Abstract
To identify therapeutic molecular targets for glioma, we performed modified serological identification of antigens by recombinant complementary DNA (cDNA) expression cloning using sera from a mouse glioma model. Two clones, kinesin family member 23 (Kif23) and structural maintenance of chromosomes 4 (Smc4), were identified as antigens through immunological reaction with sera from mice harboring synergic GL261 mouse glioma and intratumoral inoculation with a mutant herpes simplex virus. The human Kif23 homolog KIF23 is a nuclear protein that localizes to the interzone of mitotic spindles, acting as a plus-end-directed motor enzyme that moves antiparallel microtubules in vitro. Expression analysis revealed a higher level of KIF23 expression in glioma tissues than in normal brain tissue. The introduction of small interfering RNA (siRNA) targeting KIF23 into two different glioma cell lines, U87MG and SF126, downregulated KIF23 expression, which significantly suppressed glioma cell proliferation in vitro. KIF23 siRNA-treated glioma cells exhibited larger cell bodies with two or more nuclei compared with control cells. In vivo analysis using mouse xenograft showed that KIF23 siRNA/DNA chimera-treated tumors were significantly smaller than tumors treated with control siRNA/DNA chimera. Taken together, our results indicate that downregulation of KIF23 decreases proliferation of glioma cells and that KIF23 may be a novel therapeutic target in malignant glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite the development of therapeutic modalities, including surgery, chemotherapy, and radiotherapy, limited progress has been achieved in the treatment of glioblastoma (GBM), the most malignant form of glioma. Most patients with GBM die within approximately 12 months of diagnosis [1–3]. GBM has a high rate of cellular proliferation and marked propensity to invade surrounding brain parenchyma. Such aggressive and invasive growth is the hallmark feature that gives rise to its high morbidity and mortality [4]. GBM can rarely be surgically resected. Therefore, the molecular mechanisms driving tumor proliferation must be understood to exploit them as targets for new glioma therapies.
In the present study, we conducted serological identification of antigens by recombinant cDNA expression (SEREX) in an attempt to discover novel tumor antigens, and identified six genes potentially coding candidate proteins for novel tumor antigens. Among these six genes, we focused on Kif23 due to its specific immune reactivity toward serum from tumor-bearing, immune-precipitated mouse and also because of its preferable expression in glioma.
KIF23, a human homolog of mouse Kif23, is a nuclear protein that localizes to the interzone of mitotic spindles and acts as a plus-end-directed motor enzyme that moves antiparallel microtubules in vitro [5]. Previous studies have reported that depletion of KIF23 in HeLa cells induces formation of multinucleate cells, likely because of a cytokinesis defect [6, 7]. Microtubules, for which KIF23 acts as the motor enzyme [5], have been known to control cellular shape and processes such as motility, mitosis, intracellular vesicle transport, organization, and positioning of membranous organelles [8–11]. Due to these molecular functions, microtubules have been adopted as one of the major targets in cancer chemotherapy [8]. Taking together the facts that KIF23 acts as a motor enzyme of microtubules, that KIF23 showed preferable expression in gliomas compared with normal brain controls, and that SEREX identified KIF23 as a candidate for novel glioma antigen, we considered that KIF23 might perform an important role in gliomagenesis. To date, no report on the relationship between KIF23 and glioma has appeared in the literature. In this study, we continued evaluating the expression of KIF23 in glioma cells and investigated the role of KIF23 in these cells by using an RNA interference (RNAi) technique.
Materials and methods
Tissue samples and cell lines
Brain tumor samples were obtained from the Department of Neurosurgery, Keio University, School of Medicine. Written informed consent for the study was obtained from all patients, and the study was approved by the ethical review board of Keio University (no. 12-21-2). Human adult brain tissue samples were obtained from Biochain (Hayward, CA). Normal human RNA samples were obtained from Biochain and Clontech (Palo Alto, CA). The human GBM cell lines SF126, KNS81, T98G, KNS42, and U87MG, human leukemia cell line K562, mouse brain neoplasm cell line GL261, and African green monkey kidney cell line Vero were maintained in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO, Grand Island, NY) supplemented with 10% fetal bovine serum and penicillin–streptomycin in a humidified incubator at 37°C with 5% CO2.
Viruses
Herpes simplex virus-1 (HSV-1) mutant strain G207 was used in this study. The construction of G207 has been described elsewhere [12]. This mutant virus contains deletions in the ICP34.5 and ICP6 genes. Viruses were propagated and titered in Vero cells.
Glioma model and HSV vaccination
Female C57BL/6 mice (6–8 weeks old) were purchased from Japan SLC, Inc. (Tokyo, Japan). The Animal Care and Use Committee of the School of Medicine, Keio University, approved all the animal procedures. GL261 tumor cells (1 × 106) were injected subcutaneously into both flanks of the mice. Once the subcutaneous tumors reached a maximum diameter of 5–7 mm, the tumors on the right flanks of the mice (day 0) were inoculated with either G207 (1.5 × 107 plaque-forming units) in 50 μl virus buffer [150 mM NaCl and 20 mM Tris (pH 7.5)] and DMEM with 1% inactivated fetal calf serum (IFCS) (1:1) or 50 μl of a “mock-infected extract” prepared from mock-infected cells created using the same procedure as that for the preparation of virus inocula. The right flank tumors were inoculated with G207 five times, once every 3 days, for a total of 15 days. Mice treated with G207 were killed 7 days after the last virus inoculation, and sera were prepared.
