Abstract
The value of perfusion and diffusion-weighted MRI in differentiating histological subtypes according to the 2007 WHO glioma classification scheme (i.e. astrocytoma vs. oligodendroglioma) and genetic subtypes according to the 2016 WHO reclassification (e.g. 1p/19q co-deletion and IDH1 mutation status) in WHO grade II and III diffuse gliomas remains controversial. In the current study, we describe unique perfusion and diffusion MR signatures between histological and genetic glioma subtypes. Sixty-five patients with 2007 histological designations (astrocytomas and oligodendrogliomas), 1p/19q status (+ = intact/− = co-deleted), and IDH1 mutation status (MUT/WT) were included in this study. In all patients, median relative cerebral blood volume (rCBV) and apparent diffusion coefficient (ADC) were estimated within T2 hyperintense lesions. Bootstrap hypothesis testing was used to compare subpopulations of gliomas, separated by WHO grade and 2007 or 2016 glioma classification schemes. A multivariable logistic regression model was also used to differentiate between 1p19q+ and 1p19q− WHO II–III gliomas. Neither rCBV nor ADC differed significantly between histological subtypes of pure astrocytomas and pure oligodendrogliomas. ADC was significantly different between molecular subtypes (p = 0.0016), particularly between IDHWT and IDHMUT/1p19q+ (p = 0.0013). IDHMUT/1p19q+ grade III gliomas had higher median ADC; IDHWT grade III gliomas had higher rCBV with lower ADC; and IDHMUT/1p19q− had intermediate rCBV and ADC values, similar to their grade II counterparts. A multivariable logistic regression model was able to differentiate between IDHWT and IDHMUT WHO II and III gliomas with an AUC of 0.84 (p < 0.0001, 74% sensitivity, 79% specificity). Within IDHMUT WHO II–III gliomas, a separate multivariable logistic regression model was able to differentiate between 1p19q+ and 1p19q− WHO II–III gliomas with an AUC of 0.80 (p = 0.0015, 64% sensitivity, 82% specificity). ADC better differentiated between genetic subtypes of gliomas according to the 2016 WHO guidelines compared to the classification scheme outlined in the 2007 WHO guidelines based on histological features of the tissue. Results suggest a combination of rCBV, ADC, T2 hyperintense volume, and presence of contrast enhancement together may aid in non-invasively identifying genetic subtypes of diffuse gliomas.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Diffuse gliomas are a heterogeneous group of primary brain tumors with high morbidity and variable outcomes. High-grade, malignant gliomas represent a majority of primary malignant brain tumors (70–80%) [1,2,3] and have dismal prognoses of only 12–15 months for glioblastoma (WHO grade IV) and 2–5 years for anaplastic gliomas (WHO grade III) [4]. Survival, growth characteristics, and therapeutic sensitivity for lower grade diffuse gliomas (WHO II–III) are highly dependent on the dominant cell lineage represented within the tumor. In 2007, the World Health Organization (WHO) glioma classification scheme used histological appearance on light microscopy to distinguish gliomas of different glial lineages. Astrocytomas make up a majority of gliomas [4] and WHO grade III anaplastic astrocytomas have a median survival of around 2–3 years [1]. Astrocytic tumors are comprised of irregular, hyperchromatic nuclei with a glial fibrillary acidic protein (GFAP)—positive cytoplasm. On the other hand, oligodendrogliomas are characterized by round nuclei with a branching network of capillaries with possible calcification [5]. They are more sensitive to chemoradiation, and overall, WHO grade III anaplastic oligodendrogliomas have a slightly better prognosis compared with anaplastic astrocytomas [6].
Although classification of diffuse gliomas based solely on histological features of the resected tissue may be beneficial for staging disease and developing more targeted therapeutics, this strategy can be prone to sampling error during tumor resection and bias based on relatively subjective criteria. This ambiguity can often lead to a range of challenging diagnoses including mixed features of both astrocytomas and oligodendrogliomas (i.e. oligoastrocytomas). As more molecular and genetic information about these types of tumors have become mature in the literature it has became clear that sub-stratification of tumor types should likely be performed using genetic tests for common deletions and mutations. In 2016, the WHO glioma classification scheme was restructured to reflect two common molecular alterations in gliomas: 1p/19q co-deletion (1p19q−) and isocitrate dehydrogenase-1 mutation (IDHMUT) [7]. Co-deletion of 1p and 19q is most commonly associated with oligodendroglial tumors [8, 9] and is both predictive of therapeutic response and prognostic for survival [10, 11]. IDH mutations, which are known to be a driver mutation in low-grade gliomas [12, 13], are associated with more favorable outcomes as they are known to be more sensitive to chemoradiation [14, 15].
