression of PBK in the PT compared to TC
As PBK showed a significant upregulation in PT compared to TC, it was selected for protein validation using IHC. PBK expression was predominantly in cell nuclei, although it also showed considerable cytoplasmic expression (Fig. 3a). In TC, there was variable and patchy immunostaining of tumour cells (Fig. 3a–c). PBK staining was occasionally observed in the perinecrotic and perivascular tumour cells (Fig. 3b, c). The median LI of tumour cells in TC was 20 ± 15.06. In PT, infiltrating tumour cells in grey matter and white matter were positive with a median LI of 30 ± 22.56 (Fig. 3d, e). PT samples which had higher or lower than 25–30% tumour cell infiltration were also evaluated for PBK expression by IHC. 10–50% tumour cell infiltration showed a median LI of 22.5–30 (Supplementary file 2). The neuronal and glial cell population in PT and control tissues were negative for PBK staining. Taken together, PBK protein expression was higher in PT as compared to TC, given that the PT contains fewer tumour cells.
Immunohistochemistry showing staining pattern of PBK in the TC and PT of GBM tumour tissue samples. a–c show variable and patchy immunoreactivity of PBK in GBM TC. (a-inset) Nuclear and cytoplasmic positivity is seen. b shows perinecrotic and c perivascular tumour cell positivity. d and e represent PBK immunoreactivity in PT. d PT showing staining of tumour cells infiltrating white matter and e grey matter. f1, f2 PBK immunoreactvity in paired samples shows increased expression in the f2 recurrent GBM sample compared to f1 primary (newly diagnosed) sample. a, a (inset) c, f1, f2 are original magnification ×200. B, d and e are original magnification ×100
Further, PBK expression in 25 paired tissues of primary and recurrent GBM was assessed. The median LI was significantly higher in recurrent GBM (20 ± 11.86) as compared to primary (10 ± 9.45). On comparing PBK expression in paired samples, a higher expression in 64% (16/25) of recurrent as compared to their respective primary tumour was noted (p = 0.003) (Fig. 3f1, f2). However, we noted that PBK expression was not associated with either overall survival or progression free survival in our retrospective cohort (data not shown). In concurrence, PBK mRNA expression did not show association with survival in the TCGA-GBM cohort. Also, PBK protein expression did not show any changes with respect to IDH1 R132H immunopositivity, neither in the prospective cohort nor in the retrospective cohort (data not shown). Since PBK expression was higher in PT and in recurrent tumours, we further evaluated its functional role in malignant glioma cell lines.
Knock-down of PBK expression results in reduction of cell proliferation, migration and invasion
Two different shRNA constructs against PBK, which showed significant knock-down in LN229 and U251 glioma cell lines compared to Scr were used for further experiments (Fig. 4a). MTT assay was performed to ascertain if PBK-KD has any effect on proliferation. There was a significant reduction in cell proliferation in PBK-KD cells as compared to Scr (p < 0.0001), observed until 72 h (Fig. 4b). Similar results were obtained in cell counting experiment as well (p = 0.05) (Fig. 4c). On FACS analysis, we found an increase in number of cells in S phase and G2/M phase in PBK-KD cells as compared to Scr cells (Fig. S2).
