Abstract
High-grade gliomas have an aggressive clinical course and new clinical biomarkers and therapeutic targets are highly needed. WEE1 is a regulator of the G2 checkpoint in glioblastoma (GBM) cells. Inhibition of this kinase has, in experimental glioma studies, been suggested to enhance sensitivity to irradiation and temozolomide. However, expression level and prognostic potential of WEE1 protein in gliomas remain uninvestigated. In this study, glioma samples from 235 patients across all four WHO grades were analyzed by immunohistochemistry. Using image analysis, we calculated the area fraction of WEE1 positive nuclei. We found that WEE1 protein was localized in tumor cell nuclei and expressed in all glioma types and grades. Although WEE1 protein levels are higher in GBMs (mean 24.5 %) relative to grade III (mean 14,0 %, p < 0.05) and grade II (mean 6.8 %, p < 0.001) gliomas, high WEE1 protein was associated with better survival in GBMs (p = 0.002). This was confirmed in multivariate analysis (HR 0.60, p = 0.003) even when adjusted for MGMT status (HR 0.60, p = 0.005). In conclusion, we report a nuclear expression of WEE1 protein in all glioma grades and types. The WEE1 positive nuclear area was correlated with malignancy grade but it was inversely associated with prognosis in GBM. Although WEE1 is a frequently occurring protein and has been proposed as a novel target in GBM, the role of WEE1 in glioma patient survival appears to be connected to the MGMT status and is more complex than previously anticipated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gliomas of astrocytic and oligodendroglial origin account for more than 70 % of all brain tumors, and are graded by histopathologic evaluation into four tumor grades (I–IV) according to current World Health Organization (WHO) grading criteria [1]. Glioblastoma multiforme (GBM), WHO grade IV, is the most common and most malignant primary brain tumor in adults with a median survival of 15 months [2]. GBMs are particularly difficult to treat due to their infiltrative nature and high level of resistance towards radio- and chemotherapy [3]. Even though attempts are being made in understanding and targeting the underlying molecular mechanisms, there has been little improvement in clinical outcome [4] and an increasing need of useful biomarkers to support treatment decisions exists.
Kinases are a group of proteins that execute key cellular functions such as proliferation, apoptosis, cell metabolism, migration, DNA damage repair, responses to the microenvironment, and targeting of oncogenic kinase activity. Therefore kinases are widely investigated as potential targets in anticancer treatment [5]. WEE1 has recently been discovered as a nuclear regulator of the G2 checkpoint [6–8], where it serves as a mitotic inhibitor through its inhibitory phosphorylation of CDK1.
Gene expression analyses have shown that WEE1 is one of the most frequently overexpressed kinases in glioblastomas [7]. WEE1 protein has been reported to be overexpressed in 35 % of breast cancers [9], and the activity of WEE1 was found to be increased in patients with advanced hepatocellular carcinoma when compared to noncancerous liver tissue [10]. Furthermore, expression level of WEE1 was associated with prognosis in mantle cell lymphoma [11]. A recent publication suggested an association of low WEE1 mRNA expression with better patient survival in glioblastoma but no systematic studies on the association between WEE1 mRNA and protein levels or between WEE1 protein levels and survival were performed [7].
The aim of the present study was therefore to investigate WEE1 histological pattern and expression level in different gliomas as well as the association between WEE1 and survival by obtaining reliable quantitative estimates of WEE1 protein in the investigated glioma cells. We used an automated image acquisition and processing approach, replacing subjective manual scoring, and enabling continuous and precise measurements of the tumor cell area expressing WEE1. Furthermore, the tissue material used for the purpose originates from a population-based cohort circumventing selection biases [12, 13].