Diffusion- and perfusion-weighted magnetic resonance imaging (MRI), have been extensively studied as non-invasive tools for identifying glioma subtypes, characterizing the aggressiveness of gliomas, and identifying early malignant transformation. Diffusion-weighted imaging (DWI) is a physiologic imaging modality that exploits the diffusion of water molecules to create contrast between tissues. One common measurement obtained from DWI is the apparent diffusion coefficient (ADC). ADC is an estimate of the magnitude of the diffusion of water molecules within the tissue, and there is a strong negative correlation between the ADC and tumor cellularity in gliomas [16, 17]. Perfusion-weighted imaging (PWI) is an MRI modality that gives insights into the delivery of blood to tissues by monitoring a bolus of contrast agent as it passes through the blood vasculature. A common biomarker derived from PWI is the relative cerebral blood volume (rCBV). Measurements of rCBV have been shown to be higher in high-grade tumors than in low-grade tumors and may correlate with glioma vascularity [18, 19].
Several studies have illustrated differences in diffusion and perfusion MR measurements between oligodendrogliomas from astrocytomas [20, 21], 1p/19q co-deleted tumors (1p19q−) from non-1p/19q co-deleted tumors (1p19q+) [22,23,24,25], and IDH mutant (IDHMUT) from IDH wild-type (IDHWT) diffuse gliomas [26]. Presumably, these observed differences in diffusion MR measurements of apparent diffusion coefficient (ADC) and perfusion MR imaging measurements of relative cerebral blood volume (rCBV) between subtypes reflects known differences in tumor cell morphometry and aspects of vascular biology [27]. Despite these interesting observations, there is a critical gap in our current understanding of how diffusion and perfusion MRI might be used together to better understand differences between the histologic and genetic subtypes of tumors and between WHO II and III tumors of the same subtype. We hypothesize a combination of diffusion and perfusion MR measurements will better separate tumors based on their genetic characteristics (1p19q co-deletion and IDH1 mutation status) [7] than the more subjective histologic criteria. Furthermore, we hypothesize low grade gliomas (WHO II) will have similar diffusion and perfusion MR characteristics across subtypes, whereas anaplastic gliomas (WHO III) will demonstrate significantly different patterns of diffusion and perfusion MR characteristics depending on the specific histologic or genetic subtype. Finally, we hypothesize that using diffusion, perfusion, T2-enhancement volume, and contrast enhancement together will allow us to classify IDHWT from IDHMUT as well as IDHMUT/1p19q+ from IDHMUT/1p19q− tumors.
Materials and methods
Patients
All adult patients with WHO grade II or grade III gliomas between 2010 and 2016 were retrospectively reviewed (231 total). Patients were only included if they met all of the following inclusion criteria: (1) histologic diagnosis of WHO grade II or grade III gliomas [1]; (2) dynamic susceptibility contrast (DSC) perfusion-weighted MRI, diffusion-weighted MRI, T2-weighted, and post-contrast T1-weighted anatomical scan performed at initial diagnosis and prior to any surgery; (3) diagnosis of astrocytoma, mixed glioma, or oligodendroglioma via histology; and (4) known IDH1 mutation and 1p/19q co-deletion status. Patients with the rare combination of 1p/19q co-deletion positive and IDH1 wild-type were excluded from statistical analysis due to their small sample size (Grade II: n = 1; Grade III: n = 1). A total of 65 patients (38 men, 27 women; average age 46.5 ± 15.8 years; age range 21–85), with 31 WHO II and 34 WHO III gliomas, fit the inclusion criteria. All patients in this study signed institutional review board-approved informed consent to have their data stored in our neuro-oncology database and used for research purposes. The images and histology were taken from this database without new review.
MR imaging and post-processing
All patients included in this study had a T2-weighted FLAIR, a diffusion-weighted MR, and a perfusion-weighted DSC-MR that was performed on either a 1.5 or 3 T scanner prior to surgery, with the average length between MRI and surgery at 10.6 ± 11.8 days, range 0–67 days.
Diffusion MR acquisition parameters echo times (TE) varied from 67 to 100 ms, repetition times (TR) ranged from 7 to 10 s, flip angle was 90°, b-values used were 0 s/mm2 and either 700 or 1000 s/mm2 with matrix size 128 × 128. In some cases, diffusion-weighted images were diffusion tensor images with 6–12 directions. Apparent diffusion coefficients (ADC) were calculated using a log-linear fit to all available b-values in the diffusion-weighted/diffusion-tensor images using custom in-house code in MATLAB (Natick, MA).
A 0.025 mmol/kg preload dose of gadolinium contrast agent was administered prior to DSC-MRI with a 0.075 mmol/kg preload dose of gadolinium contrast agent used for DSC acquisition. For DSC-MRI, echo times (TE) ranged from 23 to 35 ms, repetition times (TR) ranged from 1250 to 2000, flip angles were 35, 60, or 90, with 40 to 120 temporal time points at a slice thickness of 4–6 mm with an interslice gap of 0–1 mm. A total of 12–25 slices were collected with matrix size ranging from 80 × 96 to 128 × 128. Relative cerebral blood volume (rCBV) was calculated using a recently introduced leakage correction algorithm using in-house custom MATLAB code that corrects for bidirectional contrast agent exchange [28, 29] and normalizing to contra-lateral normal-appearing white matter (NAWM) tissue.