Effect of PBK-KD on cell proliferation, migration and invasion. a Western blot representing PBK knockdown in LN229 and U251 cells. b MTT assay results depicted in a graph showing the OD values of PBK-KD and Scr cells at 24 h, 48 h and 72 h in LN229 and U251 cells (p ≤ 0.05, n = 3). c Cell counting assay results depicted in a graph showing the number of PBK-KD and Scr cells counted at 24 h, 48 h and 72 h in LN229 and U251 cells (p ≤ 0.05, n = 3). d and e Wound healing assay showing the migration of PBK-KD and Scr cells at 0 h and 9 h in LN229 and 0 h and 12 h in U251 (p ≤ 0.05, n = 3). f and g Matrigel invasion assay showing invaded LN229 and U251 cells with PBK-KD along with Scr cells (p ≤ 0.05, n = 3). Scr represents scrambled and clone 1 and clone 2 represent PBK-KD cells in respective cell lines. ***, ** and * represent p value ≤ 0.0001, 0.001 and 0.01 respectively
To evaluate the role of PBK in migration of glioma cells, wound healing assay was performed with the Scr and KD cells. As shown in Fig. 4d, there was a gradual closure of the wound in Scr cells as opposed to PBK-KD cells. At 9 h, Scr-LN229 cells had a 50% reduction in the area of wound whereas PBK-KD-LN229 cells closed only 15–20% of the wound area (Fig. 4e). This difference in wound closure was observed at 12 h time-point in case of U251 cells. Thus, PBK-KD significantly reduced the migratory potential of LN229 and U251 cells.
To determine the effect of PBK-KD on cell invasion, a Matrigel invasion assay was performed. On incubation of Scr and PBK-KD cells for 16 h in matrigel coated chambers with serum as the chemoattractant, a significant decrease in percentage of invading cells (represented as fold change) was observed in PBK-KD cells as compared to Scr (p < 0.0001). Therefore, PBK-KD had a significant effect on glioma cell invasion (Fig. 4f, g).
PBK knockdown showed increased sensitivity to chemotherapeutic drugs and radiation exposure
Since the PBK-KD showed a marked difference in tumour cell proliferation, invasion and migration, we further assessed its role in radio-chemo sensitivity. MTT assay was performed to assess the difference in sensitivity to TMZ, etoposide and cisplatin between Scr and PBK-KD. There was an increased sensitivity towards TMZ, Etoposide and Cisplatin observed in the PBK-KD cells as compared to Scr which is represented by decrease in IC50 (p < 0.05) (Fig. 5a–c). Similarly, to assess differences in sensitivity to radiation therapy, PBK-KD and Scr cells were exposed to gamma radiation followed by a clonogenic assay. The number of colonies formed by PBK-KD cells was fewer than Scr which is represented as percent survival (Fig. 5d). These findings suggest a role for PBK in radio-chemo resistance.
Effect of PBK-KD on chemotherapeutic drug treatment and radiation exposure. Graphs showing reduction in IC50 of Clone 1 and clone 2 cells, compared to scrambled, upon treatment with chemotherapeutic drugs—a TMZ, b Etoposide and c Cisplatin in LN229 and U251 cell line. d Graph showing reduction in percent survival of clone 1 and clone 2 compared to scrambled upon exposure to gamma irradiation in LN229 and U251 cell line (p ≤ 0.05, n = 3). ***, ** and * represent p value ≤ 0.0001, 0.001 and 0.01 respectively
Discussion
Peritumoural brain zone of GBM is the neuroparenchyma immediately adjacent to TC where there is variable infiltration of tumour cells. Tumour recurrence occurs in this PT and is mainly attributed to the tumorigenic potential of these persisting tumour cells. Therefore, we aimed to evaluate the molecular alterations of PT by employing a whole genome gene expression profiling technique of the PT tissue, although more precise techniques like Laser Capture Microdissection or single cell sequencing are known to capture the molecular alterations in individual GBM cells. The novelty of our study is the sampling of different PT regions using advanced