Abstract
Malignant gliomas are the most common and devastating primary brain tumors in adults. The rapid invasion of tumor cells into the adjacent normal brain tissues is a major cause of treatment failure, yet the mechanisms that regulate this process remain poorly understood. MicroRNAs have recently emerged as regulators of invasion and metastasis by acting on multiple signaling pathways. In this study, we found that miR-622 is significantly downregulated in glioma tissues and cell lines. Functional experiments showed that increased miR-622 expression reduced glioma cell invasion and migration, whereas decreased miR-622 expression enhanced cell invasion and migration. Moreover, activating transcription factor 2 (ATF2), an important transcription factor that regulate tumor invasion, was identified as a direct target of miR-622. Knockdown of ATF2 using small interefering RNA recapitulated the anti-invasive function of miR-622, whereas restoring the ATF2 expression attenuated the function of miR-622 in glioma cells. Furthermore, clinical data indicated that miR-622 and ATF2 were inversely expressed in glioma specimens. Our findings provide insight into the specific biological behavior of miR-622 in tumor invasion and migration. Targeting miR-622/ATF2 axis is a novel therapeutic approach for blocking glioma invasion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malignant gliomas are the most common and aggressive type of primary brain tumor in adults [1, 2]. Despite recent advances in standard therapy, including surgical resection followed by radiation and chemotherapy, the prognosis for patients with malignant gliomas remains dismal [3, 4]. A major barrier to effective treatment of gliomas is due to their highly invasive nature, yet the mechanisms governing glioma invasion remain poorly understood. [5]. Thus, there is an urgent need to investigate the novel molecular mechanisms underpinning glioma cell invasion.
MicroRNAs (miRNAs) are endogenous, small non-coding RNAs that regulate gene expression by antisense complementarity to the 3′-UTR region of specific mRNAs [6, 7]. Accumulating evidence suggests that miRNAs may regulate diverse biological processes such as cell proliferation, apoptosis, stress resistance, stem cell maintenance and cell identity [8–10], and aberrant expression of miRNAs is associated with various types of cancers, including gliomas [11–17]. Moreover, miRNAs have been shown to be involved in regulating cancer cell migration, invasion and metastasis. miR-622 is a novel member of these miRNAs, which has been characterized as a tumor suppressor in human cancers by targeting critical cancer-related pathways. It is shown that miR-622 could impact the K-Ras signal pathway and enhance the anticarcinogenic effect of resveratrol [18]. Notably, a recent study indicated that miR-622 is associated with tumor metastatic capability in gastric cancers [19]. However, the role of miR-622 in glioma invasion remains unknown.
In this study, we investigated the expression levels of miR-622 in glioma specimens and provide the first evidence for a role of miR-622 in glioma invasion and migration via directly targeting ATF2.
Materials and methods
Human tissue samples
Forty-eight glioma tissue samples were obtained postoperatively from the Department of Neurosurgery, First Affiliated Hospital, Nanjing Medical University. Five normal brain tissues were collected as negative controls from epilepsy patients undergoing lobectomy. The histological features of all of the specimens were confirmed by pathologists according to the WHO criteria. This study was reviewed and approved by the Ethical Committee of Nanjing Medical University, and written informed consent was obtained from all participants.