Materials and methods
Patients
The tissue material is from 235 patients who underwent initial surgery between 01.01.2005 and 31.12.2009 in the Region of Southern Denmark. The material includes glioblastoma (GBM) [grade IV (n = 186)]; anaplastic astrocytoma (AA, n = 15), anaplastic oligodendroglioma (AOD, n = 6), and anaplastic oligoastrocytoma (AOA, n = 4) [all grade III (n = 25)]; diffuse astrocytoma (DA, n = 12), oligodendroglioma (OD, n = 4), and oligoastrocytoma (OA, n = 5) [all grade II (n = 21)]; and pilocytic astrocytoma (PA) [grade I (n = 3)]. No treatment was received prior to surgical resection and patients had no prior history of brain tumors. The material is epidemiologically well-defined and has been histopathologically classified by two neuropathologists according to the WHO 2007 guidelines [1]. Information about clinical data, post-surgical treatment and other biomarkers has previously been reported [12–17]. Patients were followed until death or censored on November 2014. The Regional Scientific Ethical Committee and the Data Protection Authority approved the study. The Danish Tissue Application Register allowed use of all the tumors included in this study.
Tissue preparation and immunohistochemical staining
Fresh tissue biopsies were fixed in 10 % neutral buffered formalin and subsequently paraffin-embedded. Three micrometer sections were dewaxed with xylene and rehydrated with ethanol. Endogenous peroxidase activity was blocked by 1.5 % hydrogen peroxide in TBS (Tris-buffered Saline) for 10 min. Heat induced epitope retrieval was carried out in 3 steps using a microwave oven: (1) heating 9 min at 900 W in Tris-EGTA buffer, (2) boiling 15 min at 440 W, followed by (3) cooling 15 min at room temperature. Sections were incubated for 60 min with WEE1 primary antibody (Wee 1 (B-11): sc-5285, Santa Cruz Biotechnology) at a 1 + 100 concentration. Visualization of WEE1 was carried out using PowerVision (Dako, Denmark) with diaminobenzidine (DAB) as the chromogenic substrate. Sections were counterstained with Mayer’s haematoxylin. Staining and blocking protocol was carried out using a Dako Autostainer Universal Staining System (Dako, Denmark). Human placenta was used as positive control [18]. Omission of primary antibody served as negative control. Positive and negative controls were included in all staining runs. Two patients were excluded due to problems during the IHC staining process.
MGMT status
In 156 patients O6-methylguanine-DNA methyltransferase (MGMT) status was retrospectively obtained using pyrosequencing (QIAampDNA FFPE Tissue kit, Qiagen). DNA was purified from 10 µm paraffin slides using QIAamp DNA FFPE Tissue kit (Qiagen) and MGMT pyrosequencing was performed according to the kit instructions. Methylation percentages at four CpG sites were measured. If 10 % methylation or higher levels at one or more sites were measured, the patient was considered as having a methylated MGMT status.
Image acquisition and analysis
Slides were scanned on a digital slide scanner (Hamamatsu, Japan). Image analysis and quantification was carried out using TissuemorphDP (Visiopharm, Hørsholm, Denmark). The tumor tissue was manually delineated in order to exclude normal brain tissue, infiltration zone, necrosis and tissue artifacts. Thereafter sample images were collected using a 10 % systematic uniform random sampling (meander) at ×20 magnification. Prior to this, a sampling fraction study was performed with sampling values ranging from 5 to 20 %. Based on this, 10 % was found to be the lowest optimal sampling fraction giving reproducible results. Included images were required to have more than 50 % viable tumor tissue after exclusion of normal brain tissue, infiltration zone, necrosis and tissue artifacts. The WEE1 signal was measured using a trained software-based pixel classifier based on Bayesian classification. To quantify WEE1, the software classifier was trained to recognize strongly positive (yellow), weakly positive (red), and negative (green) nuclei. Identifying the nuclei in this way was found to be necessary for detecting all of the WEE1 positive (WEE1+) nuclei during training of the software. WEE1 area fraction estimates were defined as the area of strong and weak WEE1+ nuclei relative to the area of all nuclei in the tumor (area WEE1 strong + area WEE1 weak)/(area WEE1 strong + area WEE1 weak + area negative) × 100 %.
