Abstract
Glioblastoma (GBM) is among the most aggressive primary brain tumors, with a median survival rate of 12–15 months. MicroRNAs have been implicated in GBM development as oncogenes or tumor suppressors. In this study, we demonstrated that miR-519a expression was frequently downregulated in GBM specimens and cell lines, and that low-levels miR-519a expression significantly correlated with poor outcomes associated with GBM. Analysis of The Cancer Genome Atlas also demonstrated that low miR-519a expression can predict poor clinical outcomes in classical and proneural GBM subtypes. Functionally, re-expression of miR-519a effectively reduced GBM cell proliferation, migration, and invasion. Mechanistically, we confirmed that the signal transducer and activator of transcription 3 (STAT3) 3′-UTR was a putative target of miR-519a, and that re-expression of STAT3 abrogated miR-519a function in GBM cells. Furthermore, we found that STAT3 expression negatively correlated with that of miR-519a in human GBM tissues. These results elucidated the prognostic value and tumor-suppressor role of miR-519a in GBM and further suggested it as a potential therapeutic target for GBM treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma (GBM) is among the most aggressive and lethal brain tumors, with a median survival time of ~ 14 months post diagnosis [1]. Even when treated with a maximal surgical resection and concurrent radiation therapy and temozolomide (TMZ), followed by an adjuvant course of TMZ, prognosis for the GBM patient remains dismal [2]. Furthermore, the highly proliferative and invasive behavior of GBM tumors aid in the poor prognosis [3].
Signal transducer and activator of transcription 3 (STAT3) functions as a signal messenger and transcription factor regulating the expression of downstream targets in malignant transformation and tumor development. STAT3 overexpression in GBM can promote tumor progression [4–6], and specific inhibition of the STAT3 signaling pathway can suppress human GBM proliferation [7] or migration [8], sensitize GBM to TMZ treatment [9], or inhibit radiation-induced proneural-to-mesenchymal transition [10]. Therefore, an important aim in GBM research is elucidation of the mechanisms associated with STAT3 activation in tumors.
MicroRNAs (miRs or miRNAs) are small noncoding RNA molecules comprising 20–22 nucleotides that negatively regulate gene expression by binding to the 3′-untranslated region (3′UTR) of target messenger RNAs (mRNAs), leading to translational repression and/or mRNA degradation [11]. Studies demonstrated that deregulated miRNA expression contributes to tumor development [12]. While previous studies indicated that miR-9 and miR-124 negatively regulated the STAT3 signaling pathway in glioma, the miR-mediated post-transcriptional regulatory network associated with GBM remains incomplete [13, 14]. Therefore, it is important to identify new miRs that regulate STAT3 expression in order to evaluate their potential roles as new therapeutics against GBM.
miR-519a belongs to the chr19q13.41 miRNA cluster (C19MC) and was reported as either an oncogenic miR associated with ovarian epithelial tumors [15], ER + breast cancer [16], and hepatocellular carcinoma [17], or a tumor suppressor associated with kidney and lung tumors [18]. We previously demonstrated that another member of C19MC, miR-517c, was closely related to improved GBM prognosis. Moreover, the polycistronic miRNA precursors of up to 50 % of mammalian miRNAs that belong to the same cluster are often co-transcribed from one promoter [19]. Additionally, miR-519a exhibited significantly higher expression levels in supratentorial primitive neuroectodermal tumors (sPNET) [20], and that GBM with PNET-like components resulted in a better prognosis relative to primary GBM [21]. Using the MiRGen online biology prediction software(http://www.microrna.gr/mirgen/), we discovered STAT3 as a potential miR-519a target and, therefore, hypothesized that miR-519a may function as a tumor suppressor in GBM.
Here, we reported that miR-519a expression was frequently downregulated in GBM specimens and cell lines, and that low-levels of miR-519a expression significantly correlated with poor GBM-related outcomes. Our results indicated that miR-519a functions as a tumor suppressor by reducing STAT3 expression.