Cell culture and transfection
Human glioma cell lines U87, U251, A172, U118 and LN229 were purchased from American Type Culture Collection. All cells were maintained in Dulbecco’s modified eagle’s medium containing 10 % fetal bovine serum and 1 % penicillin/streptomycin. Normal human astrocytes (NHAs) were obtained from Lonza (Walkersville, MD, USA) and cultured according to the manufacturer’s instruction. miR-622 mimics, negative control (NC) and anti-miR-622 inhibitor were purchased from GenePharma (Shanghai, China). siATF2 oligonucleotides and control siNC were purchased from GenePharma. pcDNA3.1-ATF2 vector was obtained from Sangon Biotech (Shanghai, China). All the oligonucleotides and plasmids were transfected into cells using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
RNA isolation, qRT-PCR and DNA sequencing
RNA was extracted from cells or tissues using TRIzol (Invitrogen). First-strand cDNA were synthesized using the PrimerScript™ RT Master Mix (TaKaRa). The primer pairs of ATF2 were 5′-GCACAGCCCACATCAGCTATT-3′ and 5′-GGTGCCTGGGTGATTACAGT-3′. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was also amplified in the same PCR reactions as an internal control using the primers 5′-TGCACCACCAACTGCTTAGC-3′ and 5′-GGCATGGACTGTGGTCATGAG-3′. Total miRNAs were isolated using a mirVanaTM miRNA Isolation Kit (Ambion, CA, USA). qRT-PCR reactions were performed using TaqMan miRNA assays (Applied Biosystems, CA, USA). U6 was used for normalization. All the primers used for miRNA reverse transcription and qRT-PCR were purchased from RiboBio (Guangzhou, China). Relative gene expression was calculated via 2−ΔΔCt method.
IDH1 codon 132 and IDH2 codon 172 were determined by direct sequencing in all the cases. Primer sequences used were: IDH1 (forward): 5′-TGTGTTGAGATGGACGCCTATTTG-3′ and IDH1 (reverse): 5′-TGCCACCAACGACCAAGTC-3′, IDH2 (forward) 5′-GCCCGGTCTGCCACAAAGTC-3′, IDH2 (reverse): 5′-TTGGCAGACTCCAGAGCCCA-3′. PCR amplification was performed in a total of 10 µL reaction mixture containing 50 ng of tumor DNA, 1 µL of 10× PCR buffer, 0.8 µL of 10 mM dNTPs, 0.25 µL of each forward and reverse primers, and 0.2 µL of AmpliTaq Gold PCR Master Mix (Applied Biosystems, CA, USA). Initial denaturation was performed at 95 °C for 5 min. This was followed by 35 cycles of amplification consisting of denaturation at 95 °C for 1 min, annealing at 57 °C for 45 s, and extension at 72 °C for 2 min. Bidirectional sequencing was performed using ABI 3100 sequencer (Applied Biosystems).
Cell proliferation assay
Cells were seeded at 2,000 per well in 96-well plates and cultured after transfection. Cell proliferation was detected at the indicated time points using a CCK8 kit (Dojindo Laboratories) following the manufacturer’s instructions. All assays were performed in octuplicate and repeated at least three times. For colony formation assay, 5 × 102 cells were independently plated onto 60 mm tissue culture plates. After 10–14 days, visible colonies were fixed with 100 % methanol and stained with 0.1 % crystal violet in 20 % methanol for 15 min. Colony-forming efficiency was calculated as the number of colonies/plated cells × 100 %.
In situ hybridization (ISH) and Immunohistochemistry (IHC)
Paraffin-embedded tissue Sections (3–5 μM thick) were immunostained. In situ hybridization was performed using antisense locked nucleic acid (LNA)-modified probes (Boster, Wuhan, China) according to the manufacture’s instruction. Oligonucleotide sequences were: LNA-miR-622, 5′-GCTCCAACCTCAGCAGACTGT-3′. The ATF2 antibody for IHC was purchased from Abnova (Taiwan, China). The expression levels of labeling were classified according to the following grading system: staining extensity was categorized as 0 (≤5 % positive cells), 1 (>5 % and ≤25 % positive cells), 2 (>25 % and ≤50 % positive cells) or 3 (>50 % positive cells), and staining intensity was categorized as 0 (negative), 1 (weak), 2 (moderate) or 3 (strong). Each sample was examined separately and scored by two pathologists. Cases with discrepancies in the scores were discussed to reach a consensus.
