Abstract
Chordomas are low to intermediate grade malignancies that arise from remnants of embryonic notochord. They often recur after surgery and are highly resistant to conventional adjuvant therapies. Recently, the development of effective targeted molecular therapy has been investigated in chordomas that show receptors for tyrosine kinase (RTKs) activation. Expression of specific RTKs such as Epidermal Growth Factor Receptor (EGFR) and Mesenchymal-epithelial transition factor (c-MET) in chordomas may offer valuable therapeutic options. We investigated changes in copy number of chromosome 7 and correlated it with EGFR gene status and EGFR and c-MET protein expression in 22 chordoma samples. Chromosome 7 copy number was evaluated by chromogenic in situ hybridization (CISH) and protein expression of EGFR and c-MET by immunohistochemistry. Tumors mostly showed conventional histopathologic features and were found mainly in sacral (41%) and cranial sites (54.5%). Aneusomy of chromosome 7 was seen in 73% of the samples, 62% of primary tumors and in all recurrent chordomas. EGFR and c-MET were both expressed, but only c-MET protein expression was significantly correlated with chromosome 7 aneusomy (P ≤ 0.001). c-MET overexpression may represent an early chromosome 7 alteration that could play an important role during chordoma pathogenesis. c-MET overexpression shows promise as a molecular marker of response to targeted molecular therapy in the treatment of chordomas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chordomas are considered low to intermediate grade malignancies, accounting for 1–4% of all primary bone tumors. They arise from remnants of embryonic notochord, and are typically located along the axial skeleton. Cranial, spinal and sacral sites are the most common locations, although they can also be found in extra-axial locations. In the United States, the incidence rate for chordoma is 0.08 per 100,000. Chordomas are more common in males and are rare among patients 40 years old and younger [1]. The standard treatment for chordoma is surgery with clean margins, which is often difficult due to the proximity of tumor to important anatomical structures. It is well recognized that these tumors are highly resistant to radiotherapy and conventional chemotherapy, thus presenting a high metastasis rate and poor patient outcome [2–4].
Recently, molecular studies have been performed to identify chromosomal and genetic changes that may be involved in chordoma pathogenesis. More specifically, targeted molecular therapy has been investigated in tumors with tyrosine kinase receptors (RTKs) activation as a possible novel therapeutic option. Previous studies have reported that downstream tyrosine kinase signaling is activated in chordomas that express certain RTKs [5–7], which has been related to tumor development and progression [8, 9].
Expression of specific tyrosine kinase receptors such as Epidermal Growth Factor Receptor (EGFR), platelet-derived growth factor receptor (PDGFR) alpha and beta, mesenchymal-epithelial transition factor/hepatocyte growth factor receptor (c-MET) and HER2/neu have been previously reported in chordomas [10, 11]. Activation of tyrosine kinase induces signaling of several downstream proteins within MAPK, Akt and JNK pathways, which have been associated with tumorigenesis and tumor progression through effects on the cell cycle, apoptosis, angiogenesis and metastasis [8, 12]. Some RTKs, especially EGFR and c-MET [13] have been identified as potential therapeutic targets in many solid tumors [14, 15]. Previous studies in chordomas have shown that tumors that express EGFR among other RTKs may also respond to specific targeted molecular therapy inhibiting TK activation [16, 17].
Ostroumov et al. concluded that c-MET oncoprotein may play a leading role during metastasis by showing positive chemotactic and migration activities of a chordoma cell line in response to hepatocyte growth factor (HGF) [18]. On the other hand, Naka et al. have shown that positive c-MET expression in young patients with primary skull base chordomas was related to longer survival. Interestingly, it was also associated with tumor invasiveness in recurrent lesions, suggesting a role for c-MET based on clinical status. However, this association was not significantly demonstrated in spinal chordomas, [11, 19]. Therefore, it is still under investigation whether c-MET oncoprotein is related to tumor progression, metastatic potential or if its function is related to tumor location.