Materials and methods
Human tissue specimens
A total of 46 human GBM specimens and 15 normal brain (NB) tissue samples were obtained from the Department of Neurosurgery at Nanfang Hospital of Southern Medical University, Guangzhou, China. The study was approval by the Ethics Committees of Nanfang Hospital. All patients provided written informed consent in our study. The diagnoses of all specimens were based on pathological evidence.
Cell lines and transfection
The normal human astrocyte (NHA) cell line was obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The human GBM cell lines (U87-MG, T98G, A172, and U373) were purchased from the Chinese Academy of Sciences (Shanghai, China). We also established patient-derived GMB cell lines (G131212 and G141119; Fig. S1) that were cultured in neurobasal media with FBS, L-glutamate, ascorbic acid, rhEGF, insulin, and gentamicin sulfate/amphotericin B. The miR-519a mimics and mimic controls were synthesized and purified by GenePharma (Shanghai, China), and plasmids carrying human STAT3 were purchased from VigeneBio (Shandong, China). Cell transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to manufacturer instructions.
Analyses of genomic data and clinical information
miR-519a expression data and clinical information for 505 GBM patients were downloaded from The Cancer Genome Atlas (TCGA). Only 170 patients treated with newly diagnosed GBM who had undergone surgery plus standard chemoradiotherapy were included. The 170 GBM patient samples were sub-classified into four different molecular subtypes according to previously reported procedures [22], and then divided into high- and low-miR-519a groups according to the median value of miR-519a expression in all GBM patients and each subtype.
RNA isolation and reverse transcription polymerase chain reaction
The expression levels of miRNA and mRNA was detected using quantitative reverse transcription polymerase chain reaction (RT-qPCR) [23]. miRNA and mRNA expression levels were normalized to U6 and GAPDH, respectively. All primers are listed in Supplementary Table S1.
Western blot analysis
Western blotting was performed as described previously [24]. GAPDH served as the loading control.
Cell proliferation assay
Transfected GBM cells were seeded into 96-well plates for 1–5 days. Proliferation in each well was assessed at 24, 48, 72, 96, and 120 h using the Cell Counting Kit-8 (CCK-8, Dojindo; Kumamoto, Japan). Absorbance was measured at 450 nm to calculate the number of viable cells.
Cell cycle and apoptosis assays
Apoptosis and cell cycle progression were assessed 48 h after transfection. For analysis of apoptosis, transfected cells were detected using the ApopNexin FITC Apoptosis Detection Kit (Beyotime; Jiangsu, China) according to manufacturer instructions. The analysis of the apoptosis rate (%)was performed by flow cytometry (BD Biosciences, San Diego, CA, USA). The analysis of cell cycle distribution was then performed using a FACScan flow cytometer (BD Biosciences).
Wound healing and transwell assays
Cells were grown to 60 % confluence six-well plates, and then culture medium was replaced with serum-free DMEM. The monolayer cells were serum-starved for 12 h before being transfected with 50 nM miR-519a or negative control. Six hours after transfection, a scratch was made using a 20-μL pipette tip. The healing process was monitored for 24 h under a microscope, and the rate of wound closure was quantified using ImageJ software (NIH; Bethesda, MD, USA). Transfected cells in 200 μL serum-free DMEM medium were seeded onto a 1:10-diluted Matrigel-coated membrane matrix upper chamber, and 600 μL of high-glucose DMEM medium supplemented with 10 % FBS was added to the lower chamber wells as chemoattractants. The invasive cells located on the lower surface of the membrane were stained with 0.1 % crystal violet (Sigma-Aldrich) and counted.
Luciferase reporter assay
The STAT3 3′-UTR fragment containing the predicted miR-519a-binding site was amplified using primer pairs STAT3-UTR-F/R, inserted into the XbaI site of a pGL3-basic vector (Promega; Madison, WI, USA), and named pGL3-Wt STAT3 3′-UTR. Mutation in the miR-519a-seed regions of the STAT3 3′-UTR was generated using the QuikChange Multi-site-directed mutagenesis kit (Promega) and named pGL3-Mut STAT3 3′-UTR. The luciferase activity assay was performed in 96-well plates. U87MG and T98G cells were co-transfected with the pGL3 reporter vector and the miR-519a mimics or the negative control (miR-NC) using Lipofectamine 2000 (Invitrogen). As a control, the pRL-TK containing renilla luciferase was co-transfected for normalization. Forty-eight hours after transfection, firefly and renilla luciferase assays were performed using the Dual-Luciferase Reporter System (Promega) according to manufacturer instructions. The relative luciferase activity was reported was by normalizing firefly luminescence to that of renilla.