Statistical analysis
The univariate relationship between prognostic variables and death was illustrated by Kaplan–Meier plots and assessed by log-rank statistics. Overall survival (OS) was defined as time from primary surgery until death or date of censoring (November 2014). The WEE1 estimates were investigated as binary variables with the median used as a pre-specified cutoff value and as continuous variables. Patients with WHO grade I tumors were not included in the statistical analysis. Only variables that were previously reported to be significant in the univariate analysis [12, 13] were included in the multivariate analysis. 1p/19q co-deletion was not available in all patients grade II and III patients. Combined with the limited number of patients with WHO grade II and III tumors 1p/19q-codeletion was therefore not included in multivariate analysis. Multivariate analyses for WHO grade II and III gliomas included age, performance status and IDH1 status. Age, performance status, tumor crossing midline and post-surgical treatment (reported as intention to treat) as well as MGMT status was included in the multivariate analysis for grade IV gliomas. Only 4 GBM patients had a mutated IDH1 status, and therefore IDH1 status was not included. Analysis was carried out using STATA version 11 with an overall significance level of p < 0.05.
Results
Expression of WEE1 protein in gliomas
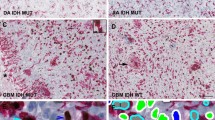
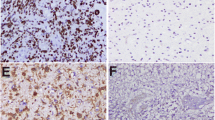
In normal brain tissue no significant positive WEE1 staining was observed (Fig. 1a) neither in cortex nor in white matter. Pilocytic astrocytomas (grade I), diffuse astrocytomas and oligodendrogliomas (grade II) showed distinct WEE1 positive tumor cell nuclei (Fig. 1b–d). Similarly, distinct WEE1 positive tumor nuclei were also evident for grade III gliomas (images not shown). Pleomorphic cells with a strong positive staining were generally present in GBMs (Fig. 2a). WEE1 positive staining was also present in GBMs with sarcomatous components consisting of spindle-shaped cells (Fig. 2b). All tumor and normal blood vessels were WEE1 negative (Fig. 2c) but WEE1 positive cells were present in areas where tumor cells infiltrated the brain parenchyma (Fig. 2d). Strong WEE1 staining was present in pseudopalisading cells around necroses (Fig. 2e–f). Grade III and IV gliomas displayed similar WEE1 tumor cell staining patterns (images not shown).
WEE1 immunohistochemically stained paraffin sections of normal brain tissue and low-grade gliomas showing WEE1 staining patterns. a Normal brain tissue showed no positive staining neither in cortex nor in white matter (control), whereas b pilocytic astrocytoma, c diffuse astrocytoma and d oligodendroglioma showed distinct WEE1 positive tumor cells. Scale bar 100 µm
WEE1 immunohistochemically stained paraffin sections of glioblastomas showing WEE1 staining patterns. a Pleomorphic cells with strong positive staining. b WEE1 positive staining was present in GBMs with sarcomatous components consisting of spindle-shaped cells. c All blood vessels showed negative staining. d Strong WEE1 positive cells were present in invasion zones. e–f A strong WEE1 staining was present in pseudopalisading cells around necroses. g–h Quantitation of WEE1 + staining was carried out using a trained pixel classifier based on Bayesian classification. The classifier successfully identified strongly positive (yellow) and weakly positive (red) nuclei as well as negative (green) nuclei. The insert in (g) represents a magnification of a distinct area illustrating software-based classification of nuclei. Scale bars 100 µm (a, b, c, and f) and 300 µm (d and e)
The software classifier successfully identified positive (yellow/red) as well as negative (green) nuclear WEE1 staining (Fig. 2g–h). To identify all positive nuclear staining it was necessary to train the classifier separately to identify both strongly positive (yellow) and weakly positive (red) nuclear staining. No additional information was obtained when separate analyses were performed compared with analyses of total staining area. The WEE1 results reported in the following are therefore results obtained by analysis of total WEE1 area fraction only. The total WEE1 area fraction increased with increasing WHO grade, and patients with grade IV had significantly higher levels of WEE1 protein (mean 24.5 %) compared to patients with grade III (mean 14 %, p < 0.05) and grade II (mean 6.8 %, p < 0.001) gliomas, respectively (Fig. 3a–b). Mean expression was higher in grade III gliomas as compared to grade II gliomas, although not significant (p = 0.9). A significantly higher WEE1 area fraction was present in GBM compared to AA (p < 0.01) and DA (p < 0.001), however this was not the case when compared to OD, OA, AOD nor AOA (p > 0.5).