Immunoscreening and characterization of immunoreactive clones
Each serum was diluted 1:100 with 5% skim milk in Tris-buffered saline with 0.05% polyoxyethylene sorbitan monolaurate. Sera were pre-absorbed with transformed Escherichia coli lysates and E. coli infected with lambda ZAP phage (Stratagene, La Jolla, CA). Three sera samples were prepared, mixed, and diluted to 1:300. A mouse testis cDNA library (Stratagene) was expressed with BL21 (DE3) pLysE, and the colonies were transferred to nitrocellulose membranes (Hybond-C; Amersham Pharmacia, Buckinghamshire, England). The membranes were incubated with the diluted sera for 4 h at room temperature, followed by incubation in 1:4,000 diluted goat anti-mouse immunoglobulin G (IgG) antibody conjugated with alkaline phosphatase (Cappel, Aurora, OH). Nitro blue tetrazolium (Boehringer Mannheim, GmbH, Germany) and 5-bromo-4-chloro-3-indolyl phosphate (Sigma Chemical Co., St Louis, MO) were used for enzymatic detection of bound secondary antibodies. Positive plaques were picked from the plates and purified through secondary and tertiary rounds of additional screening. The purified cDNAs were amplified via polymerase chain reaction (PCR) using Ex Taq (Takara Shuzo, Shiga, Japan) and sequenced using the Big Dye Terminator Cycle Sequencing Ready Reaction Kit and an ABI Prism automated sequencer (PerkinElmer, Branchburg, NJ). The sequenced DNAs were analyzed with a basic local alignment search tool (BLAST) search of genetic databases at the National Center for Biotechnology Information (NCBI).
Detection of antibodies against candidate genes using SEREX
Monoclonal phages from each clone that was reacted with IgG antibodies in the serum were mixed with nonreactive phages of the cDNA library as internal negative controls at ratio of 1:10 and expressed with BL21 (DE3) pLysE. The colonies were transferred to nitrocellulose membranes. IgG antibodies in the diluted (1:100) pre-absorbed E. coli sera from HSV-treated mice, HSV-treated GL261-harboring mice, or GL261-harboring untreated mice were screened to determine antibody responses to candidate genes.
Quantitative PCR analysis
cDNA was synthesized from 10 μg total RNA using reverse transcriptase XL (AMV) (Takara Bio, Tokyo, Japan). The primers were designed as follows: for KIF23, forward primer, 5′-CTGACCCAGAGCAAAGCTTTC-3′, and reverse primer, 5′-GTTCTAAAGTGCATTCTTGCAGC-3′; for glyceraldehyde 3-phosphate dehydrogenase (GAPDH), forward primer, 5′-CCCACTCCTCCACCTTTGAC-3′, and reverse primer, 5′-ATGAGGTCCACCACCCTGTT-3′. Quantitative reverse-transcriptase polymerase chain reaction (PCR) analysis was performed using SYBR Green (PerkinElmer, Foster City, CA) and the ABI prism 7700 Sequence Detection System (PerkinElmer). The thermal cycler conditions were as follows: 10 min at 95.0°C, 50 cycles of 95.0°C for 15 s, and 1 min at 60.0°C. The threshold cycle value was defined as the value obtained in the PCR cycle when the fluorescence signal increased above the background threshold.
Western blot analysis
Cell lysates were prepared using a radioimmunoprecipitation assay buffer (Thermo Scientific, Rockford, IL) containing protease inhibitors (Cocktail Tablet; Roche Diagnostics, Tokyo, Japan). Lysates were centrifuged at 14,000 × g for 10 min at 4°C, and the protein concentration of each sample was determined using the Bio-Rad protein assay kit (Bio-Rad Laboratories Inc., Hercules, CA) with bovine serum albumin as the standard. Identical amounts of the proteins were electrophoresed in 4–15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes. Blots were blocked with Blocking One™ (Nacalai Inc., Kyoto, Japan) at room temperature and incubated with either a rabbit anti-MKLP1 (KIF23) antibody (1:500; Santa Cruz Biotechnology Inc., Santa Cruz, CA) or a mouse anti-β-actin antibody (1:4,000; Sigma, St. Louis, MO) overnight at 4°C. After being washed three times in Tris-buffered saline Tween 20 (20 mM Tris–HCl, 150 mM NaCl, and 0.02% Tween 20; pH 7.4), the blots were incubated with the secondary antibody conjugated with horseradish peroxidase (1:4,000; anti-rabbit, MBL International Co., Woburn, MA; anti-mouse, GE Health Care Biosciences, Waukesha, WI) for 1 h at room temperature. Signals were detected with an enhanced chemiluminescence system (GE Health Care Biosciences) and exposed to Hyperfilm (GE Health Care Biosciences).