Statistical analysis
Each experiment was performed at least three times. Data are presented as mean ± SD. The IBM SPSS 19.0 (SPSS; Chicago, IL, USA) and GraphPad Prism 5 software (GraphPad Software, Inc.; San Diego, CA, USA) were used for statistical analysis. p < 0.05 was considered to be statistically significant.
Results
Downregulated miR-519a in GBM tissue and cells is inversely correlated with disease progression
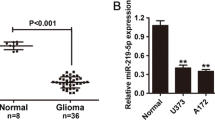
To explore the role of miR-519a in GBM, we performed qRT-PCR to investigate its expression in 46 primary GBM samples and 15 NB tissue samples and cell lines. The qRT-PCR assays showed that miR-519a expression in GBM tissues was significantly lower than that observed in NB tissues (Fig. 1a). Similarly, miR-519a expression in a panel of GBM cell lines (U87-MG, T98G, A172, and U373) was downregulated as compared with an NHA cell line (Fig. 1b). To investigate the prognostic role of miR-519a downregulation in GBM, patients were divided into high- and low-miR-519a groups according to the median value of miR-519a expression in all 46 cases. Kaplan–Meier analysis revealed that patients with high miR-519a expression had significantly longer tumor-free survival and overall survival rates (Fig. 1c, d). Using TCGA data, we analyzed the prognostic value in GBM and found that GBM patients expressing low levels of miR-519a had a significantly shorter overall survival rate. Additionally, we carried out survival analyses for each molecular subtype and found that only the proneural and classical GBM subtypes retained statistical significance (Fig. 1f, g, and Fig. S2).
miR-519a was associated with prognosis of GBM patients and the TCGA cohort a The qRT-PCR assay revealed that miR-519a expression was significantly decreased in GBM tissues (n = 46) as compared with normal brain tissues (n = 15). b The qRT-PCR assay revealed that miR-519a levels in a panel of GBM cell lines (U87-MG, T98G, A172, and U373) were downregulated as compared with an NHA cell line. c, d Kaplan–Meier curves showed the longer tumor-free survival and overall survival rates of patients with high miR-519a expression. e–g In TCGA data analysis, Kaplan–Meier plots were estimated according to different miR-519a gene expression levels related to overall survival of GBM patients in the TCGA GBM cohort. The results were presented as mean ± SD and from three independent experiments. Statistical significance was denoted by *p < 0.05 and **p < 0.01
miR-519a regulates biological behavior in GBM cell lines and patient-derived GBM cells
To investigate miR-519a function in GBM cell lines and patient-derived GBM cells, we transiently transfected miR-519a mimics or miR-NC mimics into U87-MG, T98G, G131212, and G141119 cells to assess the effects of miR-519a on cell proliferation, apoptosis, migration, and invasion. CCK-8 assays showed that miR-519a significantly decreased GBM cell viability as compared with miR-NC controls (Fig. 2a). Flow cytometry assays further revealed that miR-519a upregulation induced a sub-G1 cell cycle arrest as indicated by the increased percentage of cells in G0/G1 phase and the decreased percentage in S and G2/M phases (Fig. 2b). Additionally, apoptosis assessment by flow cytometry showed that miR-519a transfection induced apoptosis in GBM cells (Fig. 2c). Our data showed that miR-519a overexpression significantly inhibited GBM cell proliferation.