Cytogenetic and molecular genetic studies of chordomas reveal a complex karyotype, showing multiple chromosomal aberrations, either gains or losses in different chromosomes [20]. Gain of chromosome 7 is the most common chromosomal alteration reported to date [21, 22]. From a genetic perspective, gains in chromosome 7 seem to be particularly related to gene amplification in different tumors that exhibit activation of the tyrosine kinase domain. Interestingly, EGFR and c-MET are found on chromosome 7, at loci 7p12 and 7q31, respectively [23]. Although both are expressed in tumors with chromosome 7 alterations [16, 17, 24], increased copy number of chromosome 7 and their relation to RTK genes expressions have not been investigated in chordomas.
Therefore, the aim of this study was to evaluate the presence of gain or loss of chromosome 7 and to correlate it with protein expression of certain RTKs genes within this domain, specifically EGFR and c-MET oncoprotein. The association between clinicopathological features and protein expression of c-MET and EGFR was also evaluated.
Methods
Patients and samples
A total of 22 samples from 18 patients including primary and recurrent tumors were obtained from the archives of the Department of Pathology at the A.C. Camargo Hospital, São Paulo, Brazil after IRB approval. Patient clinicopathological data was retrieved from the medical records. Hematoxylin and Eosin (H&E) stained slides were reviewed independently by a neuropathologist to confirm the diagnosis and to tabulate the presence of some histopathological features as necrosis, mitotic activity or high degree of nuclear pleomorphism.
Chromogenic in situ hybridization (CISH)
CISH for the centromere region of chromosome 7 and EGFR gene status was performed on serial 5 μm thick formalin-fixed paraffin-embedded chordomas sections. Briefly, slides were deparaffinized in xylene and rehydrated in graded ethanol. Tissue sections were rinsed in deionised water and then placed in a beaker containing Tissue Pre-treatment Buffer (SpoT-Light® Tissue Pre-treatment Kit, Invitrogen, CA, USA) heated at ≥98°C for 15 min. The slides were immediately washed in deionised water followed by enzymatic digestion with pepsin (SpoT-Light® Tissue Pre-treatment Kit) for 10 min at room temperature. The slides were washed with deionised water, dehydrated with graded ethanol and air-dried. Ready-to-use biotin-labeled chromosome 7 centromere or digoxigenin-labeled EGFR DNA probes (Zymed, South San Francisco, CA) were applied to each section, respectively. Tissue sections containing the added probe were denatured at 94°C for 5 min and incubated at 37°C overnight in a slide hybridizer (StatSpin, Thermobrite®, MA, USA). The sections were washed in 0.5× standard saline citrate (SSC) at 75°C for 5 min and then with distilled water. Sections were placed in 3% H2O2 in absolute methanol for 10 min to block endogenous peroxidase. To block unspecific staining, slides were incubated with CAS-Block for 10 min. After that, for the detection of the centromere probe, the SpoT-Light® CISHTM Centromere Detection Kit (Invitrogen, CA, USA) was used, and to detect the EGFR probe, the SpoT-Light® CISHTM Polymer Detection Kit was used. The biotin-labeled chromosome 7 centromere probe was detected by incubation with HRP-conjugated streptavidin for 30 min, followed by diaminobenzidine chromogen (DAB). For the digoxigenin-labeled EGFR probe, mouse anti-digoxigenin antibody was applied for 30 min and then incubated with anti-mouse HRP-polymer and finally visualized with DAB. Sections were lightly counterstained with hematoxylin, dehydrated and mounted. CISH signals were easily visualized as dark brown scattered dots in a conventional bright-field microscope.