All images were registered to the T2-weighted image using a mutual information algorithm and a 12-degree freedom transformation using FSL (FMRIB; http://www.fmrib.ox.ac.uk/fsl/) or tkregister2 (Freesurfer, surfer.nmr.mgh.harvard.edu; Massachusetts General Hospital, Harvard Medical School).
Regions of interest
Regions of interest (ROIs) of suspected tumor and/or edema were defined by abnormal hyperintensity on T2-weighted or T2-weighted FLAIR using semi-automated segmentation techniques, followed by manual inspection and adjustment of the resulting contour as described previously [30]. The median ADC and median normalized rCBV were obtained from each tumor lesion.
Histologic characterization (2007 WHO glioma guidelines)
All tissues obtained from surgical resections were stained with hematoxylin–eosin. Tissues were classified and graded according to the 2007 WHO criteria [1] with standard hematoxylin–eosin staining. Gliomas were classified according to grade (II or III) and one of the following three categories: astrocytoma, mixed glioma, and oligodendroglioma (Fig. 1), though mixed gliomas were removed from the statistical tests because of their vague label. (Grade II: 15 astrocytomas, 10 mixed gliomas, 6 oligodendrogliomas; Grade III: 14 astrocytomas, 4 mixed gliomas, 16 oligodendrogliomas).
Diagram illustrating the 2007 and 2016 WHO classification criteria for gliomas. (Top) Categories of grade II and III gliomas under the 2007 WHO criteria based on histological features using light microscopy and hematoxylin and eosin staining. (Bottom) Categories of grade II and III gliomas under the 2016 WHO criteria based on molecular genotype using IDH1 mutation status and 1p/19q co-deletion
IDH1 mutation status and 1p/19q co-deletion (2016 WHO glioma guidelines)
IDH1 mutation status was determined by sequencing for codon 132 in the catalytic domain of IDH1 via standard genomic sequencing practices (Sanger sequencing method), as previously described [31]. In brief, tumor DNA was isolated from the frozen or formalin-fixed tissue using DNeasy Blood and Tissue Kit (Qiagen). A 236-bp fragment that included codon 132 was amplified using the primers 5′-GCGTCAAATGTGCCACTATC-3′ and 5′-GCAAAATCACATTATTGCCAAC-3′ to generate a 236 bp fragment. PCR products were sequenced by BigDye Terminator v1.1 (Applied Biosystems), and sequences were determined via a 3730 sequencer (Applied Biosystems).
1p/19q co-deletion status was determined by fluorescence in situ hybridization specific probes for the 1p36 and 19q13 loci. A deletion of >50% of the nuclei examined for both 1p and 19q constituted a co-deletion.
The gliomas were separated according to grade and one of the following categories (Fig. 1): IDH1 wild-type (IDHWT); IDH1 mutant with intact 1p or 19q (IDHMUT/1p19q+); or IDH1 mutant with 1p/19q co-deleted (IDHMUT/1p19q−). (Grade II: 8 IDHWT, 16 IDHMUT/1p19q+, 7 IDHMUT/1p19q−; Grade III: 14 IDHWT, 12 IDHMUT/1p19q+, 8 IDHMUT/1p19q−).
Statistical analyses
Due to the small sample size, bootstrap hypothesis testing was used for all statistical tests. First, using the real data, the f-score was calculated from the one-way ANOVA test if three groups were being compared or a t-score if only two groups were being compared to each other. In order to generate the null hypothesis distribution, the mean of the data for each group was subtracted from their respective groups because the null hypothesis was that the means of all groups are equal. Next, the mean-shifted data was randomly resampled with replacement such that the newly resampled data had the same number of data points as the real data for each group. The f-score or t-test was then calculated from the resampled, mean-shifted data 10,000 times to generate the null hypothesis distribution. The p value was then computed by counting the number of f-scores greater than the f-score generated from the real data or the number of t-scores that were more extreme than the t-score generated from the real data.
For analyses pertaining to both the 2007 and the 2016 WHO glioma classification, all three categories were used in the statistical comparison. All bootstrap routines were performed in MATLAB using in-house custom code, while the receiver operating characteristic statistics, i.e. area under the curve, were performed in GraphPad Prism (La Jolla, CA) using the sensitivity and specificity from the highest likelihood ratio.
A logistic regression was performed to classify IDHMUT and IDHWT gliomas (both grade II and grade III tumors) using median rCBV, median ADC, presence or absence of contrast enhancement, and volume of T2-enhancement using in-house custom MATLAB code. Sensitivity and specificity were picked based on the point on the ROC where the product was maximized. To generate the p values for the presence or absence of contrast enhancement, a Fisher’s exact test was employed. Since eight statistical tests comparing the glioma subtypes were performed (four for the 2007 analysis, four for the 2016 analysis, two tumor volume analyses, two presence of contrast enhancement analyses, and two receiver operating characteristic curve analyses), for multiple comparisons correction, a p value of 0.015/14 = 0.0036 was considered to be significant.