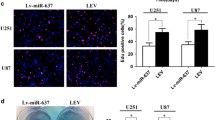
miR-519a overexpression inhibited cell proliferation, invasion, and migration, and induced apoptosis in GBM cell lines and patient-derived cells. a Cell proliferation in the miR-519a or miR-NC group was analyzed by CCK-8 assay at 24, 48, 72,96, and 120 h after transfection. b The apoptosis rate of GBM cell lines and patient-derived cells were assessed by flow cytometry following Annexin V and propidium iodide staining. c Flow cytometric analysis of cell cycle distribution. d The effect of miR-519a on the migration was determined by wound-healing assay. e Cell invasion was measured by Matrigel invasion assay. The results were presented as mean ± SD from three independent experiments. Statistical significance was denoted by *p < 0.05 and **p < 0.01
We also conducted in vitro Matrigel invasion assays to assess miR-519a effects on GBM cell invasion. To exclude the effects of miR-519a-induced dead cells on migration and invasion, we performed flow cytometry assays and verified that live-cell numbers in the miR-519a group were no different from the miR-NC group at the corresponding migration and invasion assay time points (Fig. S3). The number of cells migrating through the extracellular matrix was decreased in cells transfected with miR-519a mimics (Fig. 2d). Next, we conducted a wound-healing assay to address the role of miR-519a in GBM cell migration. miR-519a overexpression also significantly decreased wound-closure percentage (Fig. 2e). These results highlighted the potential role of miR-519a in suppressing migration and invasion.
STAT3 is a direct downstream target of miR-519a
To explore potential miR-519a-related downstream target genes, we used MiRGen software to predict possible mRNA targets. The miR-519a seed sequence matched the STAT3 3′-UTR (Fig. 3a), with STAT3 being a key oncogene associated with GBM. We then performed a dual-luciferase reporter assay in U87-MG and T98G cells to investigate whether STAT3 was a direct miR-519a target. Our results showed that miR-519a significantly inhibited firefly luciferase reporter activity with the wild-type STAT3 3′-UTR, but had no effect on mutated variations of the STAT3 3′-UTR (Fig. 3b). Moreover, the re-expression of miR-519a reduced STAT3 mRNA levels as compared to results observed from the negative control in U87-MG and T98G cells (Fig. 3c). miR-519a also regulated STAT3 protein translation levels. Concentrations of STAT3 protein and phosphorylated STAT3 (p-STAT3) decreased after re-expression of miR-519a. Since Mcl-1, Bcl-2, cyclin D1, and MMP2 are downstream molecules involved in the STAT3 signaling pathway, we further investigated the expression of these proteins in GBM cells following miR-519a overexpression. miR-519a coordinately downregulated Mcl-1, Bcl-2, cyclin D1, and MMP2 (Fig. 3d) expression. These results demonstrated that miR-519a downregulated STAT3 expression by inhibiting its translation, indicating that STAT3 was a direct target for miR-519a in GBM cells.
STAT3 was a direct downstream target of miR-519a in GBM cell lines. a miR-519a and its putative binding sequence in the STAT3 3′-UTR. The mutant miR-519a-binding sites were generated in the complementary site for the miR-519a seed region (wt, wild type; mut, mutant). b miR-519a overexpression suppressed luciferase activity for wild-type, but not mutant STAT3 3′-UTR in U87-MG and T98G cells. c miR-519a re-expression reduced STAT3 mRNA levels as compared with the negative control in U87-MG and T98G cells. d miR-519a upregulation in U87-MG and T98G cells inhibited STAT3, p-STAT3, Mcl-1, Bcl-2, Cyclin D1, and MMP2 expression. The results were presented as mean ± SD from three independent experiments. Statistical significance was denoted as *p < 0.05 and **p < 0.01
STAT3 overexpression attenuated miR-519a tumor-suppressor functions in vitro
To determine whether STAT3 re-expression could reverse the tumor-suppressor function of miR-519a in GBM, we performed a series of rescue experiments using an expression construct encoding STAT3, but lacking the 3′-UTR. GBM cells were transfected with this construct together with miR-519a mimics or with control mimics. Our results indicated that enforced STAT3 expression recapitulated the inhibitory effects of miR-519a in GBM cells, and endogenous STAT3 expression levels were detected by western blot in U87-MG and T98G cells (Fig. 4a). CCK-8 (Fig. 4b), cell apoptosis (Fig. 4c), and cycle (Fig. 4d) assays all revealed that miR-519-related decreases in cell proliferation were reversed by restoration of STAT3 expression. To further determine the role of STAT3 in miR-519a–mediated antitumor activity in GBM cells, we performed a wound-healing assay to assess GBM cell migration. As shown in Fig. 4e, ectopic expression of STAT3 rescued inhibition of cell migration associated with miR-519a upregulation. Consistently, the transwell assay showed that decreased invasion capability in miR-519a-overexpressing cells was rescued by STAT3 overexpression (Fig. 4f).Restoration of STAT3 expression in miR-519a-overexpressed cells reverses the reduction of downstream genes (Bcl-2, Mcl-l, cyclinDl and MMP2)in STAT3 signaling pathways (Fig.S4).These results indicated that STAT3 was crucial for miR-519a-mediated tumor-suppressor activity in GBM.