Interpretation of CISH signals
In situ hybridization signals were evaluated under light microscopy using a 40× objective. Signals within the nuclei in three different areas of the sample were counted. As previously reported [14], interpretation of chromosome 7 centromere signals resulted in two categories as follows: disomy (≤2 copies in >90% of cells) and aneusomy (three and more copies). The aneusomy category was further divided into trisomy (≥3 copies in >90% of cells or ≥4 copies in less than 50% of cells) and polysomy (≥4 copies in ≥50% of cells). Hence, the disomy group included only normal samples, while the polysomy group consisted of samples that showed a high degree of chromosomal alterations in most of the tumor cells. To evaluate the EGFR gene, gene amplification was defined by the presence of ≥15 copies of the gene in more than 10% of tumor cells, as has been previously described [14].
Immunohistochemistry
Five micron-thick slides were deparaffinized, rehydrated in graded ethanol and rinsed in water. Antigen retrieval was performed for EGFR staining using enzyme digestion (Pepsin, Digest All 3, Zymed) applied for 10 min at 37°C. For c-MET staining, heat-induced antigen retrieval was performed with 1 mM EDTA buffer, pH 8.0 in a microwave oven for 20 min. Endogenous peroxidase blocking solution was applied for 10 min followed by incubation with the primary antibodies for 1 h at room temperature (either mouse anti-EGFR monoclonal antibody (31G7, Zymed, San Francisco, CA, 1:100 dilution) or rabbit polyclonal anti-MET (C-28, SC-161, Santa Cruz Biotechnology, Santa Cruz, CA, 1:1000 dilution). Incubation with the EnVisionTM Dual Link System-HRP (DAB+) (Dako North America, Carpinteria, CA, USA) for 30 min and DAB chromogen was used for detection. Tissue sections were subsequently counterstained, dehydrated and mounted. During all the staining procedures, positive and negative controls were run simultaneously.
Evaluation of immunohistochemistry (IHC)
EGFR and C-MET cytoplasmic/membranous expression was determined by consensus evaluation of three observers. Interpretation of IHC was scored based on staining intensity and distribution as previously described [25, 26].
The intensity score was defined as follows: 0 = nondetectable staining, 1 = weak staining, 2 = moderate staining and 3 = strong staining in cytoplasm. Samples with a score of two or more were considered positive. The staining pattern was determined as the presence of positive staining in some tumor areas (focally) or in the entire tumor (diffuse) within a tissue section.
Statistical analysis
Correlation between protein expression, chromosome 7 status and clinicopathological data were estimated with either Chi-square or Fisher’s exact tests using SPSS software version 13 (Chicago, IL). Statistical significance was defined as a 2-sided P value < 0.05. A sample size of N = 22 allowed for an 80% power to detect at least a 70% change between any two categories in the Chi-square tests.
Results
Clinical findings
Twenty-two samples from 18 different patients diagnosed with chordomas were evaluated, which included 19 primary conventional chordomas and 3 recurrent tumors. Case 7 presented two primary tumors at different locations. In cases 5, 7 and 8, both primary and recurrent tumors were evaluated (Table 1).
Patient age at diagnosis ranged from 18 to 77 years (median 47 years). Fifty-nine percent of the patients were 40 years old or more and female preponderance was observed, representing 59% of the patients. Of the 18 patients studied, 3 cases presented recurrent disease (16.6%). Ninety-five percent of the tumors were located in the axial skeleton, distributed either in cranial (41%) or sacral (54.5%) locations. Within the cranial sites, clivus, nasopharynx and mandible were the most common. One recurrent tumor was found extra-axially within the soft tissue of the lower limb. In young patients (<26 years old), there was also a female preponderance (P = 0.049). This group of patients showed mostly cranial chordomas (P = 0.010), while patients older than 40 years presented more often with tumors in the sacral region (P < 0.001).
Microscopic findings
Histologically, tumors contained characteristic physaliphorous cells with vacuolated cytoplasm, or in some instances, cells with a clear or signet-ring appearance organized in a chord-like manner. The diagnosis of chordoma was supported by the positive expression of cytokeratin (AE1/3), epithelial membrane antigen (EMA) and S-100 antibodies. Tumors with necrotic areas were seen in 9% of the cases; however, only a few mitotic figures were identified. Forty-five percent of cases showed variable nuclear pleomorphism. In one case, the tumor consisted of two types of cells: small uniform non-vacuolated cells with hyperchromatic nucleus arranged in nests, and in other areas, large physaliphorous cells arranged in groups. However, on statistical analysis, none of the histological features correlated with clinical data, tumor type or recurrence.