WEE1 protein levels increase with grade, vary in glioma subtypes, and are associated with survival. a Scatter plot showing WEE1 protein levels relative to WHO grade. b Scatter plot showing WEE1 protein levels relative to subtype. c–e Overall survival is shown for WHO grade II (c), grade III (d), and IV (e), respectively. Each WHO grade was dichotomized at the median. Overall survival when MGMT promotor methylation status is included (f). Asterisks are defined as follows: * < 0.05, ** < 0.01 and *** < 0.001. Abbreviations: PA pilocytic astrocytoma, DA diffuse astrocytoma, OD oligodendroglioma, OA oligoastrocytoma, AA anaplastic astrocytoma, AOD anaplastic oligodendroglioma, AOA anaplastic oligoastrocytoma, GBM glioblastoma. Un-MGMT un-methylated MGMT promotor, m-MGMT methylated MGMT promotor
WEE1 expression level and survival
WEE1 area fraction was not associated with survival in grade II gliomas, neither in univariate analyses (HR 1.4, 95 % CI 0.5–4.0, p = 0.5) (Fig. 3c) nor in multivariate analyses (HR 1.9, 95 % CI 0.6–6.2, p = 0.3).
Patients with high WEE1 area fraction had poorer survival than did patients with low WEE1 area fraction (HR 2.6, 95 % CI 1.03–6.7, p = 0.04) in grade III gliomas (Fig. 3d). In multivariate analyses high expression of WEE1 was associated with poor OS (HR 4.9, 95 % CI 1.5–16.1, p = 0.009) in grade III gliomas.
Patients with WHO grade IV tumors and high WEE1 area fraction had a better survival than did patients with low WEE1 area fraction (HR 0.6, 95 % CI 0.5–0.8, p = 0.002) (Fig. 3e). In multivariate analyses high expression of WEE1 was associated with an improved OS (HR 0.6, 95 % CI 0.4–0.8, p = 0.003) in grade IV gliomas (Table 1). No additional prognostic information was obtained when the WEE1 estimates were investigated as a continuous variable (data not shown), and we did not discover a more optimal cutoff value than the median when dichotomizing the WEE1 area fraction.
WEE1 expression level, MGMT status and survival
MGMT status was identified in 156 patients. In these patients median WEE1 was 22.5 (0.0–79.6). Median WEE1 expression in patients with methylated and un-methylated MGMT status was 23.6 and 21.5, respectively. In patients with methylated MGMT status high levels of WEE1 was associated with improved OS (HR 0.44, p = 0.003), whereas the effect was less pronounced in patients with un-methylated MGMT status (HR 0.77, p = 0.23).
When MGMT status was included in multivariate analysis high levels of WEE1 was associated with improved survival (HR 0.60, 95 % CI 0.42–0.86, p = 0.005) (Table 2). No interaction between WEE1 and MGMT status was identified (p = 0.78). However, in univariate analysis patients with high level of WEE1 and methylated MGMT promotor had a better OS than patients with low WEE1 and un-methylated MGMT status (p = 0.007) (Fig. 3f). This was also significant in multivariate analyses (HR 0.59, 95 % CI 0.36–0.97, p = 0.04). The better survival effect having high WEE1 was more pronounced in the methylated than in the un-methylated MGMT patients.