RNA interference
Two short interference RNA (siRNA) oligonucleotide sequences for KIF23 were used: siRNA1 (sense strand, 5′-GACUAUAUCUAGAUCAUGUCU-3′; antisense strand, 5′-ACAUGAUCUAGAUAUAGUCUU-3′) and siRNA2 (sense strand, 5′-GAGUGUUGCAUAGAAGUGAAA-3′; antisense strand, 5′-UCACUUCUAUGCAACACUCAA-3′). Predesigned RNAi (sense strand, 5′-GUACCGCACGUCAUUCGUAUC-3′; antisense strand, 5′-UACGAAUGACGUGCGGUACGU-3′) was used as the negative control. The final concentration of 10 nM siRNA was incubated with Lipofectamine RNAiMax (Invitrogen, CA) according to the manufacturer’s instructions. Cells were maintained for 24–120 h before experimentation.
Cell viability assay
Cell viability was evaluated using the CellTiter Glo® luminescent cell viability assay (Promega, Madison, WI) according to the manufacturer’s instructions, using a luminometer (Wallac ARVO 1420 multilabel counter; WALLAC OY, Truku, Finland). Each experiment was performed in triplicate.
Immunofluorescence
SF126 cells were seeded on chamber slides (Nunc, Kamstrup, Denmark) in DMEM containing 10% fetal bovine serum and placed in eight-well plates at density of 1 × 104/well. Twelve hours later, KIF23 and control siRNAs were transfected, and the cells were cultured for 48 h. The cells were then fixed, permeabilized, blocked, and incubated with 1:50 diluted anti α/β-tubulin antibody (Cell Signaling Technology, Beverly, MA). The cells were then incubated with Alexa-Fluor® anti-rabbit IgG. Finally, they were treated with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min for nuclear staining. The slides were washed, mounted, and imaged using fluorescence microscopy (Axioplan 2 imaging; Carl Zeiss, Japan).
In vivo analyses of tumors administered siRNA/DNA
A total of 2.5 × 106 U87MG cells suspended in 0.2 ml saline were subcutaneously inoculated into the lower flank of 7-week-old nude mice [BALB/c (nu/nu)] (CLEA, Tokyo, Japan). When the tumors reached approximately 5 mm in size (day 0), the mice were randomly divided into two groups and treated with either KIF23-targeting or control siRNA/DNA (2.5 nM each). The same amount of siRNA/DNA was injected into the same region 7 and 14 days after the first injection. The modified siRNAs (DNA-RNA chimeras) were designed according to the method of Ui-Tei et al. [13]. Each target sequence was as follows: KIF23, 5′-rGrArGUrGUUrGrCrAUrArGAAGTGAAA; control, 5′-rGUrArCrCrGrCrArCrGUrCrATTCGTATC. Tumor diameters were measured at regular intervals with digital calipers, and the tumor volume in cubic mm3 was calculated according to the following formula: volume = 4/3π(width/2)2 × length/2. The tumors were removed surgically at 3 weeks after the first injection of siRNA/DNA and were embedded in Tissue-Tek OCT compound (Sakura Finetechnical Corp., Tokyo, Japan) and then frozen. Cryostat sections (20 μm) were prepared for hematoxylin and eosin (H&E) staining.
In silico REMBRANDT analysis
The relationship between patients’ prognosis and expression intensity of the target gene was analyzed in silico using National Cancer Institute’s (NCI’s) REMBRANDT database [14] (http://rembrandt.nci.nih.gov, accessed 9 July 2011).
Statistical analysis
Data are expressed as mean ± standard error (SE). Statistical significance was determined with one-way analysis of variance and a Tukey post-test method or Student’s t-test using Prism software (GraphPad, San Diego, CA). P values lower than 0.05 were regarded as statistically significant.
Results
Immunoreactive clones
Approximately 1 × 106 clones from the mouse testis cDNA library were screened with SEREX, using mixed sera samples of three GL261-implanted mice after intratumoral inoculation with HSV. After subsequent sequencing of positive clones, six clones that encoded an open reading frame were identified using the BLAST database (NCBI; Table 1). We further evaluated the immune response of the clones to the following mouse sera: normal male or female mice, mice with subcutaneously implanted GL261, and mice with subcutaneously implanted GL261 and HSV treatment. The screening showed that two clones, Kif23 (gene bank accession number Mm.259374) and Smc4 (Mm.206841), reacted exclusively with sera from GL261-bearing mice treated with HSV (Table 2). The expression of SMC4, a human homolog of mouse SMC4, was ubiquitous in the NCBI expressed sequence tags database (http://www.ncbi.nlm.nih.gov/UniGene, accessed 25 March 2011). In contrast, the expression of KIF23, a human homolog of mouse Kif23, in the NCBI database was higher in glioma tissue (9 transcripts/million) than in normal brain tissue (1 transcript/million) (http://www.ncbi.nlm.nih.gov/UniGene, accessed 25 March 2011). Previous reports have indicated the functional relation of KIF23 to cytokinesis [7, 15–19]. Therefore, we focused on further analysis of KIF23 in this study.