Expression of EGFR and c-MET
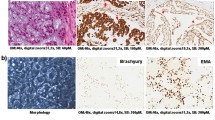
Both antibodies showed membranous or cytoplasmic immunoreactivity within tumor cells. Positive EGFR expression was seen in 32% (7/22) of the cases (Fig. 1).
Representative tissue sections showing EGFR expression detected by immunohistochemistry. a EGFR negative sample (score 0). No brown staining is seen in tumor cells. b Moderately positive section (score 2). Staining is seen delineating cell membrane and disperses throughout the cytoplasm of the cells. Insert shows EGFR expression in a skin tissue used as positive control
There was a significant correlation between EGFR immunohistochemical reactivity and female gender (P = 0.01). Chromosome 7 status or c-MET gene expression did not correlate with EGFR either gene amplification or protein expression.
c-MET expression was evaluated using the same score applied for EGFR, including intensity of staining and staining distribution. Weak staining (score 1) was seen only in 5% of the cases (1/22), moderate expression (score 2) in 59% of the cases (13/22), and strong staining (score 3) was found in 18% (4/22) (Fig. 2). Negative c-MET expression approached 20% of the cases (4/22, 18%).
c-MET expression evaluated by immunoperoxidase staining in chordoma. a Negative sample (score 0). No appreciable staining is observed within the tissue. b Score 1 = weak. Some brown reaction product is observed delineating the membrane of the cells and weakly within some tumor cells. c Moderately positive sample (score 2). Membranous and cytoplasmic brown staining is easily seen in the tumor cells. d Intense brown staining is appreciated in the tumor cells (score 3). Small insert shows breast tissue used as a control
Most of the tumor samples showed positive c-MET expression (77% of the cases) (score ≥2), which was diffusely distributed in 82% of the samples, and focally in only a few cases.
Positive expression of c-MET correlated with increased copy number of chromosome 7, being significantly correlated with aneusomic (trisomy or polysomy) samples (P ≤ 0.001). c-MET or EGFR expression were not found to correlate with any other clinicopathological features.
Copy number of chromosome 7 by chromogenic in situ hybridization (CISH)
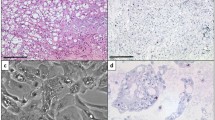
Twenty-seven percent (6/22) of cases showed normal disomy for chromosome 7. Trisomy was seen in 59% (13/22) and polysomy in 14% (3/22) of samples, accounting for almost 73% of aneusomy (Table 2). No significant correlation was found between aneusomy and EGFR expression (Table 3). However, aneusomy (trisomy and polysomy combined) correlated with positive c-MET expression with 84% of diploid cases showing negative expression, whereas all trisomy and polysomy cases were positive for c-MET expression (P < 0.001) (Table 4, Fig. 3).
Chromogenic in situ hybridization (CISH) of chromosome 7 in chordomas. a and c Hematoxylin and Eosin staining. a Nest and cluster of cells, with vacuolated and clear cytoplasm that represent tumor cells is shown. b Note the high grade of nuclear pleomorphism in the chordoma cells that are distributed within a myxoid matrix. b and d CISH technique was performed to evaluate copy number for chromosome 7. Dark brown dots can be seen within the nuclei of the cells. b Representative diploid cells showing two dots within each nucleus. d High degree of aneusomy is appreciable within the nuclei of the cells, where three or more dots are observed
Although not statistically significant, 87.5% of the cases that demonstrated polysomy of chromosome 7 were in patients 26 years old or older (P = 0.100) and 62% of the patients that presented sacral chordomas had chromosome 7 polysomy (P = 0.513). According to tumor status, 68.4% of primary tumors showed polysomy of chromosome 7; all recurrent cases had this chromosomal alteration (P = 0.346). There was no correlation between histological features, diploidy, trisomy or polysomy of chromosome 7 and the presence of necrosis or mitoses in the tumor cells. Interestingly, nuclear pleomorphism was seen in 80% of cases where tumor cells showed an increase in copy number for chromosome 7.