Discussion
We show, for the first time, that WEE1 protein is expressed in different types of gliomas and that WEE1 protein expression increases with malignancy grade. Moreover we found that WEE1 protein levels are associated with prognosis in glioma patients.
Expression of WEE1 protein in gliomas
WEE1 protein levels correlate with tumor malignancy grade, and patients with GBMs have significantly higher levels of WEE1 protein compared to patients with grade III and grade II gliomas. This is similar to results obtained for mRNA expression data [7]. Additionally another study found WEE1 protein to be overexpressed in pediatric high-grade gliomas, with increasing expression positively correlated with grade [19].
When dividing into subtypes in order to estimate WEE1 protein level relative to histology, a significantly lower level of WEE1 protein was present in anaplastic astrocytoma (p < 0.01) and diffuse astrocytoma (p < 0.001) compared to GBM. However, no significant differences were observed between GBM and the other subtypes or among the other subtypes. A thorough analysis revealing possible significance of differential expression levels between subtypes would require inclusion of more grade II and III glioma patients in the study cohort. In a recent study, the WEE1 mRNA expression was investigated in 90 medulloblastoma patient samples and the authors showed a significant increase in WEE1 expression compared to normal adult cerebellum. However, they found no significant difference in WEE1 expression between four subgroups of medulloblastoma [20]. Together the high expression in GBMs and medulloblastomas, which both are highly malignant tumors, suggest that WEE1 is closely associated with aggressive tumor biology. This is supported by the high protein expression found in the invasion zones in the present study.
The high WEE1 protein expression level found in many GBMs but also some of the other types of gliomas suggest that WEE1 is a frequently occurring target in these tumors. Future clinical studies are required to identify whether a certain level of WEE1 protein expression is necessary to obtain effect of WEE1 inhibitors.
WEE1 and tumor necrosis
Strong WEE1 staining was consistently found in pseudopalisading cells around necrotic areas in GBMs, thereby suggesting an association of mechanisms to hypoxia with high WEE1 expression. Although no endothelial expression of WEE1 was found in neither tumor vessels nor brain vessels in our study, induction of WEE1 by hypoxia was found in a pancreatic endothelial cell line, MS-1 [21], thereby suggesting a potential association of hypoxia with high WEE1 expression also in GBMs.
WEE1 protein levels and survival
The present study highlights the potential of WEE1 as a prognostic biomarker in GBMs. However, our results are in contradiction to the study by Mir et al. who found high tumor WEE1 mRNA expression to be associated with worse patient survival in GBM [7]. Surprisingly, our results clearly show that high WEE1 protein levels correlated with better patient survival in GBM (p = 0.002). Yoshida et al. show a similar correlation in non-small cell lung cancer (NSCLC), as patients with high WEE1 (more than 30 %) had a lower recurrence rate and a better survival. Furthermore, a multivariate analysis suggested that both cyclin B1 and WEE1 were significant prognostic factors in NSCLC [22]. In GBMs, further analysis including MGMT methylation status showed that WEE1 protein levels were correlated with better survival in MGMT methylated patients being sensitive to chemotherapy, while in MGMT un-methylated patients the survival benefit in patients with high levels of WEE1 was less pronounced. This suggests that high WEE1 may enhance the effect of chemotherapy. Although WEE1 inhibitors have been suggested as potential anti-cancer candidates [6, 8, 23–25], the obtained results suggest that combinations of WEE1 inhibitors and temozolomide should be avoided in patients with MGMT methylated GBMs.