Invasion assay
Invasion was examined using 24-well BD Matrigel invasion chambers (BD Biosciences) according to the manufacturer’s instructions. 2 × 104 cells were seeded in the upper well of the invasion chamber in DMEM without serum. The lower chamber well contained DMEM supplemented with 10 % FBS to stimulate invasion. After incubation for 24 h, non-invading cells were removed from the top well with a cotton swab while the bottom cells were fixed with 100 % methanol, and stained with 0.1 % crystal violet, and photographed in three independent 10× magnification fields.
Wound healing assay
About 3 × 105 cells were seeded in 6-well dishes and an incision was made in the central area of the confluent culture to create an artificial wound. Images of the wound area were captured by microscope (Leica, Wetzlar, Germany) 24 h after injury.
Luciferase reporter assay
The putative binding sites of miR-622 and its homologous mutation sites in the 3′-UTR region of ATF2 mRNA were amplified and cloned into pGL3-contral luciferase reporter plasmid (Invitrogen, Carlsbad, CA). The pRL vector constitutively expressing Renilla luciferase was used to normalize for trasfection efficiency. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, USA).
Western-blot assay
Cells were lysed in RIPA buffer, and equal amounts of protein were separated by 10 % SDS-PAGE followed by electrotransfer onto a polyvinylidene difluoride membrane (Millipore, MA, USA). The membranes were blocked for 1 h with 5 % nonfat milk and then incubated at room temperature with primary antibodies, followed by incubation with a horseradish peroxidase-conjugated secondary antibody. The bound antibodies were detected by using the enhanced chemiluminescence method (Amersham Biosciences, Uppsala, Sweden). GAPDH was used as an internal control. ATF2 and GAPDH primary antibodies were purchased from Bethyl Laboratories (TX, USA).
Statistical analysis
Data were analyzed via one-way analysis of variance (ANOVA) for 3-group comparisons and two tail student’s t-tests for 2-group comparisons, using SPSS 13.0 soft package. Differences were considered statistically significant at p < 0.05.
Results
miR-622 is decreased in glioma tissues and cell lines
To explore the expression and significance of miR-622 in glioma, we initially measured the expression of miR-622 in 48 glioma (Grade I 7; Grade II 8; Grade III 15; Grade IV 18) and five normal brain tissues using qRT-PCR. The clinicopathologic characteristics are summarized in Supplementary Tables 1 and 2. As shown in Fig. 1a, miR-622 expression was significantly decreased in glioma specimens, especially in GBM, compared with norma brain tissues. To further confirm this, we analyzed the expression of miR-622 in different grades of glioma using ISH staining. Consistently, the results showed a reduction of miR-622 in the glioma tissues compared with normal brain tissues (Fig. 1c, d). Moreover, we assessed the expression levels of endogenous miR-622 in five glioma cell lines (U87, U251, A172, U118 and LN229) and normal human astrocytes (NHAs). miR-622 expression levels in all five glioma cell lines were much lower than those in the NHAs (Fig. 1b). Together, these results suggest that miR-622 was reduced in glioma samples and cell lines.