EGFR gene amplification by chromogenic in situ hybridization (CISH)
CISH of the EGFR gene was performed in 13 samples of this series. Fifty-three percent (8/15) of cases that lacked protein expression for EGFR by immunohistochemistry also lacked amplification of the EGFR gene.
Discussion
The presence of multiple chromosomal alterations in chordomas has been reported previously [27–29], but this observation has not been implicated in the pathogenesis of chordomas [30, 31]. In a study of using comparative genomic hybridization (CGH) and fluorescence in situ hybridization (FISH) to detect chromosomal alterations in six sacral chordomas, the most frequent gain (66%) was found on chromosome 7 [21]. Sheil et al. reported that gain on chromosome arm 7q [22] was a common finding in 69% of chordomas studied. The authors concluded that the 7q gain may be related to overexpression of c-MET via 7q amplification and that it may represent an early event during chordoma progression [16].
In our study, we investigated changes in copy number of chromosome 7 by chromogenic in situ hybridization (CISH). This technique has advantages over FISH, because it uses conventional peroxidase reactions that can be observed under light microscopy at the sites where the probe hybridized, thus allowing simultaneous visualization of the signal and surrounding tissue morphology. Using this technique, we detected an increase in copy number of chromosome 7. Trisomy was found in around 59% of the cases and polysomy in 14%. Our results confirm previously published findings that gain of chromosome 7 is a frequent alteration in chordomas, supporting its possible pathogenetic role [20]. Molecular cytogenetic techniques have mapped the most frequent gains at the following loci: 7p15 [29], 7p21–p22 and portions of arms 7q [22]; 7q22 [22], 7q33 [32], 7q34 and 7q36 [22]. However, candidate genes present in these chromosomal areas have not being positively correlated with chordoma formation.
Interestingly, gains of chromosome 7 have been reported in primary and recurrent chordomas [32]. No significant differences among tumors at different anatomic sites were found [22]. We observed gain of chromosome 7 in primary and recurrent tumors, with a high percentage (83%) of sacral tumors showing polysomy of chromosome 7.
Despite our limited sample size, we believe that both primary and recurrent chordomas exhibit a high incidence of polysomy of chromosome 7. In addition, we found that this alteration is more frequent in patients older than 26 years and that it is common in sacral tumors. These facts imply that this chromosomal aberration is not only an early effect for chordoma formation, but also, that it persists during tumor progression.
In the present study, we focused our investigation on the differential expression of receptors TKs genes on chromosome 7 that may be related to the presence of the chromosomal aberration. Since the proto-oncogene c-MET and EGFR are currently being investigated as potential therapeutic targets and biomarkers for chordoma, we selected to study their expression [7, 33]. Previous reports on EGFR expression in chordomas have not achieved a consensus regarding its role in disease onset and progression; however, it has been noted that chordomas overexpress EGFR and c-MET. In addition, HER2/neu and EGFR expression has been reported between 48 and 66% of the cases, and c-MET expression in 90% of them in previous studies [6, 16, 17].
The EGF pathway is involved in tumor growth and cell survival. Drugs such as cetuximab and gefitinib have demonstrated reduced tyrosine kinase activity and its downstream signals. Different types of tumors, which have been treated with these agents have shown encouraging response rates. In a patient with progressive sacral chordoma, a partial response was achieved inhibiting the EGF pathway with a combination of two monoclonal antibodies [6]. In the present study, we found that 32% of the cases showed EGFR protein overexpression but none of the cases showed EGFR gene amplification. The role of c-MET proto-oncogene in many cellular processes and during tumor progression and invasion has been reported in different types of tumors [9, 16, 34]. In chordomas, c-MET expression is seen in up to 94.5% of primary lesions and 88% of recurrent lesions, without much distinction between skull-base and non-skull base chordomas [5, 11, 19]. In a human chordoma cell line established from a primary sacral tumor in which c-MET expression was studied, the authors suggested that in vivo, c-MET oncoprotein plays a leading role during tumor growth and metastasis [18]. Furthermore, different investigators have reported that c-MET expression may either correlate with tumor invasion and progression, or alternatively, with favorable prognosis. These conflicting results suggest that its role may vary with the clinical status of the disease; however, further investigation is warranted to resolve the issue of this apparent differential function.