Restoration of STAT3 expression rescued miR-519a–induced proliferation, migration, and invasion in GBM cells. a Endogenous STAT3 expression levels were detected by western blot in U87-MG cells and T98G cells transfected with miR-519a or miR-NC mimics in the presence of STAT3 or vector control. b CCK-8 assay showed that STAT3 overexpression promoted cell growth and abrogated the reduction in cell viability induced by miR-519a. c Apoptosis assays showed that restoration of STAT3 expression rescued the acceleration of apoptosis caused by miR-519a. d Cell cycle analysis showed that STAT3 overexpression restored G0/G1 cell-cycle progression arrested by miR-519a. e Cell migration was analyzed by wound-healing assays. f Cell invasion was analyzed by transwell assays. The results were presented as mean ± SD from three independent experiments. Statistical significance was denoted as *p < 0.05 and **p < 0.01
Clinical associations of miR-519a with STAT3 expression
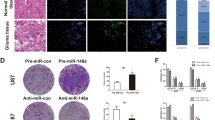
To determine an association between miR-519a expression and STAT3 expression in human GBM tissues, STAT3 expression was detected in tissues from 46 GBM patients using qRT-PCR. We used Pearson rank correlation analysis to reveal a negative correlation between miR-519a expression and STAT3 mRNA levels (Fig. 5a). GBM tissues were then segregated into two groups according to median miR-519a expression. We founded that STAT3 expression in GBM tissues expressing high levels of miR-519a was significantly lower than that observed in cases involving lower levels of miR-519a expression (Fig. 5b). Furthermore, immunohistochemistry assays revealed that STAT3 expression was significantly reduced in patients with higher miR-519a levels, while STAT3 levels were increased in patients with lower miR-519a levels (Fig. 5c).
miR-519a expression negatively correlated with STAT3 levels in GBM tissues. a Pearson’s correlation analysis showed a statistically significant inverse correlation (R = − 0.8325, p < 0.001) between miR-519a and STAT3 expression in human GBM tissues. b STAT3 expression in the group expressing high levels of miR-519a was significantly lower than that in the group expression lower levels of miR-519a (p < 0.01). c Immunohistochemistry assays showed an inverse relationship between miR-519a expression of and STAT3 levels in human GBM specimens. d Model explaining miR-519a functions as a tumor suppressor in GBM through downregulation of the STAT3-signaling pathway
Discussion
miRNAs are closely involved in the regulation of cellular functions through post-transcriptional regulation of gene expression via binding to 3′-UTRs in mRNA, resulting in translational repression and/or targeted degradation [11]. Accumulating evidence indicates that miRNAs function as oncomiRs or tumor suppressors in different tumors by targeting protein-coding genes. Here, we demonstrated that miR-519a acted as a tumor suppressor in GBM, and that miR-519a expression was frequently downregulated in GBM specimens and cell lines. Additionally, low levels of miR-519a expression significantly correlated with poor GBM-related outcomes. The prognostic value of miR-519a was also validated in the TCGA cohort. In survival analyses for each molecular subtype, we found that patients in the proneural and classical GBM subtypes with high miR-519a levels survived longer. Neural and mesenchymal GBM subtypes exhibited no significant differences in clinical prognosis. This analysis showed that the relationship of miR-519a with overall survival in GBM was highly subtype specific. A previous study also reported that miR-222 could predict prognosis in classical and neural GBM, miR-370 could predict prognosis in neural GBM, and miR-34a, miR-145, and miR-182 could predict prognosis in a proneural non-G-CIMP group [25]. These results may be related to the different genetic backgrounds associated with each subtype. Furthermore, tumor microenvironments could also be associated with the differences. The neural and mesenchymal subtypes have been reported to harbor similar gene expression characteristics within normal neural and stromal tissues, respectively [26, 27]. Studies involving neural and stromal cells need to be undertaken to verify these results. miR-519a could function as a marker for predicting proneural and classical GBM subtypes.