The elliptical error bounds in Figs. 3 and 4 were calculated using custom code in MATLAB. The major and minor axes were determined by the eigenvectors, and the lengths of the axes represent one standard deviation in those directions. Ellipses with rotated axes were chosen over horizontal/vertical axes to better represent the trends that are observed in the different glioma subtype populations.
Results
Using the 2007 WHO glioma classification scheme, we observed a substantial overlap in perfusion and diffusion MR measurements between WHO grade and histological subtype (Fig. 2). No significant differences in rCBV or ADC were observed between pure astrocytomas and pure oligodendrogliomas within WHO grade II or III tumors. (No p values were less than 0.05/14 = 0.0036). A joint plot of median rCBV versus median ADC (Fig. 2a, d) for the different tumor subtypes revealed an increase in rCBV with increasing tumor grade, but further highlighted the noteworthy overlap in imaging measurements between histologically classified astrocytomas and oligodendrogliomas.
Comparisons of rCBV and ADC between histologic subtypes of WHO grade II and III gliomas using the 2007 WHO classification. a Scatter plot of median rCBV and ADC for populations of grade II glioma subtypes with elliptical error bars. b Comparison of rCBV between grade II gliomas. c Comparison of ADC between grade II gliomas. d Scatter plot of median rCBV and ADC for populations of grade III glioma subtypes with elliptical error bars. e Comparison of rCBV between grade III gliomas. f Comparison of ADC between grade III gliomas
Perfusion and diffusion was also not found to be statistically significant in grade II gliomas with respect to genotypes (Fig. 3). WHO grade II gliomas has substantial overlap between joint rCBV and ADC measurement (Fig. 3a). We observed no statistically significant differences in rCBV (Fig. 3b, P = 0.0875) or ADC between subtypes (Fig. 3c, P = 0.1217).
Comparisons of rCBV and ADC between genetic subtypes of WHO grade II and III gliomas using the 2016 WHO classification. a Scatter plot of median rCBV and ADC for populations of grade II glioma subtypes with elliptical error bars. b Comparison of rCBV between grade II gliomas. c Comparison of ADC between grade II gliomas. d Scatter plot of median rCBV and ADC for populations of grade III glioma subtypes with elliptical error bars. e Comparison of rCBV between grade III gliomas. f Comparison of ADC between grade III gliomas
In contrast to WHO II gliomas, joint rCBV and ADC characteristics of WHO grade III gliomas revealed distinct clustering of genetic subtypes (Fig. 3d). In particular, IDHMUT gliomas appeared to have relatively small variance and formed well-defined clusters exhibiting higher ADC and moderate rCBV compared with IDHWT anaplastic gliomas. Looking at perfusion and diffusion measurements independently exposed a trend toward significantly lower ADC (Fig. 3f, p = 0.0016) particularly between IDHWT and IDHMUT/1p19q+ tumors (p = 0.0013).
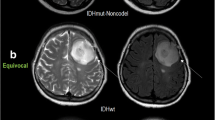
In Fig. 4, we illustrate specific examples of rCBV and ADC maps extracted from three representative patients in WHO grade III gliomas with different histological or genetic subtypes. Using the 2007 WHO glioma grading criteria, the two astrocytoma examples have highly variable ADC and rCBV compared to the oligodendroglioma patient. On the other hand, the addition of the IDH1 mutation status differentiates the two astrocytomas, with the IDH wild-type (IDHWT) tumors having high rCBV and low ADC (rCBV = 1.21, ADC = 0.9 μm2/ms), and IDH mutant tumors with intact 1p or 19q (IDHMUT/1p19q+) having lower rCBV and much higher ADC (rCBV = 0.60, ADC = 1.7 μm2/ms). Additionally, we observed an intermediate ADC and rCBV in tumors with IDHMUT/1p19q− (rCBV = 0.87, ADC = 1.2 μm2/ms).