Expression pattern of KIF23
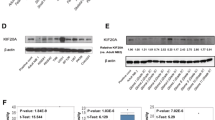
Quantitative PCR analysis using a cDNA panel containing normal human tissue (Fig. 1a) showed that KIF23 was more highly expressed in the testis and stomach than in normal brain. Western blot analysis was performed using lysate from K562 human leukemia cells as a positive control. KIF23 has two isoforms: a 110-kDa isoform 1 and a 98-kDa isoform 2 (http://www.uniprot.org/uniprot/, accessed 11 February 2011) [20, 21]. According to the manufacturer of the antibody for KIF23 used in this study, both isoforms can be detected. Our results showed two clear bands, corresponding to the two isoforms. KIF23 was expressed in all 11 glioma tissue samples evaluated, but was expressed to a lesser extent in control samples from normal brain tissue (Fig. 1b). Although it cannot be statistically evaluated, no correlation is apparent between the World Health Organization tumor grading and level of expression. We also conducted a Western blot analysis using five glioma cell lines (U87MG, KNS42, T98G, KNS81, and SF126) and confirmed the expression of KIF23 in all five glioma cell line samples evaluated (Fig. 1c). Among the five cell line samples evaluated, the expression of KIF23 was especially high in samples from SF126 and U87MG, and we conducted functional analyses of KIF23 in glioma using those two cell lines.
Expression of KIF23 in gliomas. Quantitative PCR analysis of the KIF23 gene in normal human tissues (a). The relative KIF23 expression level was normalized to the GAPDH level in each sample and calculated as the threshold cycle (CT) value in each sample divided by the CT value in normal brain. Mean ± standard error of the mean (SEM) for at least three experiments are shown. Western blot analysis of the KIF23 protein in normal brain, glioma tissues (b), and glioma cell lines (c) with β-actin as the control. d Effects of KIF23 short interfering RNA (siRNA) on KIF23 expression. Western blot analysis of KIF23 protein expression in U87MG and SF126 cells treated with mock, negative control siRNA, and two KIF23 siRNAs. Cell lysate of K562 cells was used as a positive control
Effect of KIF23 siRNA on KIF23 expression in glioma cells
To evaluate the function of KIF23 protein in glioma cells, we designed two different siRNAs to downregulate the gene expression of KIF23. To confirm the effect of KIF23 siRNA on KIF23 protein expression in the glioma cell lines U87MG and SF126, we performed Western blot analysis (Fig. 1d). Transfection with two independent siRNAs targeting KIF23 was capable of downregulating the expression of KIF23 in these glioma cells 72 h after transfection in both glioma cell lines evaluated.
Inhibition of glioma cell proliferation by siRNA-mediated downregulation of KIF23 expression
Cell number was observed after siRNA transfection into SF126 and U87MG cells. There seemed to be fewer cells present after transfection with either of the two KIF23 siRNAs compared with their control counterpart (data not shown). So, to evaluate the relationship between cell proliferation and KIF23 expression, we conducted a cell viability assay using the CellTiter Glo® luminescent cell viability assay. After KIF23 siRNA treatment, the proliferation of both glioma cell lines, U87MG and SF126, was significantly suppressed compared with control cells at 5 days after transfection (Fig. 2a, b).
Effect of KIF23 siRNAs on glioma cell proliferation in vitro: a U87MG cells and b SF126 cells. Cells were treated with siRNAs for 24–120 h, and cell viability was determined using a CellTiter Glo luminescent cell viability assay. Results shown are mean ± SEM (bars) of three experiments. n.s. not significant, ***P < 0.001 using one-way analysis of variance with a Tukey post-test method
Change in glioma cell morphology by downregulation of KIF23
Previous reports showed that KIF23 is a motor enzyme that moves antiparallel microtubules [5] and depletion of KIF23 in HeLa cells induces the formation of multinucleate cells, likely because of a cytokinesis defect [6, 7]. To identify the function of KIF23 in glioma cells, we examined the effect of downregulation of KIF23 on cellular shapes in SF126 cells by staining microtubule with α/β-tubulin and nucleus by DAPI. Typical fluorescent images of the stained cells (blue, nucleus; green, α/β-tubulin) are shown in Fig. 3. More KIF23 siRNA-treated glioma cells than control cells exhibited large cell bodies with two or more nuclei. KIF23 siRNA-treated glioma cells can be seen as not able to complete cytokinesis properly and enlarged.
Effect of KIF23 siRNA/DNA chimeras on the growth of a U87MG mouse xenograft tumor model
To examine the effect of KIF23 depletion on tumor growth in vivo, we used a xenograft model in which the U87MG human glioma cells were subcutaneously implanted into nude mice. When the average tumor size reached approximately 5 mm in size, KIF23 siRNA/DNA or control siRNA/DNA was injected into the tumors weekly for 3 weeks, and the tumor size was measured periodically (Fig. 4a). Under control siRNA/DNA treatment, U87MG cells developed into prominent tumors, and the average tumor volume at 3 weeks from initial injection was 908 mm3. The administration of KIF23 siRNA/DNA statistically significantly inhibited the growth of U87MG tumors (Student t-test; P = 0.0354). The average tumor volume at 3 weeks was 529 mm3 (Fig. 4).