MR imaging parameters and navigation assistance. On histopathological analysis, we observed that PT tissues (grey or white matter) taken from different points around TC of the same patient have variable number of infiltrating tumour cells indicating the arbitrary pattern of infiltration. This has been observed in few other studies as well [4, 5]. Also, Tamura et al. showed that microvessel density in PT was lower than TC [8]. Interestingly, we noted that TC and PT show a significant overlap in differentially expressed genes/pathways despite PT having lesser number of tumour cells. Among the genes commonly differentially expressed in TC and PT compared to control, top up-regulated genes included UBE2C, NUSAP1, IGFBP2 and PBK which are involved in cell growth and proliferation of cancers of brain, breast, ovaries and others [9,10,11,12]. Among these, IGFBP2 has been reported to have a dual status in PT of GBM: whilst Hoelzinger et al. showed down-regulation of this gene in PT, Mangiola et al. have noted its’ up-regulation [7, 13]. Genes associated with Neuropile and its structures like NRP1, NTN1 and ROBO1 with known role in cell survival, migration and axonal guidance during neural development were also up-regulated in PT and TC. Other notable up-regulated genes included VEGF and HIF1α which regulate angiogenesis [14, 15], and ECM associated genes like collagens and matrix metalloproteinases [19,20,21] and microglial marker CD68. The top down-regulated genes included SLC17A7 and CHD5 which are known tumour suppressor genes in GBM and other systemic cancers respectively [16,17,18,19]. Some up-regulated genes specific to PT compared to control included genes known to be associated with invasion and migration like Scinderin (SCIN) and stem cell markers like aldehyde dehydrogenase 1 family member A1 (ALDH1A1) [20,21,22]. TGFα, a neurotropic factor secreted by astrocytes and known to promote invasion of tumour cells was also up-regulated in PT. Among the genes specific to TC compared to control, up-regulated genes included S100A8 and S100A9-known serum biomarkers in GBM; IGFBP7—involved in angiogenesis; DKK1—hypoxia induced gene in GBM [23,24,25]; CCL2, a known chemo-attractant for Tumour Associated Macrophage (TAM) recruitment. Some important down-regulated genes included NKAIN2 (tumour suppressor gene) and ULK2 (role in autophagy) [26, 27]. Thus, as shown previously, we also observed an increase in expression of genes associated with cell growth, survival, proliferation and motility in both TC and PT compared to control [7]. In addition, we also show an up-regulation of stem cell markers specifically in PT and down-regulation of tumour suppressor genes in both TC and PT.
When we compared PT with TC, genes related to neurogenesis like MAL, MOG, MAG, MOBP and others were up-regulated in PT, probably contributed by the native neuroparenchyma. Similar observation was made by Mangiola et al. where genes associated with CNS development were up-regulated in Brain Adjacent Tumour (BAT) compared to TC. Interestingly, in spite of limited number of tumour cells in PT, we also noted up-regulation of genes such as KLK6, SH3GL3, ENPP2 and few others which are known to confer chemoresistance and promote invasion in GBM and other cancers [28,29,30], pointing out to the aggressive nature of the infiltrating cells. Notable genes up-regulated in TC compared to PT are NNMT, LGALS3, SOD2, TGFBI and IL8 which are known to be up-regulated and associated with poor patient prognosis in GBM and other cancers [31,32,33,34,35,36]. Taken together, gene expression analysis of TC and PT, show that cells in PT are probably in the process of undergoing active remodelling of the extra-cellular matrix, thus promoting cell invasion and migration. However, TC is mostly enriched with genes and pathways related to tumour cell survival and growth, hypoxia and angiogenesis.