miR-622 is decreased in glioma tissues and glioma cell lines. a Comparison of miR-622 levels among normal brain tissues (NBTs) and different pathological grades of gliomas using qRT-PCR analysis. *p < 0.05; ***p < 0.001. b qRT-PCR analysis of miR-622 in normal human astrocytes (NHAs) and a series of glioma cell lines. *p < 0.05; **p < 0.01. c In situ hybridization and immunohistochemistry staining analysis of miR-622 levels and ATF2 protein levels in the tissue microarray contained 48 primary glioma specimens and five normal brain tissues. d e Semi-quantitative expression analysis of miR-622 and ATF2 levels in the tissue microarray. *p < 0.05; **p < 0.01
miR-622 suppresses glioma cell invasion, migration and growth
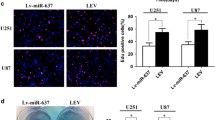
A recent study has reported that miR-622 is associated with tumor metastatic capability in human cancers [19]. To examine whether miR-622 could regulate glioma cell invasion and migration, we performed in vitro gain-of-function analysis by overexpressing miR-622 with a lentiviral vector in U87 and U251 cells. qRT-PCR analysis verified that miR-622 was remarkably increased in cells transfected with miR-622 mimics compared to the scramble control (Fig. 2a). Transwell invasion assays were performed on the miR-622-transduced cells. We found that ectopic expression of miR-622 significantly reduced the invasion of U87 and U251 in transwell assays with matrigel (Fig. 2b, c). Moreover, the wound healing assay was carried out to demonstrate the function of miR-622 on glioma cell migration. Overexpression of miR-622 markedly suppressed the migration of U87 and U251 cells (Fig. 2d, e). By contrast, depleting endogenous miR-622 with antisense oligonucleotides increased the invasion and migration of U87 and U251 cells (Fig. 2a–e). Parallel experiments were performed in A172, U118 and LN229 cells, and similar trends were observed (Supplementary Figs. 1, 2). Furthermore, we examined the effect of miR-622 on proliferation and observed that forced expression of miR-622 in U87 and U251 cells induced inhibition of cell growth (Supplementary Fig. 3a, b). Collectively, these results suggest that miR-622 may suppress glioma cell invasion, migration and growth in vitro.
miR-622 suppresses glioma cells invasion and migration in vitro. a qRT-PCR analysis of the expression level of miR-622 in U87 and U251 cells transfected with miR-622 mimics, miR-NC, anti-miR-622 oligonucleotide, or anti-miR-NC. *p < 0.05; **p < 0.01. b Representative images of transwell invasion assay using U87 and U251 cells stably expressing miR-622 or mi-NC or transiently transfected with the miR-622 inhibitor or NC. c The quantification of transwell invasion assay is shown. *p < 0.05; **p < 0.01; d Representative images of wound healing assay using U87 and U251 cells stably expressing miR-622 or mi-NC or transiently transfected with the miR-622 inhibitor or NC inhibitor. e The quantification of wound healing assay is shown.*p < 0.05; **p < 0.01
ATF2 is a direct target of miR-622. a Predicted miR-622 target sequence in wild-type and mutant (red) ATF2-3′-UTR. b Luciferase assay of the indicated cells transfected with pGL3-ATF2-3′-UTR-WT or pGL3- ATF2-3′-UTR-mutant reporter with miR-622 mimic. **p < 0.01. c Western blot analysis of lysates from miR-NC-transfected, miR-622-transfected, anti-miR-NC-treated or anti-miR-622-treated U87 and U251 cells probed with ATF2 antibody. GAPDH served as the loading control
miR-622 downregulates ATF2 through interaction with its 3′-untranslated region
To explore the underlying molecular mechanism by which miR-622 suppresses glioma cell invasion and migration, we searched for miR-622 targets using three algorithms, including TargetScan, miRanda, and miRGen. We found that ATF2, an important transcriptional factor relating to its positive roles in cancer cell invasion and.
Metastasis [20], was a putative target of miR-622. To determine whether ATF2 is a direct target of miR-622, we cloned the 3′-UTR fragments of ATF2 containing the potential miR-622 binding sites or mutant binding sites into a luciferase construct (Fig. 3a). A remarkable reduction of luciferase activity upon miR-622 transfection was observed in U87 and U251 cells when the ATF2 plasmid containing wild-type 3′-UTR was present (Fig. 3b). However, mutations in the potential miR-622 binding sites in the ATF2 3′-UTR abrogated this suppressive function of miR-622. Moreover, western blot analysis showed that the ectopic expression of miR-622 substantially suppressed ATF2 expression in U87 and U251 cells (Fig. 3c). In contrast, depleting endogenous miR-622 with anti-miR-622 oligonucleotides enhanced the expression of ATF2 in the two glioma cells (Fig. 3c). Taken together, these results suggested that ATF2 is a bona fide target of miR-622.