Effect of KIF23 siRNA/DNA chimeras on the growth of a U87MG mouse xenograft tumor model. a Tumor volume measurement. Results shown are mean ± SEM (bars) (control, n = 4; KIF23 siRNA/DNA-chimeras treated, n = 6). Arrows: KIF23 siRNA/DNA chimera injection. After implantation, one set of mice was injected with control siRNA/DNA and another set with KIF23 siRNA/DNA, at day 0, 7, and 14. *P < 0.05 using Student’s t-test. b 2.5 × 106 U87MG cells were subcutaneously inoculated in nude mice (3 weeks after initial injection). c Representative H&E staining of tumor developed in nude mice from each set. Scale bar 1 mm
Relationship between clinical prognosis of glioma patients and expression value of KIF23
To examine the relationship between clinical prognosis of glioma patients and KIF23 expression value, we used the NCI REMBRANDT database [14]. According to the result obtained from the REMBRANDT database, patients with highly KIF23 messenger RNA (mRNA)-expressing glioma (n = 171) tend to show statistically poorer prognosis compared with patients with intermediately KIF23 mRNA-expression glioma (n = 165, P = 0.00321). Patients with highly KIF23 mRNA-expressing glioma also tend to show poorer prognosis compared with patients with low KIF23 mRNA-expression glioma; however, this was not statistically significant (P = 0.357), probably due to small sample size of patients with low KIF23 mRNA expression (n = 7) (Fig. 5).
Correlation between glioma patient survival and KIF23 mRNA expression, red indicates high KIF23 expression, yellow indicates intermediate KIF23 expression, green indicates low KIF23 expression, and blue indicates all patients (http://rembrandt.nci.nih.gov. accessed 2011 July 9)
Discussion
Serological identification of antigens by recombinant expression cloning (SEREX) has been developed for the discovery of novel tumor antigens [22]. SEREX can isolate antigens recognized by both B and T cells; thus, it can be used to identify antigenic targets [22]. For example, NY-ESO-1, a tumor-specific antigen in the cancer/testis antigen group that is expressed in various types of tumors, was identified using SEREX [23]. Several candidate anticancer vaccines using NY-ESO-1-based immunogens are now under clinical trial [24–26]. Several candidate glioma antigens including Sox6 [27] have also been isolated using SEREX. Thus, SEREX is an effective method for the identification of useful tumor antigens.
Several lytic viruses have been used in an attempt to induce antitumor immunoreactions by modifying tumor cells [28]. Most of these cancer vaccines have included oncolysates or membranes from tumor cells infected with the vaccinia virus [29], parainfluenza virus [30], Newcastle disease virus [31], etc. Accumulating evidence suggests, however, that the immunogenicity of cellular material such as membranes is lower than that of whole tumor cells [31, 32]. To induce a strong antitumor response, we have used a replication-conditional herpes simplex virus (HSV) mutant, G207, to modify tumor cells directly in situ [33–37]. We have demonstrated that intratumoral inoculation with G207 elicits strong immune responses not only to HSV but also to the tumor antigen [34, 35].
In this study, we used intratumoral administration of G207 to vaccinate mice harboring a syngenic mouse glioma cell line, GL261, which is widely used for the study of immunotherapy against brain tumors [38–41]. We applied a modified SEREX method to the mouse glioma GL261 model in an attempt to identify molecular therapeutic targets for glioma.
Using SEREX, we identified six clones that encode an open reading frame confirmed by BLAST analysis (NCBI; Table 1). We further evaluated the specific immunoreactivity of these clones against glioma by using sera from normal male and female mice, GL261-implanted mice, and mice harboring GL261 treated with HSV. Two of the six clones (Kif23 and Smc4) reacted with only the sera from GL261-harboring mice treated with G207. The fact that we found no positive reaction to the two clones with sera from GL261-implanted mice indicates that GL261 glioma cells may not be immunogenic themselves and that HSV treatment could activate an antitumor response against GL261 in mice. Of the two clones identified, we focused on Kif23, since immunoreaction against Kif23 was detected in all five sera from GL261-bearing mice given HSV treatment, while immunoreaction against Smc4 was detected in two out of five sera from GL261-bearing mice given HSV treatment, and since KIF23, a human homolog of mouse Kif23, showed higher expression in glioma compared with normal brain according to NCBI database (http://www.ncbi.nlm.nih.gov/dbEST, accessed 25 March 2011).
Human Kif23 homolog, KIF23, was firstly identified by Nislow et al. [5] in 1992 as a plus-end-directed motor enzyme that moves antiparallel microtubules in vitro. After this first report, others were published, and KIF23 has been identified as a key regulator of cytokinesis [7, 15–19]. In this study, the KIF23 protein was expressed in all 11 glioma tissue samples evaluated but not in the normal brain controls. We also showed that KIF23 was expressed in five glioma cell lines (U87MG, KNS42, T98, KNS81, and SF126) but not in normal control brains. These findings demonstrate high KIF23 expression in glioma compared with that in the normal brain. The recruitment of KIF23 to the spindle midzone/midbody by a chromosomal passenger protein is essential for midbody formation and completion of cytokinesis in human cells [7]. Zhu et al. [7] have shown that downregulation of KIF23 abrogated midbody formation and completion of cytokinesis. Liu and Erikson have reported that a mutant Kif23 without nuclear localization signals leads to cell cycle arrest, further demonstrating that Kif23 is essential for cytokinesis [42]. These studies suggest that KIF23 is essential for cytokinesis. In addition, microtubules, for which KIF23 acts as the motor enzyme, have been known to control cellular shape and processes such as motility, mitosis, intracellular vesicle transport, organization, and positioning of membranous organelles [8–11], and they have been one of the major targets in cancer chemotherapy [8]. Natural microtubule-targeting agents such as paclitaxel, vinblastine, or vincristine [43–46] have been used in clinical practice for years. In this study, the results of the cell viability assay using CellTiter Glo® luminescent and evaluation of in vivo tumor growth analysis as well showed that downregulation of KIF23 suppressed the growth of glioma cells both in vitro and in vivo. In addition, KIF23 siRNA-treated glioma cells showed larger cell bodies, and were binuclear/multinuclear, likely because of a cytokinesis defect as demonstrated in HeLa cells by others in previous reports [6, 7]. Taken together, downregulation of KIF23 may inhibit glioma proliferation by interrupting cytokinesis via modification of microtubules. Targeting microtubules for treating cancer has been shown to be a promising strategy [43–46]; however, clinical use is limited by hematological and neurological toxicities and by tumor resistance that leads to tumor recurrence [47]. Although further evaluation is required, molecular therapy targeting KIF23 may overcome this problem.