PBK, also known as T-LAK cell originated protein kinase (TOPK), is a protein related to the mitogen-activated protein kinase kinase (MAPKK) family. Physiologically, it is involved in activation of lymphoid cells and is specifically expressed in testes with possible role in spermatogenesis [37]. It is implicated in tumorigenesis of several systemic cancers including breast, prostate and lung cancers [38,39,40]. PBK is also known to be upregulated in GBM, especially in glioblastoma stem cells (GSC) [41]. Joel et al. showed that PBK knockdown and treatment with PBK inhibitor (HI-TOPK) in glioma initiating cells (GIC) cultures reduced cell viability and sphere formation [12]. We noted that PBK is expressed in nucleus and cytoplasm of GBM tissues in accordance with its reported expression pattern in prostate and cervical cancers [38, 42]. We also noted a higher PBK immunoreactivity in perivascular and perinecrotic regions of GBM indicating that PBK could probably contribute towards tumour cell invasion through a perivascular conduit. Our observation of PBK staining of perinecrotic tumour cells probably suggests that this gene is induced by hypoxia in GBM. For the first time, we report PBK mRNA and protein expression to be significantly up-regulated in PT as compared to TC. This observation has not been made in previous studies on PT, probably due to variation in the sampling of PT when compared to ours [4, 7, 13]. Further, we also noted an increased expression of PBK in recurrent GBM in a significant number of paired GBM samples. From these observations, we imply that tumour cells with higher PBK expression probably have an advantage over others in migrating and invading into the normal neuroparenchyma, thereby contributing to tumour recurrence in GBM. Although high PBK expression is known to be associated with poor patient prognosis in GBM and other cancers [40, 43, 44], we could not establish this in our cohort. However, our survival data is in line with TCGA data which also did not show correlation of PBK mRNA with patient prognosis. PBK protein expression did not show any correlation with IDH1 R132H mutation. This could be because of fewer cases showing IDH1 R132H mutation which is expected as per WHO 2016 classification of GBM. Our in vitro studies showed that PBK significantly contributes to tumour cell proliferation, migration and invasion which is line with other studies on PBK in GBM and other systemic cancers [12, 38, 45]. These observations further strengthened the significance of PBK up-regulation in PT. We demonstrate a significant role for PBK in conferring resistance to both radiation and TMZ treatment along with other chemotherapeutic drugs like cisplatin and etoposide. Only one study till date showed a role for PBK along with miR-770-5p in conferring radiation resistance [46]. Studies have shown that treatment of glioma cells with PBK inhibitor brings down the tumour volume in vivo [12, 47]. In support of our data on the functional role of PBK in conferring tumour aggressiveness in GBM, there are few studies that have shown that treatment of glioma cells with PBK inhibitor brings down the tumour volume in vivo [12, 47]. However, we were not able to establish a prognostic significance in our cohort. Hence, further studies and clinical validation of the efficacy of PBK inhibitors are warranted.
References
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820. https://doi.org/10.1007/s00401-016-1545-1
Weller M, Fisher B, Taphoorn MJB et al (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. Cancer/Radiothér 9:196–197. https://doi.org/10.1016/j.canrad.2005.05.001
De Bonis P, Maira G, Balducci M, Sica G, Lauriola L (2008) Invasive tumor cells and prognosis in a selected population of patients with glioblastoma multiforme. Cancer 113:841–846. https://doi.org/10.1002/cncr.23624