Inhibition of ATF2 is involved in the reduced invasive capability of miR-622 overexpression
Previous studies have suggested that ATF2 plays a key role in modulating invasion and migration of human cancers [21, 22]. To examine the effect of ATF2 on glioma cell invasion and migration, U251 cells were infected with lentiviral constructs containing siRNA against ATF2 or the negative control (Fig. 4a). Downregulation of ATF2 repressed the invasion and migration of U251 cells (Fig. 4b–e). Of note, the reduced invasive capacity of ATF2 knockdown mediated by siRNA was similar to those of ectopic miR-622 expression, suggesting that downregulation of ATF2 might be a mechanism of the reduction of invasion and migration ability of miR-622 in glioma cells. To test this hypothesis, we co-transfected U251 cells with miR-622 and ATF2 plasmid (without 3′-UTR) (Fig. 4f). Both transwell assay and wound healing assay indicated that restoration of ATF2 expression could partially abrogate miR-622-mediated suppression of cell invasion and migration (Fig. 4g–j). Moreover, the increased invasion and migration capability were rescued by suppression of ATF2 in miR-622-depleted cells (Fig. 5a–e). These results further support the notion that ATF2 is a direct and functional target of miR-622.
Knockdown of ATF2 is involved in the miR-622-induced inhibition of glioma cell invasion and migration. a Western blot analysis of ATF2 expression in U251 cells after knockdown of ATF2. GAPDH served as the loading control. b Representative images of transwell invasion assay with U251 cells transfected with siATF2 or siNC. c The quantification of transwell invasion assay is shown. **p < 0.01. d The quantification of wound healing assay is shown.*p < 0.05. e Representative images of wound healing assay using U251 cells transfected with siATF2 or siNC. f The rescue experiment was performed by introducing pcDNA3.1-ATF2 or Mock (empty vector, pcDNA3.1) in the presence or absence of ectopic miR-622 or miR-NC expression in U251 cells. Western blot analysis of ATF2 in the indicated cells. GAPDH was used as the loading control. g, h Transwell invasion assay of the above cells were analyzed. Columns are the average of three independent experiments. *p < 0.05, **p < 0.01. i, j Wound healing assay of the above cells were analyzed. Columns are the average of three independent experiments. *p < 0.05
ATF2 is involved in the miR-622 inhibition-induced increased invasion and migration of glioma cells. a The rescue experiment was performed by introducing siATF2 or siCtrl in the presence or absence of miR-622 inhibitor or control inhibitor in U251 cells. Western blot analysis of ATF2 in the indicated cells. GAPDH was used as the loading control. b, c Transwell invasion assay of the above cells were analyzed. Columns are the average of three independent experiments. *p < 0.05, **p < 0.01. d, e Wound healing assay of the above cells were analyzed. Columns are the average of three independent experiments. **p < 0.01
miR-622 and ATF2 are inversely expressed in glioma specimens
To address the clinical significance of the miR-622-ATF2 interaction in gliomas, we performed immunohistochemistry using the tissue microarrays contained 48 glioma specimens and five normal brain tissues. Immunohistochemistry staining revealed that ATF2 was significantly higher in glioma tissues than in non-tumor brain tissues (Fig. 1c, e). Moreover, statistical analysis revealed that ATF2 expression was inversely correlated with miR-622 expression in glioma specimens (Supplementary Table 3). Taken together, these results suggest that ATF2 is elevated in glioma tissues and that its enhancement is correlated with reduced miR-622.
Discussion
The invasion of malignant glioma cells into regions of the normal brain is a major cause for the dismal prognosis of malignant gliomas, but the underlying mechanisms remain incompletely understood [23]. Recent studies have reported that miRNAs play a critical role in invasion and metastasis of human cancers, including gliomas. In the present study, we investigated the biological role of miR-622 in human glioma invasion and migration.