miRNAs from C19MC were reported to mediate oncogenic functions, whereas others were associated with tumor-suppressor functions [28]. Previous studies suggested that miR-519a upregulation correlated significantly with advanced clinical stages and poor prognosis in ovarian epithelial tumors. miR519a was also considered a putative target of TGFBR2, a known tumor suppressor associated with many human cancers [15]. Moreover, Ward et al. [16] identified miRNA-519a as a novel oncomiR in ER + breast cancer cells, and validated three central tumor-suppressor genes (CDKN1A, RB1, and PTEN) as direct miRNA-519a targets [16]. Furthermore, Wang et al. [17] identified that miRNA-519a was upregulated in hepatitis B virus-associated hepatocellular carcinoma [17]. Interestingly, Abdelmohsen et al. [18] recently reported that miR-519 inhibited tumorigenesis by repressing expression of the RNA-binding protein HuR, a key regulator of gene expression [18]. Our previous reports [20] indicated that miR-519a may function as a tumor suppressor in GBM. Here, our results indicated that re-expression of miR-519a exhibited inhibitory effects on GBM cell lines and patient-derived GBM cells.
STAT3 is a signal mediator that can be activated by a variety of cytokines, growth factors, and interferons [29], and is persistently active in approximately 70 % of human malignancies, including GBM [4, 30]. In GBM, aberrant STAT3 activation is associated with poor prognosis [31, 32], and miR-mediated inhibition of the STAT3 signaling pathway is an effective strategy for GBM therapy [13]. Constitutive STAT3 activation is involved in cell proliferation, apoptosis, angiogenesis, migration, and invasion in glioma [4]. STAT3 activation requires phosphorylation of its tyrosine residue (p-STAT3-Y705) [33], resulting in p-STAT3 forming dimers with partner proteins in order to translocate into the nucleus to activate expression of downstream targets, including Bcl-2, Mcl-l, cyclinDl, and MMP2. The downregulation of cyclin Dl can result in cell cycle arrest at the G1 phase [34]. Furthermore, downregulation of p-STAT3 can promote apoptosis through downregulation of Mcl-1 and Bcl-2 [35]. MMP2 is overexpressed in GBM tissues [36] and plays a pivotal role in cell migration and invasion through degradation of the extracellular matrix [37]. Here, we showed that STAT3 was a direct target of miR-519a, and that miR-519a overexpression significantly reduced the levels of both STAT3 protein and mRNA. We also demonstrated that miR-519a expression levels negatively correlated with STAT3 mRNA levels in human GBM specimens.
To further understand the underlying mechanisms by which miR-519a exerts its biological effects on GBM cells, it is necessary to identify its downstream functional targets. In our study, western blot analysis revealed that miR-519a overexpression significantly reduced STAT3 concentrations, resulting in the downregulation of p-STAT3,Bcl-2, Mcl-l, cyclin Dl, and MMP2 expression levels. Furthermore, STAT3 re-expression rescued these downstream genes expression in miR-519a-overexpressing cells. These data displayed evidence of miR-519a-mediated suppression of the STAT3 signaling pathway in GBM cells.
In conclusion, our results showed that miR-519a expression was frequently downregulated in GBM specimens and cell lines, and that low levels of miR-519a expression correlated with poor GBM-related outcomes. Furthermore, we demonstrated the significant prognostic value of miR-519a in specific molecular GBM subtypes, and that miR-519a functions as a tumor suppressor responsible for reductions in STAT3 expression and translation. Further studies of a larger population is required to validate the effectiveness of miR-519a as a biomarker for GBM diagnosis and therapy.