Van Meter T, Dumur C, Hafez N, Garrett C, Fillmore H, Broaddus WC (2006) Microarray analysis of MRI-defined tissue samples in glioblastoma reveals differences in regional expression of therapeutic targets. Diagn Mol Pathol 15(4):195–205. https://doi.org/10.1097/01.pdm.0000213464.06387.36
Gill BJ, Pisapia DJ, Malone HR et al (2014) MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.1405839111
Ruiz-Ontañon P, Orgaz JL, Aldaz B et al (2013) Cellular plasticity confers migratory and invasive advantages to a population of glioblastoma-initiating cells that infiltrate peritumoral tissue. Stem Cells 31(6):1075–1085. https://doi.org/10.1002/stem.1349
Mangiola A, Saulnier N, De Bonis P et al (2013) Gene expression profile of glioblastoma peritumoral tissue: an ex vivo study. PLoS ONE. https://doi.org/10.1371/journal.pone.0057145
Tamura R, Ohara K, Sasaki H, Morimoto Y, Yoshida K, Toda M (2018) Histopathological vascular investigation of the peritumoral brain zone of glioblastomas. J Neurooncol 136(2):233–241. https://doi.org/10.1007/s11060-017-2648-9
Wagner KW, Sapinoso LM, El-rifai W et al (2004) Overexpression, genomic amplification and therapeutic potential of inhibiting the UbcH10 ubiquitin conjugase in human carcinomas of diverse anatomic origin. Oncogene. https://doi.org/10.1038/sj.onc.1207861
Iyer J, Moghe S, Furukawa M, Tsai M (2011) What’ s Nu (SAP) in mitosis and cancer ? Cell Signal 23(6):991–998. https://doi.org/10.1016/j.cellsig.2010.11.006
Han S, Li Z, Master LM, Master ZW, Wu A (2014) Exogenous IGFBP-2 promotes proliferation, invasion, and chemoresistance to temozolomide in glioma cells via the integrin b 1-ERK pathway. Br J Cancer. https://doi.org/10.1038/bjc.2014.435
Joel M, Mughal AA, Grieg Z et al (2015) Targeting PBK/TOPK decreases growth and survival of glioma initiating cells in vitro and attenuates tumor growth in vivo. Mol Cancer 14:121. https://doi.org/10.1186/s12943-015-0398-x
Hoelzinger DB, Mariani L, Weis J et al (2005) Gene expression profile of glioblastoma multiforme invasive phenotype points to new therapeutic targets. Neoplasia 7(1):7–16. https://doi.org/10.1593/neo.04535
Zhang M, Ye G, Li J, Wang Y (2015) Recent advance in molecular angiogenesis in glioblastoma: the challenge and hope for anti-angiogenic therapy. Brain Tumor Pathol 32(4):229–236. https://doi.org/10.1007/s10014-015-0233-5
Kaur B, Khwaja FW, Severson EA, Matheny SL, Brat DJ, Van Meir EG (2005) Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis 1. Neuro-oncology. https://doi.org/10.1215/S1152851704001115
Lin B, Lee H, Yoon J et al (2015) Global analysis of H3K4me3 and H3K27me3 profiles in glioblastoma stem cells and identification of SLC17A7 as a bivalent tumor suppressor gene. Oncotarget. https://doi.org/10.18632/oncotarget.3030
Ma Z, Song J, Liu S et al (2016) Decreased expression of the CHD5 gene and its clinicopathological significance in breast cancer: correlation with aberrant DNA methylation. Oncol Lett 12:4021–4026. https://doi.org/10.3892/ol.2016.5147
Baykara O, Tansarikaya M, Bulut P, Demirkaya A, Buyru N (2017) CHD5 is a potential tumor suppressor in non small cell lung cancer (NSCLC). Gene 618:65–68. https://doi.org/10.1016/j.gene.2017.04.010
Du Z, Li L, Huang X, Jin J, Huang S, Zhang Q et al (2016) The epigenetic modifier CHD5 functions as a novel tumor suppressor for renal cell carcinoma and is predominantly inactivated by promoter CpG methylation. Oncotarget. https://doi.org/10.18632/oncotarget.7822
Wang D, Sun S-Q, Yu Y-H, Wu W-Z, Yang S-L, Tan J-M (2014) Suppression of SCIN inhibits human prostate cancer cell proliferation and induces G0/G1 phase arrest. Int J Oncol 44(1):161–166. https://doi.org/10.3892/ijo.2013.2170