Imaging features extracted from three representative patients with different histologic and genetic subtypes. Post-contrast T1-weighted anatomical, T2-weighted FLAIR anatomical, rCBV, and ADC maps are shown. The scatter plot shown on the right illustrates voxel-wise plots of rCBV versus ADC within the red region of interest outlining the T2 hyperintense lesion. Top row A patient with an AA subsequently characterized as IDHWT exhibited relatively high rCBV and low ADC. Middle row an AA patient subsequently characterized as IDHMUT/1p19q+ exhibiting a low rCBV and high ADC. Bottom row an AO oligodendroglioma characterized as IDHMUT/1p19q− exhibiting intermediary rCBV and ADC characteristics. (AA anaplastic astrocytoma, AO anaplastic oligodendroglioma)
Formulation of a combined, multivariate logistic model that included median rCBV, median ADC, T2 hyperintense lesion volume, and presence or absence of contrast enhancement allowed for further improvement in differentiation of tumor genotypes, when pooled across tumor grade (Fig. 5). After multiple comparisons correction, ADC (Fig. 5b, p = 0.0030) alone was significantly different between IDHMUT and IDHWT, while presence or absence of contrast enhancement trended toward significance (Fig. 5d, p = 0.0060). The combined multivariate model including all four parameters significantly improved the ability to differentiate IDHMUT and IDHWT WHO II–III tumors with a high sensitivity and specificity (Fig. 5e; ROC Analysis, p < 0.0001, AUC = 0.84 ± 0.05; 74% sensitivity, 79% specificity). We then evaluated performance of a similar model to further differentiate 1p19q+ from 1p19q− gliomas within the IDHMUT subtype. Univariate results suggested ADC was significant between these subtypes (Fig. 5g, p = 0.0018). The composite logistic regression model further improved subclassification (Fig. 5j, p = 0.0015; AUC = 0.80 ± 0.07; 64% sensitivity, 82% specificity). The coefficients for these models are outlined Table 1.
Classification of glioma genetic subtypes in combined WHO II–III tumors using rCBV, ADC, T2 hyperintense lesion volume, and presence of contrast enhancement. a Comparison of rCBV between IDHWT and IDHMUT WHO II–III gliomas. b Comparison of ADC between IDHWT and IDHMUT WHO II–III gliomas. c Comparison of T2 hyperintense tumor volume between IDHWT and IDHMUT WHO II–III gliomas. d Proportion of contrast enhancing (CE) and non-enhancing (NE) tumors between IDHWT and IDHMUT WHO II–III gliomas. e ROC curve using all four biomarkers (rCBV, ADC, Volume, CE/NE) to differentiate IDHWT and IDHMUT WHO II–III gliomas. f Comparison of rCBV between IDHMUT/1p19q+ and IDHMUT/1p19q− WHO II–III gliomas. g Comparison of ADC between IDHMUT/1p19q+ and IDHMUT/1p19q− WHO II–III gliomas. h Comparison of T2 hyperintense tumor volume between IDHMUT/1p19q+ and IDHMUT/1p19q− WHO II–III gliomas. i Proportion of contrast enhancing (CE) and non-enhancing (NE) tumors between IDHMUT/1p19q+ and IDHMUT/1p19q− WHO II–III gliomas. j ROC curve using all four biomarkers (rCBV, ADC, Volume, CE/NE) to differentiate IDHMUT/1p19q+ and IDHMUT/1p19q− WHO II–III gliomas
Lastly, we used the same model approach to differentiate between IDHMUT and IDHWT gliomas within each respective tumor grade (Fig. 6). Univariate results did not demonstrate an association between any of the MR biomarkers and IDH status within WHO II gliomas (Fig. 6a–d). However, the multivariable logistic regression model was able to differentiate between WHO II IDHMUT and IDHWT with reasonable sensitivity and specificity (Fig. 6e, P = 0.0470, AUC = 0.74; 75% sensitivity and 74% specificity). Within WHO III tumors, IDHMUT gliomas had significantly higher ADC (Fig. 6H, P = 0.0166) and a significantly higher proportion of tumors without enhancing disease (Fig. 6I, p = 0.0035) compared with IDHWT WHO III gliomas. A multivariable logistic regression model created by combining all four MR biomarkers was able to stratify WHO III gliomas based on their IDH status with high sensitivity and specificity (Fig. 6j, p < 0.0001, AUC = 0.91; 94% sensitivity and 74% specificity).
Classification of IDH status in WHO II and III gliomas using rCBV, ADC, T2 hyperintense lesion volume, and presence of contrast enhancement. a Comparison of ADC between IDHWT and IDHMUT grade II gliomas. b Comparison of rCBV between grade II gliomas. c Comparison of tumor volume between grade II gliomas. d Presence and absence of contrast enhancement among grade II gliomas. e ROC curve analysis using all four biomarkers in grade II tumors. f Comparison of ADC between IDHWT and IDHMUT grade III gliomas. b Comparison of rCBV between grade III gliomas. c Comparison of tumor volume between grade III gliomas. d Presence and absence of contrast enhancement among grade III gliomas. e ROC curve analysis using all four biomarkers in grade III tumors. The open circles within the IDHMUT groups indicate 1p/19q co-deletion
Discussion
In the current study, we explored whether diffusion and perfusion MR signatures could identify histologic or genetic subtypes of WHO II or III gliomas that form the basis for the 2007 and 2016 WHO glioma classification schemes, respectively. Results suggest no significant differences in median rCBV or ADC in T2 hyperintense regions between histologic subtypes of tumors (2007) within the same WHO grade and much overlap between the primary subtypes with respect to joint rCBV and ADC characteristics. On the other hand, the joint rCBV and ADC characteristics have much smaller variation in both IDH1 mutation subtypes. ADC was found to be highly discriminative among the three different genetic subtypes, particularly between the IDHWT and IDHMUT/1p19q+, the two groups that would have originally been classified as being astrocytomas. rCBV was not statistically significant in grade III tumors, in part due to the high variability in grade III tumors, i.e. some have characteristics that more closely resemble grade II, while others may have characteristics more similar to grade IV. Nevertheless, the data does demonstrate that the highest rCBV values belong to the IDHWT group.