References
Das S, Marsden PA (2013) Angiogenesis in glioblastoma. N Engl J Med 369:1561–1563
Ng K, Kim R, Kesari S, Carter B, Chen CC (2012) Genomic profiling of glioblastoma: convergence of fundamental biologic tenets and novel insights. J Neurooncol 107:1–12
Louis DN (2006) Molecular pathology of malignant gliomas. Annu Rev Pathol 1:97–117
Brantley EC, Benveniste EN (2008) Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res 6:675–684
Lee K, Byun K, Hong W, Chuang HY, Pack CG, Bayarsaikhan E, Paek SH, Kim H, Shin HY, Ideker T, Lee B (2013) Proteome-wide discovery of mislocated proteins in cancer. Genome Res 23:1283–1294
Dasgupta A, Raychaudhuri B, Haqqi T, Prayson R, Van Meir EG, Vogelbaum M, Haque SJ (2009) Stat3 activation is required for the growth of U87 cell-derived tumours in mice. Eur J Cancer 45:677–684
Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ (2002) Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene 21:8404–8413
Senft C, Priester M, Polacin M, Schroder K, Seifert V, Kogel D, Weissenberger J (2011) Inhibition of the JAK-2/STAT3 signaling pathway impedes the migratory and invasive potential of human glioblastoma cells. J Neurooncol 101:393–403
Lo HW, Cao X, Zhu H, Ali-Osman F (2008) Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin Cancer Res 14:6042–6054
Lau J, Ilkhanizadeh S, Wang S, Miroshnikova YA, Salvatierra NA, Wong RA, Schmidt C, Weaver VM, Weiss WA, Persson AI (2015) STAT3 blockade inhibits radiation-induced malignant progression in glioma. Cancer Res 75:4302–4311
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6:857–866
Wei J, Wang F, Kong LY, Xu S, Doucette T, Ferguson SD, Yang Y, McEnery K, Jethwa K, Gjyshi O, Qiao W, Levine NB, Lang FF, Rao G, Fuller GN, Calin GA, Heimberger AB (2013) miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res 73:3913–3926
Kim TM, Huang W, Park R, Park PJ, Johnson MD (2011) A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res 71:3387–3399
Kim TH, Kim YK, Kwon Y, Heo JH, Kang H, Kim G, An HJ (2010) Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology 57:734–743
Ward A, Shukla K, Balwierz A, Soons Z, Konig R, Sahin O, Wiemann S (2014) MicroRNA-519a is a novel oncomir conferring tamoxifen resistance by targeting a network of tumour-suppressor genes in ER+ breast cancer. J Pathol 233:368–379
Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT (2012) Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol 18:5442–5453
Abdelmohsen K, Kim MM, Srikantan S, Mercken EM, Brennan SE, Wilson GM, Cabo R, Gorospe M (2010) miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle 9:1354–1359
Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14:1902–1910
Li M, Lee KF, Lu Y, Clarke I, Shih D, Eberhart C, Collins VP, Van Meter T, Picard D, Zhou L, Boutros PC, Modena P, Liang ML, Scherer SW, Bouffet E, Rutka JT, Pomeroy SL, Lau CC, Taylor MD, Gajjar A, Dirks PB, Hawkins CE, Huang A (2009) Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell 16:533–546
Song X, Andrew Allen R, Terence Dunn S, Fung KM, Farmer P, Gandhi S, Ranjan T, Demopoulos A, Symons M, Schulder M, Li JY (2011) Glioblastoma with PNET-like components has a higher frequency of isocitrate dehydrogenase 1 (IDH1) mutation and likely a better prognosis than primary glioblastoma. Int J Clin Exp Pathol 4:651–660
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN, Cancer Genome Atlas Research N (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17:98–110
Uhlmann S, Zhang JD, Schwager A, Mannsperger H, Riazalhosseini Y, Burmester S, Ward A, Korf U, Wiemann S, Sahin O (2010) miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 29:4297–4306
Lu Y, Wang L, He M, Huang W, Li H, Wang Y, Kong J, Qi S, Ouyang J, Qiu X (2012) Nix protein positively regulates NF-kappaB activation in gliomas. PLoS ONE 7:e44559