Surprisingly, prognostic results obtained in grade III gliomas and GBMs were opposite. A high WEE1 protein level was associated with worse patient survival in grade III gliomas (p = 0.04). There is no obvious explanation to this but it may be speculated that GBM therapy as well as therapy given to grade III tumor may influence the results. Neither age nor the presence of oligodendrogliomas can explain the results. One might have hypothesized that the poor survival for patients with high WEE1 protein was associated with a higher age, as it is known that incidence rates increases with age for most cancers including gliomas [26–28]. However, age was equally distributed in both groups. Moreover, it is known that oligodendrogliomas are slowly growing with prolonged survival compared to astrocytomas [29] but since 80 % of the oligodendrogliomas were present in the group with high WEE1, this could not explain the tendency. In line with this, co-deletion of 1p/19q in oligodendrogliomas and presence of mutated IDH1 most likely do not affect the results, since the frequency of these molecular changes were equally distributed in the groups. A drawback of our study is the retrospective nature. Recent publications have thus shown that TERT mutational status is important for survival in GBM patients but due to the lack of sufficient tissue, it was not possible to achieve this status in the present study. However, the prognostic impact of TERT mutational status has recently been shown to be absent in patients receiving temozolomide chemotherapy [30] suggesting that our findings are independent of TERT mutational status. One might also speculate that tumor location and whether the tumor was completely or partly removed, could explain this opposite relationship compared to GBMs. In order to obtain more reliable results for low-grade gliomas more patients must be included.
Software-based classification of WEE1 staining
The WEE1 expression estimates were obtained by replacing subjective manual scoring with advanced image analysis. We used an automated image acquisition and processing approach, enabling continuous measurements of the WEE1 staining area. Previous studies investigating protein expression have primarily used conventional semi-quantitative and pathologist-based scoring. Calculating WEE1 area fractions using an observer independent approach, avoids biases due to e.g. increases in cellularity. The association between tumor grade and WEE1 protein levels is therefore not explained by a simple increase in cellularity but is instead explained by a higher fraction of positive cells being WEE1 positive. Moreover tumor diagnosis-induced bias is avoided, since the software-classifier make estimates without knowing the diagnosis. This is in contrast to a human observer easily identifying e.g. oligo and GBM histological patterns.
Conclusion
In conclusion, we found WEE1 protein to be expressed in different types of gliomas having an increased expression with increasing malignancy grade. Moreover high WEE1 protein expression in GBMs was found to be associated with longer survival especially in patients with methylated MGMT status. Although, the high WEE1 protein expression in GBMs and other types of gliomas suggest that WEE1 is a frequently occurring target, the role of WEE1 in glioma patient survival appears to be more complex than previously anticipated.
References
Louis DN, Ohgaki H, Wiestler OD et al (2007) WHO classification of tumours of the central nervous system, 4th edn. International Agency for Research on Cancer (IARC), Lyon
Stupp R, Hegi ME, Mason WP et al (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10:459–466
Kesari S (2011) Understanding glioblastoma tumor biology: the potential to improve current diagnosis and treatments. Semin Oncol 38:S2–S10
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359:492–507
Manning G, Whyte DB, Martinez R et al (2002) The protein kinase complement of the human genome. Science 298:1912–1934
De Witt Hamer PC, Mir SE, Noske D et al (2011) WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin Cancer Res 17:4200–4207