In the present in vivo tumor growth analysis, we used siRNA/DNA, since it has been reported that siRNA/DNA can overcome the disadvantages of usual siRNAs such as the problems of instability [48], induction of silencing of unintended genes (off-target effects) [49], or induction of immune response in vivo [50]. On the other hand, it has previously been reported that gene silencing induced by siRNA/DNA is likely to be less effective than that induced by its unmodified counterparts, because siRNA/DNA exhibits weaker activity for formation of am RNA-induced silencing complex [13, 51]. Considering these facts, we adopted a multiple siRNA/DNA injection protocol (in the present study, thrice, at days 0, 7, and 14) as described elsewhere [51, 52].
As the next step of in vivo study, we also should conduct histological analysis of in vivo experiments in an attempt to identify if there is any morphological or immunohistological difference between KIF23-knockdown tumors and control tumors as shown at the cellular level in in vitro experiments. KIF23 siRNA-treated glioma cells more frequently exhibited large cell bodies with two or more nuclei when compared to control cells. For now, it is not clear whether cells in the KIF23-knockdown tumors also present similar morphological change observed in the in vitro analysis or not. We will conduct further research to reveal the point.
The results of this study show that downregulation of KIF23 by specific RNAi could inhibit glioma proliferation in vitro and in vivo. Although RNAi rescue experiments could facilitate the establishment of a direct link between KIF23 expression and the cellular functions demonstrated in the present study, we designed two siRNA sequences for KIF23 instead and proved that their effects were similar. The present study suggests that KIF23 is indispensable for glioma cytokinesis and, consequently, glioma growth. The expression analysis demonstrated high expression of KIF23 in glioma cells compared with normal brain cells. Therefore, KIF23 may be a novel therapeutic target for GBM.
References
Prados MD, Levin V (2000) Biology and treatment of malignant glioma. Semin Oncol 27(3 Supplement 6):1–10
Takahashi S, Hirose Y, Ikeda E, Fukaya R, Kawase T (2007) Chromosome arm 1q gain associated with good response to chemotherapy in a malignant glioma case report. J Neurosurg 106(3):488–494. doi:10.3171/jns.2007.106.3.488
Takahashi S, Yamada-Okabe H, Hamada K, Ohta S, Kawase T, Yoshida K, Toda M (2010) Downregulation of uPARAP mediates cytoskeletal rearrangements and decreases invasion and migration properties in glioma cells. J Neurooncol 103(2):267–276. doi:10.1007/s11060-010-0398-z
Tabuse M, Ohta S, Ohashi Y, Fukaya R, Misawa A, Yoshida K, Kawase T, Saya H, Thirant C, Chneiweiss H, Matsuzaki Y, Okano H, Kawakami Y, Toda M (2011) Functional analysis of HOXD9 in human gliomas and glioma cancer stem cells. Mol Cancer 10:60. doi:10.1186/1476-4598-10-60
Nislow C, Lombillo VA, Kuriyama R, McIntosh JR (1992) A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature 359(6395):543–547. doi:10.1038/359543a0
Liu X, Zhou T, Kuriyama R, Erikson RL (2004) Molecular interactions of Polo-like-kinase 1 with the mitotic kinesin-like protein CHO1/MKLP-1. J Cell Sci 117(Pt 15):3233–3246. doi:10.1242/jcs.01173
Zhu C, Bossy-Wetzel E, Jiang W (2005) Recruitment of MKLP1 to the spindle midzone/midbody by INCENP is essential for midbody formation and completion of cytokinesis in human cells. Biochem J 389(Pt 2):373–381. doi:10.1042/BJ20050097
Calligaris D, Verdier-Pinard P, Devred F, Villard C, Braguer D, Lafitte D (2010) Microtubule targeting agents: from biophysics to proteomics. Cell Mol Life Sci 67(7):1089–1104. doi:10.1007/s00018-009-0245-6
Vasiliev JM, Gelfand IM, Domnina LV, Ivanova OY, Komm SG, Olshevskaja LV (1970) Effect of colcemid on the locomotory behaviour of fibroblasts. J Embryol Exp Morphol 24(3):625–640
Kline-Smith SL, Walczak CE (2004) Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol Cell 15(3):317–327. doi:10.1016/j.molcel.2004.07.012
Wittmann T, Hyman A, Desai A (2001) The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol 3(1):E28–E34. doi:10.1038/35050669
Mineta T, Rabkin SD, Yazaki T, Hunter WD, Martuza RL (1995) Attenuated multi-mutated herpes simplex virus-1 for the treatment of malignant gliomas. Nat Med 1(9):938–943
Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K (2008) Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res 36(7):2136–2151. doi:10.1093/nar/gkn042
National Cancer Institute (2005) REMBRANDT home page. http://rembrandt.nci.nih.gov. Accessed July 7, 2011
Seguin L, Liot C, Mzali R, Harada R, Siret A, Nepveu A, Bertoglio J (2009) CUX1 and E2F1 regulate coordinated expression of the mitotic complex genes Ect2, MgcRacGAP, and MKLP1 in S phase. Mol Cell Biol 29(2):570–581. doi:10.1128/MCB.01275-08
Li WM, Webb SE, Chan CM, Miller AL (2008) Multiple roles of the furrow deepening Ca2+ transient during cytokinesis in zebrafish embryos. Dev Biol 316(2):228–248. doi:10.1016/j.ydbio.2008.01.027
Neef R, Klein UR, Kopajtich R, Barr FA (2006) Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr Biol 16(3):301–307. doi:10.1016/j.cub.2005.12.030
Kuriyama R, Gustus C, Terada Y, Uetake Y, Matuliene J (2002) CHO1, a mammalian kinesin-like protein, interacts with F-actin and is involved in the terminal phase of cytokinesis. J Cell Biol 156(5):783–790. doi:10.1083/jcb.200109090
Chen MC, Zhou Y, Detrich HW 3rd (2002) Zebrafish mitotic kinesin-like protein 1 (Mklp1) functions in embryonic cytokinesis. Physiol Genomics 8(1):51–66. doi:10.1152/physiolgenomics.00042.2001
Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E (2009) Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics 10:136. doi:10.1186/1471-2105-10-136
The UniProt Consortium (2011) Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res 39 (database issue):D214–219. doi:10.1093/nar/gkq1020
Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M (1995) Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci USA 92(25):11810–11813
Chen YT, Scanlan MJ, Sahin U, Tureci O, Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old LJ (1997) A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci USA 94(5):1914–1918
Ayyoub M, Pignon P, Dojcinovic D, Raimbaud I, Old LJ, Luescher I, Valmori D (2010) Assessment of vaccine-induced CD4 T cell responses to the 119–143 immunodominant region of the tumor-specific antigen NY-ESO-1 using DRB1*0101 tetramers. Clin Cancer Res 16(18):4607–4615. doi:10.1158/1078-0432.CCR-10-1485
Jager E, Karbach J, Gnjatic S, Neumann A, Bender A, Valmori D, Ayyoub M, Ritter E, Ritter G, Jager D, Panicali D, Hoffman E, Pan L, Oettgen H, Old LJ, Knuth A (2006) Recombinant vaccinia/fowlpox NY-ESO-1 vaccines induce both humoral and cellular NY-ESO-1-specific immune responses in cancer patients. Proc Natl Acad Sci USA 103(39):14453–14458. doi:10.1073/pnas.0606512103
Valmori D, Souleimanian NE, Tosello V, Bhardwaj N, Adams S, O’Neill D, Pavlick A, Escalon JB, Cruz CM, Angiulli A, Angiulli F, Mears G, Vogel SM, Pan L, Jungbluth AA, Hoffmann EW, Venhaus R, Ritter G, Old LJ, Ayyoub M (2007) Vaccination with NY-ESO-1 protein and CpG in Montanide induces integrated antibody/Th1 responses and CD8 T cells through cross-priming. Proc Natl Acad Sci USA 104(21):8947–8952. doi:10.1073/pnas.0703395104
Ueda R, Iizuka Y, Yoshida K, Kawase T, Kawakami Y, Toda M (2004) Identification of a human glioma antigen, SOX6, recognized by patients’ sera. Oncogene 23(7):1420–1427. doi:10.1038/sj.onc.1207252
Iizuka Y, Kojima H, Kobata T, Kawase T, Kawakami Y, Toda M (2006) Identification of a glioma antigen, GARC-1, using cytotoxic T lymphocytes induced by HSV cancer vaccine. Int J Cancer 118(4):942–949. doi:10.1002/ijc.21432
Boone CW, Paranjpe M, Orme T, Gillette R (1974) Virus-augmented tumor transplantation antigens: evidence for a helper antigen mechanism. Int J Cancer 13(4):543–551
Freedman RS, Bowen JM, Herson JH, Wharton JT, Edwards CL, Rutledge FN (1983) Immunotherapy for vulvar carcinoma with virus-modified homologous extracts. Obstet Gynecol 62(6):707–714
Cassel WA, Murray DR, Phillips HS (1983) A phase II study on the postsurgical management of Stage II malignant melanoma with a Newcastle disease virus oncolysate. Cancer 52(5):856–860
Kobayashi H, Gotohda E, Hosokawa M, Kodama T (1975) Inhibition of metastasis in rats immunized with xenogenized autologous tumor cells after excision of the primary tumor. J Natl Cancer Inst 54(4):997–999
Toda M, Martuza RL, Kojima H, Rabkin SD (1998) In situ cancer vaccination: an IL-12 defective vector/replication-competent herpes simplex virus combination induces local and systemic antitumor activity. J Immunol 160(9):4457–4464