Chen XM, Guo JM, Chen P et al (2014) Suppression of scinderin modulates epithelial mesenchymal transition markers in highly metastatic gastric cancer cell line SGC 7901. Mol Med Rep 10(5):2327–2333. https://doi.org/10.3892/mmr.2014.2523
Schäfer A, Teufel J, Ringel F et al (2012) Aldehyde dehydrogenase 1A1-a new mediator of resistance to temozolomide in glioblastoma. Neuro Oncol 14(12):1452–1464. https://doi.org/10.1093/neuonc/nos270
Popescu ID, Codrici E, Albulescu L et al (2014) Potential serum biomarkers for glioblastoma diagnostic assessed by proteomic approaches. Proteom Sci 12(1):1–14. https://doi.org/10.1186/s12953-014-0047-0
Pen A, Moreno MJ, Durocher Y, Deb-Rinker P, Stanimirovic DB (2008) Glioblastoma-secreted factors induce IGFBP7 and angiogenesis by modulating Smad-2-dependent TGF-β signaling. Oncogene 27(54):6834–6844. https://doi.org/10.1038/onc.2008.287
Guo KT, Fu P, Juerchott K et al (2014) The expression of Wnt-inhibitor DKK1 (Dickkopf 1) is determined by intercellular crosstalk and hypoxia in human malignant gliomas. J Cancer Res Clin Oncol 140(8):1261–1270. https://doi.org/10.1007/s00432-014-1642-2
Zhao S-C, Zhou B-W, Luo F, Mao X, Lu Y-J (2015) The structure and function of NKAIN2-a candidate tumor suppressor. Int J Clin Exp Med 8(10):17072–17079
Shukla S, Patric IRP, Patil V et al (2014) Methylation silencing of ULK2, an autophagy gene, is essential for astrocyte transformation and tumor growth. J Biol Chem 289(32):22306–22318. https://doi.org/10.1074/jbc.M114.567032
Drucker KL, Paulsen AR, Giannini C et al (2013) Clinical significance and novel mechanism of action of kallikrein 6 in glioblastoma. Neuro Oncol 15(3):305–318. https://doi.org/10.1093/neuonc/nos313
Delic S, Lottmann N, Jetschke K, Reifenberger G, Riemenschneider MJ (2012) Identification and functional validation of CDH11, PCSK6 and SH3GL3 as novel glioma invasion-associated candidate genes. Neuropathol Appl Neurobiol 38(2):201–212. https://doi.org/10.1111/j.1365-2990.2011.01207.x
Chen M, O’Connor KL (2005) Integrin α6β4 promotes expression of autotaxin/ENPP2 autocrine motility factor in breast carcinoma cells. Oncogene 24(32):5125–5130. https://doi.org/10.1038/sj.onc.1208729
Win KT, Lee S-W, Huang H-Y et al (2013) Nicotinamide N-methyltransferase overexpression is associated with Akt phosphorylation and indicates worse prognosis in patients with nasopharyngeal carcinoma. Tumour Biol 34(6):3923–3931. https://doi.org/10.1007/s13277-013-0980-z
Le Mercier M, Fortin S, Mathieu V, Kiss R, Lefranc F (2010) Galectins and gliomas. Brain Pathol 20(1):17–27. https://doi.org/10.1111/j.1750-3639.2009.00270.x
Shwetha SD, Shastry AH, Arivazhagan A (2016) Manganese superoxide dismutase (MnSOD) is a malignant astrocytoma specific biomarker and associated with adverse prognosis in p53 expressing glioblastoma. Pathol Res Pract 212(1):17–23. https://doi.org/10.1016/j.prp.2015.11.002
Pajares MJ, Agorreta J, Salvo E, Behrens C, Wistuba II, Montuenga LM et al (2014) TGFBI expression is an independent predictor of survival in adjuvant-treated lung squamous cell carcinoma patients. Br J Cancer 110(6):1545–1551. https://doi.org/10.1038/bjc.2014.33
Zhu J, Chen X, Liao Z, He C, Hu X (2015) TGFBI protein high expression predicts poor prognosis in colorectal cancer patients. Int J Clin Exp Pathol 8(1):702–710
Gales D, Clark C, Manne U, Samuel T (2013) The chemokine CXCL8 in carcinogenesis and drug response. ISRN Oncol. https://doi.org/10.1155/2013/859154
Abe Y, Matsumoto S, Kito K, Ueda N (2000) Cloning and expression of a novel MAPKK-like protein kinase, lymphokine-activated killer T-cell-originated protein kinase, specifically expressed in the testis and activated lymphoid cells. J Biol Chem 275(28):21525–21531. https://doi.org/10.1074/jbc.M909629199