Previous studies have suggested that miR-622 is significantly decreased in lung cancer and working as a tumor suppressor via targeting K-ras [18]. Moreover, it has been reported that miR-622 has been associated with aggressiveness and metastasis in gastric cancers [19]. In this study, we analyzed the expression of miR-622 in several glioma cell lines and NHAs using qRT-PCR. miR-622 expression was significantly decreased in all tested glioma cell lines compared with NHAs. Furthermore, the results obtained from clinical glioma tissues showed that miR-622 was downregulated in glioma specimens. Subsequently, we showed that restoration of miR-622 caused a significant reduction in cell migration and invasion abilities, suggesting it functions as a tumor suppressor and is involved in glioma invasion.
ATF2 is one of 16 members of the ATF and CREB group of bZIP transcription factors which contribute to multiple cellular functions, such as cell cycle, proliferation, and invasion [24]. ATF2 contains Sp1 elements and a CRE-like element in promoter region −50 to +90 that is essential for basal promoter activity. Recently, ATF2 has been implicated in a transcriptional response leading to tumor cell migration, invasion and progression. Moreover, ATF2 is crucial for MMP-2 promoter activity as well as induction of invasive and migrative phenotypes in breast epithelial cells [22]. However, whether ATF2 is involved in glioma invasion remains unknown. In our study, we demonstrated that ATF2 is upregulated in glioma tissues. Knockdown of ATF2 by siRNA significantly suppresses the invasion and migration potentials of glioma cell lines, which is similar with the role of miR-622 overexpression. Furthermore, exogenous expression of miR-622 in glioma cells downregulated the expression of ATF2 protein. Additionally, luciferase reporter assays revealed that miR-622 could directly target the 3′-UTR of ATF2 mRNA. Finally, ATF2-induced cell migration and invasion were reversed by miR-622. Our findings establish a functional connection between miR-622 and ATF2, and confirm that miR-622 functions as an anti-invasive miRNA in glioma cells by targeting ATF2.
In conclusion, our study demonstrates, for the first time, the role of miR-622 in suppressing glioma invasion and migration mediated by ATF2. Moreover, miR-622 expression inversely correlates with ATF2 in glioma patients. Our findings reveal that loss of miR-622 consequences in gaining expression of oncogene ATF2, which in turn favors glioma invasion. This novel miR-622/ATF2 axis provides new insight into the mechanisms underlying glioma invasion, and targeting miR-622/ATF2 may be a potential therapeutic strategy for the treatment of patients with malignant gliomas.
References
Behin A, Hoang-Xuan K, Carpentier AF, Delattre JY (2003) Primary brain tumours in adults. Lancet 361:323–331
Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21:2683–2710
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359:492–507
Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO, Treatment EOR, Grp CBT, Grp RO, N.C.I.C.C. Trials (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10:459–466
Giese A, Bjerkvig R, Berens ME, Westphal M (2003) Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol 21:1624–1636
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
MacFarlane LA, Murphy PR (2010) MicroRNA: biogenesis function and role in cancer. Curr Genomics 11:537–561
Hwang HW, Mendell JT (2006) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 94:776–780
Yang LQ, Li Q, Wang QX, Jiang Z, Zhang L (2012) Silencing of miRNA-218 promotes migration and invasion of breast cancer via Slit2-Robo1 pathway. Biomed Pharmacother 66:535–540
Kloosterman WP, Plasterk RHA (2006) The diverse functions of MicroRNAs in animal development and disease. Dev Cell 11:441–450