However, using rCBV and ADC along with the more traditional biomarkers of contrast enhancement and volume of T2-enhancement allows us to better identify the IDH1 mutation as well as 1p19q co-deletion within IDHMUT tumors. Since survival is similar between grade II and grade III IDHMUT tumors, but may differ between IDHWT tumors, we chose only to look at grade II and grade III comparing IDH mutation [32]. With respect to these four parameters, grade II gliomas from the different genotypes look somewhat similar, whereas there is much more spread in grade III gliomas. Interestingly, tumor volume was lower in IDHWT tumors of the same grade. Furthermore, volumes were similar between grade II and grade III IDHWT tumors, whereas the volumes were higher in grade III IDHMUT tumors. This is likely because grade IV tumors, which have much higher volumes, were excluded from this study. However, it does indicate that perfusion and diffusion changes likely precede volume changes in IDHWT tumors, underscoring the importance of using diffusion and perfusion to monitor gliomas.
Our results are not consistent with previous studies reporting a higher rCBV in oligodendrogliomas [20, 21]. These inconsistencies, however, may be explained through differences in methodology and interpretation. For example, Cha et al. [20] reported a four-fold higher maximum rCBV within WHO II oligodendrogliomas compared with astrocytomas; however, authors noted more cortical involvement in oligodendrogliomas in their study sample (N = 25; 11 astrocytomas and 14 oligodendrogliomas), which has an inherently higher CBV. Additionally, authors used maximum CBV instead of median CBV and they did not perform post-hoc contrast agent leakage correction, which can lead to over or underestimation of true CBV. Since the percentage signal recovery (PSR) after bolus injection, a measure related to contrast extravasation, is known to be altered in different types of tumors [33], lack of correction for contrast agent extravasation alone may have altered their results. Similarly, Lev et al. [21] observed a higher maximum rCBV in oligodendrogliomas compared with astrocytomas in a similar sized cohort of patients (N = 30; 8 oligodendrogliomas; 4 WHO II and 4 WHO III) even after leakage correction; however, the relatively low number of patients and use of maximum rCBV measurements likely biased their results toward more cortically-based lesions or tumors near vascular structures. Lastly, our current results demonstrated no difference in median ADC between histologic subtypes, which corroborate the findings by Fellah et al. [22], but are inconsistent with the findings from Bian et al. [34], who found a lower ADC in oligodendrogliomas compared with astrocytomas. Some of the differences may have stemmed from our smaller sample size in grade II gliomas and the differences in criteria used for distinguishing oligoastrocytomas from oligodendrogliomas and astrocytomas. In particular, the latter may help explain the inconsistency in the literature in being able to distinguish the histologic subtypes of oligodendrogliomas, oligoastrocytomas, and astrocytomas from each other based on diffusion and perfusion characteristics.
Consistent with previous studies, we observed significant differences in diffusion MR measurements between 1p/19q co-deleted tumors from non-1p/19q co-deleted tumors [22,23,24,25] and IDH1 mutant from IDH1 wild-type diffuse gliomas [26]. Our findings did not appear to corroborate those of Tan et al. [26], who reported a significantly higher rCBV in IDHWT compared with IDHMUT gliomas, though differences may stem from our smaller sample size as well as our more conservative statistical methods. However, we identified a significantly higher ADC in IDHMUT gliomas compared with IDHWT tumors and a significantly higher ADC in IDHMUT/1p19q+ compared with IDHWT tumors. In total, results suggest that diffusion MR signatures may better differentiate tumors based on their genetic characteristics compared with histological features.
There have been a few studies that have tried to identify IDH1 mutations using MR spectroscopy. The short-echo MRS is widely available on clinical scanners, though the false-positive rate is approximately 22% [35]. There have been other, more complex MRS techniques that have been more successful, such as Choi et al. [36], who identified all patients with IDH mutations without false-positive results. This compares to a sensitivity and specificity of 75 and 70% (AUC = 0.84) using median ADC alone. However, diffusion-weighted imaging is more commonly acquired than spectroscopy, and the smaller voxel-wise resolution certainly can lend itself to more complex histogram analyses.