Mir SE, De Witt Hamer PC, Krawczyk PM et al (2010) In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell 18:244–257
Wang Y, Li J, Booher RN et al (2001) Radiosensitization of p53 Mutant Cells by PD0166285, a Novel G2 Checkpoint Abrogator. Cancer Res 61:8211–8217
Iorns E, Lord CJ, Grigoriadis A et al (2009) Integrated functional, gene expression and genomic analysis for the identification of cancer targets. PLoS One 4:e5120
Masaki T, Shiratori Y, Rengifo W et al (2003) Cyclins and cyclin-dependent kinases: comparative study of hepatocellular carcinoma versus cirrhosis. Hepatology 37:534–543
Blenk S, Engelmann JC, Pinkert MWS et al (2008) Explorative data analysis of MCL reveals gene expression networks implicated in survival and prognosis supported by explorative CGH analysis. BMC Cancer 8:106
Dahlrot RH, Kristensen BW, Hjelmborg J et al (2013) A population-based study of high-grade gliomas and mutated isocitrate dehydrogenase 1. Int J Clin Exp Pathol 6:31–40
Dahlrot RH, Kristensen BW, Hjelmborg J et al (2013) A population-based study of low-grade gliomas and mutated isocitrate dehydrogenase 1 (IDH1). J Neurooncol 114:309–317
Hermansen SK, Dahlrot RH, Nielsen BS et al (2013) MiR-21 expression in the tumor cell compartment holds unfavorable prognostic value in gliomas. J Neurooncol 111(1):71–81
Dahlrot RH, Hansen S, Herrstedt J et al (2013) Prognostic value of Musashi-1 in gliomas. J Neurooncol 115:453–461
Lathia JD, Li M, Sinyuk M (2014) High-throughput flow cytometry screening reveals a role for junctional adhesion molecule a as a cancer stem cell maintenance factor. Cell Rep 6(1):117–129
Dahlrot RH, Hansen S, Jensen SS et al (2014) Clinical value of CD133 and nestin in patients with glioma: a population-based study. Int J Clin Exp Pathol 7(7):3739–3751
PosthumaDeBoer J, Wurdinger T, Graat H et al (2011) WEE1 inhibition sensitizes osteosarcoma to radiotherapy. BMC Cancer 11:156
Mueller S, Hashizume R, Yang X et al (2013) Targeting Wee1 for the treatment of pediatric high-grade gliomas. Neuro-Oncology 16(3):352–360
Harris PS, Venkataraman S, Alimova I et al (2014) Integrated genomic analysis identifies the mitotic checkpoint kinase WEE1 as a novel therapeutic target in medulloblastoma. Mol Cancer 13:1–14
Hong KS, Kim HS, Kim SH et al (2011) Hypoxia induces WEE1 expression and attenuates hydrogen peroxide-induced endothelial damage in MS1 cells. Exp Mol Med 43:653–659
Yoshida T, Tanaka S, Mogi A et al (2004) The clinical significance of cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann Oncol 15:252–256
Guertin AD, Li J, Liu Y et al (2013) Preclinical evaluation of the WEE1 inhibitor MK-1775 as single-agent anticancer therapy. Mol Cancer Ther 12:1442–1452
Kreahling JM, Gemmer JY, Reed D et al (2011) MK1775, a selective Wee1 inhibitor, shows single-agent antitumor activity against sarcoma cells. Mol Cancer Ther 11(1):174–182
Sarcar B, Kahali S, Prabhu AH et al (2011) Targeting radiation-induced G2 checkpoint activation with the Wee-1 inhibitor MK-1775 in glioblastoma cell lines. Mol Cancer Ther 10(12):2405–2414
Cancer Incidence by Age. Cancer Research UK. http://www.cancerresearchuk.org/cancer-info/cancerstats/incidence/age/. Accessed 26 Nov 2014
Do Cancer Rates Differ with Age? Australian Institute of Health and Welfare http://www.aihw.gov.au/cancer/. Accessed 26 Nov 2014
Finkel T, Serrano M, Blasco MA (2007) The common biology of cancer and ageing. Nature 448:767–774
Ohgaki H, Kleihues P (2005) Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol 64:479–489
Simon M, Hosen I, Gousias K et al (2015) TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro-Oncology 17:45–52
Hegi ME, Diserens AC, Gorlia T et al (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352(10):997–1003
Acknowledgments
We acknowledge the excellent laboratory work performed by technicians Helle Wohlleben and Tanja Dreehsen Højgaard.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Music, D., Dahlrot, R.H., Hermansen, S.K. et al. Expression and prognostic value of the WEE1 kinase in gliomas. J Neurooncol 127, 381–389 (2016). https://doi.org/10.1007/s11060-015-2050-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-015-2050-4