Delic S, Lottmann N, Stelzl A, Liesenberg F, Wolter M, Gotze S, Zapatka M, Shiio Y, Sabel MC, Felsberg J, Reifenberger G, Riemenschneider MJ (2014) MiR-328 promotes glioma cell invasion via SFRP1-dependent Wnt-signaling activation. Neuro Oncol 16:179–190
Wang XR, Luo H, Li HL, Cao L, Wang XF, Yan W, Wang YY, Zhang JX, Jiang T, Kang CS, Liu N, You YP (2013) Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro Oncol 15:1491–1501
Yan-nan B, Zhao-yan Y, Li-xi L, Jiang Y, Qing-jie X, Yong Z (2014) MicroRNA-21 accelerates hepatocyte proliferation in vitro via PI3K/Akt signaling by targeting PTEN. Biochem Biophys Res Commun 443:802–807
Li S, Xu X, Hu Z, Wu J, Zhu Y, Chen H, Mao Y, Lin Y, Luo J, Zheng X, Xie L (2013) MicroRNA-490-5p inhibits proliferation of bladder cancer by targeting c-Fos. Biochem Biophys Res Commun 441:976–981
Wu H, Xiao Z, Wang K, Liu W, Hao Q (2013) MiR-145 is downregulated in human ovarian cancer and modulates cell growth and invasion by targeting p70S6K1 and MUC1. Biochem Biophys Res Commun 441:693–700
Ma Y, Qin H, Cui Y (2013) MiR-34a targets GAS1 to promote cell proliferation and inhibit apoptosis in papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem Biophys Res Commun 441:958–963
Deng J, Lei W, Fu JC, Zhang L, Li JH, Xiong JP (2014) Targeting miR-21 enhances the sensitivity of human colon cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys Res Commun 443:789–795
Han Z, Yang Q, Liu B, Wu J, Li Y, Yang C, Jiang Y (2012) MicroRNA-622 functions as a tumor suppressor by targeting K-Ras and enhancing the anticarcinogenic effect of resveratrol. Carcinogenesis 33:131–139
Guo XB, Jing CQ, Li LP, Zhang L, Shi YL, Wang JS, Liu JL, Li CS (2011) Down-regulation of miR-622 in gastric cancer promotes cellular invasion and tumor metastasis by targeting ING1 gene. World J Gastroenterol 17:1895–1902
Lopez-Bergami P, Lau E, Ronai Z (2010) Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat Rev Cancer 10:65–76
Kim ES, Sohn YW, Moon A (2007) TGF-beta-induced transcriptional activation of MMP-2 is mediated by activating transcription factor (ATF)2 in human breast epithelial cells. Cancer Lett 252:147–156
Song H, Ki SH, Kim SG, Moon A (2006) Activating transcription factor 2 mediates matrix metalloproteinase-2 transcriptional activation induced by p38 in breast epithelial cells. Cancer Res 66:10487–10496
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Vlahopoulos SA, Logotheti S, Mikas D, Giarika A, Gorgoulis V, Zoumpourlis V (2008) The role of ATF-2 in oncogenesis. BioEssays 30:314–327
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81201978, No. 81172389, No. 81200362, No. 81372709, No. 81272792), the Jiangsu Province’s Natural Science Foundation (BK2012483, BK2010580), the Program for Advanced Talents within Six Industries of Jiangsu Province (2012-WSN-019), the National High Technology Research and Development Program of China (863) (2012AA02A508), International Cooperation Program (2012DFA30470), the Jiangsu Province’s Key Provincial Talents Program (RC2011051), the Jiangsu Province’s Key Discipline of Medicine (XK201117), the Jiangsu Provincial Special Program of Medical Science (BL2012028), the Program for Development of Innovative Research Team in the First Affiliated Hospital of NJMU, the Provincial Initiative Program for Excellency Disciplines, Jiangsu Province and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Rui Zhang, Hui Luo, Shuai Wang, Zhengxin Chen and Lingyang Hua have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.



Rights and permissions
About this article
Cite this article
Zhang, R., Luo, H., Wang, S. et al. miR-622 suppresses proliferation, invasion and migration by directly targeting activating transcription factor 2 in glioma cells. J Neurooncol 121, 63–72 (2015). https://doi.org/10.1007/s11060-014-1607-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-014-1607-y