Toda M, Rabkin SD, Kojima H, Martuza RL (1999) Herpes simplex virus as an in situ cancer vaccine for the induction of specific anti-tumor immunity. Hum Gene Ther 10(3):385–393. doi:10.1089/10430349950018832
Toda M, Iizuka Y, Kawase T, Uyemura K, Kawakami Y (2002) Immuno-viral therapy of brain tumors by combination of viral therapy with cancer vaccination using a replication-conditional HSV. Cancer Gene Ther 9(4):356–364. doi:10.1038/sj.cgt.7700446
Endo T, Toda M, Watanabe M, Iizuka Y, Kubota T, Kitajima M, Kawakami Y (2002) In situ cancer vaccination with a replication-conditional HSV for the treatment of liver metastasis of colon cancer. Cancer Gene Ther 9(2):142–148. doi:10.1038/sj.cgt.7700407
Iizuka Y, Suzuki A, Kawakami Y, Toda M (2004) Augmentation of antitumor immune responses by multiple intratumoral inoculations of replication-conditional HSV and interleukin-12. J Immunother 27(2):92–98
Ausman JI, Shapiro WR, Rall DP (1970) Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res 30(9):2394–2400
O I Blaszczyk-Thurin M, Shen CT, Ertl HC (2003) A DNA vaccine expressing tyrosinase-related protein-2 induces T-cell-mediated protection against mouse glioblastoma. Cancer Gene Ther 10(9):678–688. doi:10.1038/sj.cgt.7700620
Plautz GE, Touhalisky JE, Shu S (1997) Treatment of murine gliomas by adoptive transfer of ex vivo activated tumor-draining lymph node cells. Cell Immunol 178(2):101–107. doi:10.1006/cimm.1997.1140
Aoki H, Mizuno M, Natsume A, Tsugawa T, Tsujimura K, Takahashi T, Yoshida J (2001) Dendritic cells pulsed with tumor extract-cationic liposome complex increase the induction of cytotoxic T lymphocytes in mouse brain tumor. Cancer Immunol Immunother 50(9):463–468
Liu X, Erikson RL (2007) The nuclear localization signal of mitotic kinesin-like protein Mklp-1: effect on Mklp-1 function during cytokinesis. Biochem Biophys Res Commun 353(4):960–964. doi:10.1016/j.bbrc.2006.12.142
Rowinsky EK, Calvo E (2006) Novel agents that target tublin and related elements. Semin Oncol 33(4):421–435. doi:10.1053/j.seminoncol.2006.04.006
Parness J, Horwitz SB (1981) Taxol binds to polymerized tubulin in vitro. J Cell Biol 91(2 Pt 1):479–487
Derry WB, Wilson L, Jordan MA (1995) Substoichiometric binding of taxol suppresses microtubule dynamics. Biochemistry 34(7):2203–2211
Dhamodharan R, Jordan MA, Thrower D, Wilson L, Wadsworth P (1995) Vinblastine suppresses dynamics of individual microtubules in living interphase cells. Mol Biol Cell 6(9):1215–1229
Fojo AT, Ueda K, Slamon DJ, Poplack DG, Gottesman MM, Pastan I (1987) Expression of a multidrug-resistance gene in human tumors and tissues. Proc Natl Acad Sci USA 84(1):265–269
Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, Janke O, Endruschat J, Durieux B, Roder N, Loffler K, Lange C, Fechtner M, Mopert K, Fisch G, Dames S, Arnold W, Jochims K, Giese K, Wiedenmann B, Scholz A, Kaufmann J (2008) Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res 68(23):9788–9798. doi:10.1158/0008-5472.can-08-2428
Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A (2006) 3’ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 3(3):199–204. doi:10.1038/nmeth854
Judge AD, Bola G, Lee AC, MacLachlan I (2006) Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther 13(3):494–505. doi:10.1016/j.ymthe.2005.11.002
Ueyama K, Ikeda K, Sato W, Nakasato N, Horie-Inoue K, Takeda S, Inoue S (2010) Knockdown of Efp by DNA-modified small interfering RNA inhibits breast cancer cell proliferation and in vivo tumor growth. Cancer Gene Ther 17(9):624–632. doi:10.1038/cgt.2010.19
Nogawa M, Yuasa T, Kimura S, Tanaka M, Kuroda J, Sato K, Yokota A, Segawa H, Toda Y, Kageyama S, Yoshiki T, Okada Y, Maekawa T (2005) Intravesical administration of small interfering RNA targeting PLK-1 successfully prevents the growth of bladder cancer. J Clin Invest 115(4):978–985. doi:10.1172/jci23043
Acknowledgments
We thank Ms. Y. Aikawa, S. Teramoto, T. Muraki, and M. Kokubo for technical assistance. This work was supported by grants from the Ministry of Education, Science, Sports, Science, and Technology, Japan, the Keio University Grant-in-Aid for Encouragement of Young Medical Scientists to S.T., and the Keio Medical Association to S.T.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Takahashi, S., Fusaki, N., Ohta, S. et al. Downregulation of KIF23 suppresses glioma proliferation. J Neurooncol 106, 519–529 (2012). https://doi.org/10.1007/s11060-011-0706-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-011-0706-2