Li R, Gao K, Luo H, Wang X, Shi Y, Dong Q, Luan W, You Y (2014) Identification of intrinsic subtype-specific prognostic microRNAs in primary glioblastoma. J Exp Clin Cancer Res 33:9
Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H (2011) The brain tumor microenvironment. Glia 59:1169–1180
Lathia JD, Heddleston JM, Venere M, Rich JN (2011) Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell Stem Cell 8:482–485
Flor I, Bullerdiek J (2012) The dark side of a success story: microRNAs of the C19MC cluster in human tumours. J Pathol 227:270–274
Wagner KU, Schmidt JW (2011) The two faces of Janus kinases and their respective STATs in mammary gland development and cancer. J Carcinog 10:32
Brantley EC, Nabors LB, Gillespie GY, Choi YH, Palmer CA, Harrison K, Roarty K, Benveniste EN (2008) Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clin Cancer Res 14:4694–4704
Lin GS, Yang LJ, Wang XF, Chen YP, Tang WL, Chen L, Lin ZX (2014) STAT3 Tyr705 phosphorylation affects clinical outcome in patients with newly diagnosed supratentorial glioblastoma. Med Oncol 31:924
Birner P, Toumangelova-Uzeir K, Natchev S, Guentchev M (2010) STAT3 tyrosine phosphorylation influences survival in glioblastoma. J Neurooncol 100:339–343
Zhong Z, Wen Z, Darnell JE Jr (1994) Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264:95–98
Lee MH, Yang HY (2003) Regulators of G1 cyclin-dependent kinases and cancers. Cancer Metastasis Rev 22:435–449
Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, Hess K, Conrad C, Madden T, Sawaya R, Kondo S, Priebe W, Kondo Y (2007) A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 26:2435–2444
Kessenbrock K, Plaks V, Werb Z (2010) Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141:52–67
Li Z, Du L, Li C, Wu W (2013) Human chorionic gonadotropin beta induces cell motility via ERK1/2 and MMP-2 activation in human glioblastoma U87MG cells. J Neurooncol 111:237–244
Acknowledgments
This project was supported by the National Natural Science Foundation of China (Grant Nos. 81101921), Natural science Foundation of Guangdong Province (Grant Nos. 2014A030313298), and the National Key Clinical Specialist Construction Program of China.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11060_2016_2095_MOESM1_ESM.tif
Fig. S1 The morphology of U87-MG, T98G, G131212, and G141119 was observed under an inverted microscope. Scale bars, 50 μm. Supplementary material 1 (TIFF 45543 kb)
11060_2016_2095_MOESM2_ESM.tif
Fig. S2 The prognostic value of miR-519a in neural and mesenchymal subtypes of GBM patients in the TCGA cohort. Kaplan–Meier curves indicated that there was no statistical significance in overall survival for patients with higher miR-519a expression in the a neural and b mesenchymal subtypes. Supplementary material 2 (TIFF 32992 kb)
11060_2016_2095_MOESM3_ESM.tif
Fig. S3 The apoptosis rate of GBM cell lines (U87-MG, T98G) at 30 h after transfection with miR-519a mimics or miR-NC mimics was assessed by flow cytometry following Annexin V and propidium iodide staining. Supplementary material 3 (TIFF 1115 kb)
11060_2016_2095_MOESM4_ESM.tif
Fig. S4 Restoration of STAT3 expression in miR-519a-overexpressing cells reversed the reduction of downstream genes associated with the STAT3 signaling pathways. Western blot analysis of the expression and translation of downstream genes, including Bcl-2, Mcl-l, cyclin Dl, and MMP2. GAPDH served as a loading control. Values represented the relative band intensity normalized to GAPDH using ImageJ software. Supplementary material 4 (TIFF 12298 kb)
Rights and permissions
About this article
Cite this article
Hong, L., Ya-wei, L., Hai, W. et al. MiR-519a functions as a tumor suppressor in glioma by targeting the oncogenic STAT3 pathway. J Neurooncol 128, 35–45 (2016). https://doi.org/10.1007/s11060-016-2095-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-016-2095-z