Brown-clay JD, Shenoy DN, Timofeeva O, Bhaskar V, Nandi AK, Banerjee PP (2015) PBK/TOPK enhances aggressive phenotype in prostate cancer via β-Catenin-TCF/LEF-mediated matrix metalloproteinases production and invasion. Oncotarget 6(17):15594–15609. https://doi.org/10.18632/oncotarget.3709
Park J, Lin M, Nishidate T, Nakamura Y, Katagiri T (2006) PDZ-binding kinase/T-LAK cell-originated protein kinase, a putative cancer/testis antigen with an oncogenic activity in breast cancer. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-06-1601
Shih M, Chen J, Wu Y, Jan Y, Yang B, Lu P, Lai J (2012) TOPK/PBK promotes cell migration via modulation of the PI3K/PTEN/AKT pathway and is associated with poor prognosis in lung cancer. Oncogene 31:2389. https://doi.org/10.1038/onc.2011.419
Stangeland B, Mughal AA, Grieg Z, Sandberg CJ, Joel M, Nygård S, Langmoen IA et al (2015) Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells. Oncotarget 6(28):26192–26215. https://doi.org/10.18632/oncotarget.4613
Luo Q, Lei B, Liu S, Chen Y, Sheng W, Lin P et al (2014) Expression of PBK/TOPK in cervical cancer and cervical intraepithelial neoplasia. Int J Clin Exp Med 7(11):8059–8064
Hayashi T, Hayakawa Y, Koh M, Tomita T, Nagai S, Kashiwazaki D, Kuroda S et al (2018) Impact of a novel biomarker, T-LAK cell-originating protein kinase (TOPK) expression on outcome in malignant glioma. Neuropathology 38(2):144–153. https://doi.org/10.1111/neup.12446
Quan C, Xiao J, Duan Q, Yuan P, Xue P, Dogan S, Liu L et al (2018) T-lymphokine-activated killer cell-originated protein kinase (TOPK) as a prognostic factor and a potential therapeutic target in glioma. Oncotarget 9(8), 7782–7795. https://doi.org/10.18632/oncotarget.23674
Dou X, Wei J, Sun A, Shao G, Childress C, Yang W, Lin Q (2015) PBK/TOPK mediates geranylgeranylation signaling for breast cancer cell proliferation. Cancer Cell Int 15(1):1–9. https://doi.org/10.1186/s12935-015-0178-0
Lee HC, Her N-G, Kang D, Jung SH, Shin J, Lee M, Lee J-S, et al (2017) Radiation-inducible miR-770-5p sensitizes tumors to radiation through direct targeting of PDZ-binding kinase. Cell Death Dis 8(3):e2693. https://doi.org/10.1038/cddis.2017.116
Sugimori M, Hayakawa Y, Koh M, Hayashi T, Tamura R, Kuroda S (2018) Targeting the T-Lak cell originated protein kinase by OTS964 shrinks the size of power-law coded heterogeneous glioma stem cell populations. Oncotarget 9(3):3043–3059. https://doi.org/10.18632/oncotarget.23077
Acknowledgements
The results presented here are in part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov/. We acknowledge Prof. Kumarvel Somasundaram, Indian Institute of Science, for his support and guidance. We thank Prof. Subba Rao, Department of Microbiology and Cell Biology, Indian Institute of Science, for providing shPBK constructs as a kind gift. This work was carried out as a part of the project under the umbrella of Centre of Excellence in Neuro-Oncology funded by Department of Biotechnology, Government of India; University Grants Commission is acknowledged for fellowship to BSK. PK’s lab received core support from the IISc-DBT partnership program and DST-FIST infrastructure funding. All the project investigators and project assistants of DBT-COE are acknowledged. We acknowledge Mr. K. Manjunath, Department of Neuropathology for the preparation of figure montages.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The experiments conducted for this manuscript comply with current laws of the country.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kruthika, B.S., Jain, R., Arivazhagan, A. et al. Transcriptome profiling reveals PDZ binding kinase as a novel biomarker in peritumoral brain zone of glioblastoma. J Neurooncol 141, 315–325 (2019). https://doi.org/10.1007/s11060-018-03051-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-018-03051-5