Currently, the treatment of low-grade gliomas is controversial. One strategy that is often employed is the “watch-and-wait” approach. The biopsy is not necessarily required initially, but neuro-radiological follow-up is certainly warranted. Although not specifically tested, the current data in both WHO II and III gliomas may suggest tumors with different genetic subtypes evolve with either changes in rCBV or ADC, depending on the genotype. For example, Fig. 3a demonstrates significant overlap between genetic subtypes in WHO II gliomas, whereas Fig. 3d suggests IDHWT tumors may shift to a higher median rCBV and lower ADC, IDHMUT/1p19q+ tumors may increase in median rCBV and increase in median ADC, while IDHMUT/1p19q− tumors may not change rCBV or ADC appreciably between grades. Future studies aimed at testing these specific hypotheses through serial imaging in tumors are warranted to further test how shifts in rCBV and ADC may reflect increasing tumor malignancy within a certain genotype at a time in which biopsy has not yet been performed. Future validation of these shifts could underscore the importance of using perfusion- and diffusion-weighted imaging as the standard of care for low-grade gliomas, rather than simply the volume of T1- or T2-enhancement.
This study has a few limitations. In total, the sample size is small, and a larger validation cohort is needed to determine if diffusion and perfusion could help with monitoring gliomas over time when the “wait-and-watch” approach is used. We also employed simple metrics, the median rCBV and ADC, to determine if perfusion and diffusion may be useful given the new glioma classification guidelines. However, in a larger dataset, much more complex 2D histogram analyses can be performed to boost classification schemes that may try to focus on the mutational status of tumors, rather than the histological phenotype.
Conclusion
In summary, the current study suggests that ADC better correlates with genetic subtypes of gliomas according to the 2016 WHO guidelines and imaging measurements were varied less when tumors were stratified based on histological features using the 2007 criteria. Furthermore, using ADC in combination with rCBV, T2 volume enhancement, and contrast enhancement allowed us to distinguish between IDHWT and IDHMUT gliomas as well as between IDHMUT/1p19q+ and IDHMUT/1p19q− gliomas.
References
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114:97–109
Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS (2015) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol 17(Suppl 4):iv1–iv62
Sathornsumetee S, Rich JN, Reardon DA (2007) Diagnosis and treatment of high-grade astrocytoma. Neurol Clin 25:1111–1139
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359:492–507
Sharma S, Deb P (2011) Intraoperative neurocytology of primary central nervous system neoplasia: a simplified and practical diagnostic approach. J Cytol 28:147–158
Omuro A, DeAngelis LM (2013) Glioblastoma and other malignant gliomas: a clinical review. JAMA 310:1842–1850
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820
van den Bent MJ, Looijenga LH, Langenberg K, Dinjens W, Graveland W, Uytdewilligen L, Sillevis Smitt PA, Jenkins RB, Kros JM (2003) Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer 97:1276–1284
Felsberg J, Erkwoh A, Sabel MC, Kirsch L, Fimmers R, Blaschke B, Schlegel U, Schramm J, Wiestler OD, Reifenberger G (2004) Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol 14:121–130
Smith JS, Perry A, Borell TJ, Lee HK, O’Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB (2000) Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol 18:636–645
Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, Flynn H, Passe S, Felten S, Brown PD, Shaw EG, Buckner JC (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66:9852–9861
Watanabe T, Nobusawa S, Kleihues P, Ohgaki H (2009) IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 174:1149–1153
Ichimura K, Pearson DM, Kocialkowski S, Backlund LM, Chan R, Jones DT, Collins VP (2009) IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol 11:341–347
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773
Nobusawa S, Watanabe T, Kleihues P, Ohgaki H (2009) IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res 15:6002–6007
Ellingson BM, Malkin MG, Rand SD, Connelly JM, Quinsey C, LaViolette PS, Bedekar DP, Schmainda KM (2010) Validation of functional diffusion maps (fDMs) as a biomarker for human glioma cellularity. J Magn Reson Imaging 31:538–548
Chen L, Liu M, Bao J, Xia Y, Zhang J, Zhang L, Huang X, Wang J (2013) The correlation between apparent diffusion coefficient and tumor cellularity in patients: a meta-analysis. PLoS ONE 8:e79008
Boxerman JL, Schmainda KM, Weisskoff RM (2006) Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol 27:859–867
Aronen HJ, Gazit IE, Louis DN, Buchbinder BR, Pardo FS, Weisskoff RM, Harsh GR, Cosgrove GR, Halpern EF, Hochberg FH et al (1994) Cerebral blood volume maps of gliomas: comparison with tumor grade and histologic findings. Radiology 191:41–51
Cha S, Tihan T, Crawford F, Fischbein NJ, Chang S, Bollen A, Nelson SJ, Prados M, Berger MS, Dillon WP (2005) Differentiation of low-grade oligodendrogliomas from low-grade astrocytomas by using quantitative blood-volume measurements derived from dynamic susceptibility contrast-enhanced MR imaging. AJNR Am J Neuroradiol 26:266–273
Lev MH, Ozsunar Y, Henson JW, Rasheed AA, Barest GD, Harsh GRt, Fitzek MM, Chiocca EA, Rabinov JD, Csavoy AN, Rosen BR, Hochberg FH, Schaefer PW, Gonzalez RG (2004) Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas. AJNR Am J Neuroradiol 25:214–221
Fellah S, Caudal D, De Paula AM, Dory-Lautrec P, Figarella-Branger D, Chinot O, Metellus P, Cozzone PJ, Confort-Gouny S, Ghattas B, Callot V, Girard N (2013) Multimodal MR imaging (diffusion, perfusion, and spectroscopy): is it possible to distinguish oligodendroglial tumor grade and 1p/19q codeletion in the pretherapeutic diagnosis? AJNR Am J Neuroradiol 34:1326–1333
Jenkinson MD, Smith TS, Brodbelt AR, Joyce KA, Warnke PC, Walker C (2007) Apparent diffusion coefficients in oligodendroglial tumors characterized by genotype. J Magn Reson Imaging 26:1405–1412
Cui Y, Ma L, Chen X, Zhang Z, Jiang H, Lin S (2014) Lower apparent diffusion coefficients indicate distinct prognosis in low-grade and high-grade glioma. J Neurooncol 119:377–385
Chawla S, Krejza J, Vossough A, Zhang Y, Kapoor GS, Wang S, O’Rourke DM, Melhem ER, Poptani H (2013) Differentiation between oligodendroglioma genotypes using dynamic susceptibility contrast perfusion-weighted imaging and proton MR spectroscopy. AJNR Am J Neuroradiol 34:1542–1549
Tan W, Xiong J, Huang W, Wu J, Zhan S, Geng D (2016) Noninvasively detecting Isocitrate dehydrogenase 1 gene status in astrocytoma by dynamic susceptibility contrast MRI. J Magn Reson Imaging 45:492–499
Schiffer D, Bosone I, Dutto A, Di Vito N, Chio A (1999) The prognostic role of vessel productive changes and vessel density in oligodendroglioma. J Neurooncol 44:99–107
Leu K, Boxerman JL, Cloughesy TF, Lai A, Nghiemphu PL, Liau LM, Pope WB, Ellingson BM (2016) Improved leakage correction for single-echo dynamic susceptibility contrast perfusion MRI estimates of relative cerebral blood volume in high-grade gliomas by accounting for bidirectional contrast agent exchange. AJNR Am J Neuroradiol 37:1440–1446
Leu K, Boxerman JL, Lai A, Nghiemphu PL, Pope WB, Cloughesy TF, Ellingson BM (2016) Bidirectional Contrast agent leakage correction of dynamic susceptibility contrast (DSC)-MRI improves cerebral blood volume estimation and survival prediction in recurrent glioblastoma treated with bevacizumab. J Magn Reson Imaging 44:1229–1237
Ellingson BM, Cloughesy TF, Lai A, Nghiemphu PL, Mischel PS, Pope WB (2011) Quantitative volumetric analysis of conventional MRI response in recurrent glioblastoma treated with bevacizumab. Neuro Oncol 13:401–409
Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, Forrest WF, Pujara K, Carrillo JA, Pandita A, Ellingson BM, Bowers CW, Soriano RH, Schmidt NO, Mohan S, Yong WH, Seshagiri S, Modrusan Z, Jiang Z, Aldape KD, Mischel PS, Liau LM, Escovedo CJ, Chen W, Nghiemphu PL, James CD, Prados MD, Westphal M, Lamszus K, Cloughesy T, Phillips HS (2011) Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol 29:4482–4490
Olar A, Wani KM, Alfaro-Munoz KD, Heathcock LE, van Thuijl HF, Gilbert MR, Armstrong TS, Sulman EP, Cahill DP, Vera-Bolanos E, Yuan Y, Reijneveld JC, Ylstra B, Wesseling P, Aldape KD (2015) IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II–III diffuse gliomas. Acta Neuropathol 129:585–596
Aprile I, Giovannelli G, Fiaschini P, Muti M, Kouleridou A, Caputo N (2015) High- and low-grade glioma differentiation: the role of percentage signal recovery evaluation in MR dynamic susceptibility contrast imaging. Radiol Med 120:967–974
Bian W, Khayal IS, Lupo JM, McGue C, Vandenberg S, Lamborn KR, Chang SM, Cha S, Nelson SJ (2009) Multiparametric characterization of grade 2 glioma subtypes using magnetic resonance spectroscopic, perfusion, and diffusion imaging. Transl Oncol 2:271–280
Pope WB, Lai A, Mehta R, Kim HJ, Qiao J, Young JR, Xue X, Goldin J, Brown MS, Nghiemphu PL, Tran A, Cloughesy TF (2011) Apparent diffusion coefficient histogram analysis stratifies progression-free survival in newly diagnosed bevacizumab-treated glioblastoma. AJNR Am J Neuroradiol 32:882–889
Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, Pascual JM, Madden CJ, Mickey BE, Malloy CR, Bachoo RM, Maher EA (2012) 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 18:624–629
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Leu, K., Ott, G.A., Lai, A. et al. Perfusion and diffusion MRI signatures in histologic and genetic subtypes of WHO grade II–III diffuse gliomas. J Neurooncol 134, 177–188 (2017). https://doi.org/10.1007/s11060-017-2506-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-017-2506-9