Nonetheless, our results confirm that c-MET expression is a common event in both primary and recurrent lesions. These results confirm previous observations by Ostroumov, et al. that c-MET overexpression may be a result of chromosome 7 alteration during an early stage of chordoma formation, which persists during disease progression until recurrent lesions occur. We hypothesize that further accumulation of c-MET gene in chordomas may lead to a break-point in which loss of c-MET gene may occur, thus promoting aggressive and invasive behavior.
In conclusion, polysomy of chromosome 7 in primary and all recurrent tumors confirmed by CISH is associated with c-MET expression alterations, thus suggesting a role in chordomas progression. These results contribute to the understanding of the pathogenesis of chordoma and support the development of targeted therapies that inhibit c-MET activation. Further studies are needed to evaluate the effectiveness of treatment, especially focusing on therapies inhibiting different TK receptors.
Abbreviations
- CISH:
-
Chromogenic in situ hybridization
- RTKs:
-
Receptors tyrosine kinase
- EGFR:
-
Epidermal growth factor receptor
- c-Met:
-
Mesenchymal-epithelial transition factor
References
McMaster ML, Goldstein AM, Bromley CM, Ishibe N, Parry DM (2001) Chordoma: incidence and survival patterns in the United States, 1973–1995. Cancer Causes Control 12:1–11
Yonemoto T, Tatezaki S, Takenouchi T, Ishii T, Satoh T, Moriya H (1999) The surgical management of sacrococcygeal chordoma. Cancer 85:878–883
Forsyth PA, Cascino TL, Shaw EG, Scheithauer BW, O’Fallon JR, Dozier JC, Piepgras DG (1993) Intracranial chordomas: a clinicopathological and prognostic study of 51 cases. J Neurosurg 78:741–747
McPherson CM, Suki D, McCutcheon IE, Gokaslan ZL, Rhines LD, Mendel E (2006) Metastatic disease from spinal chordoma: a 10-year experience. J Neurosurg Spine 5:277–280
Naka T, Iwamoto Y, Shinohara N, Ushijima M, Chuman H, Tsuneyoshi M (1997) Expression of c-met proto-oncogene product (c-MET) in benign and malignant bone tumors. Mod Pathol 10:832–838
Hof H, Welzel T, Debus J (2006) Effectiveness of cetuximab/gefitinib in the therapy of a sacral chordoma. Onkologie 29:572–574
Tamborini E, Miselli F, Negri T, Lagonigro MS, Staurengo S, Dagrada GP, Stacchiotti S, Pastore E, Gronchi A, Perrone F, Carbone A, Pierotti MA, Casali PG, Pilotti S (2006) Molecular and biochemical analyses of platelet-derived growth factor receptor (PDGFR) B, PDGFRA, and KIT receptors in chordomas. Clin Cancer Res 12:6920–6928
Chu SH, Feng DF, Zhang H, Chen ET, Duan ZX, Li XY, Li J, Ma YB, Zhu ZA, Qiu JH (2009) c-Met-targeted RNA interference inhibits growth and metastasis of glioma U251 cells in vitro. J Neurooncol 93:183–189
Hirose Y, Kojima M, Sagoh M, Murakami H, Yoshida K, Shimazaki K, Kawase T (1998) Immunohistochemical examination of c-Met protein expression in astrocytic tumors. Acta Neuropathol 95:345–351
Lamszus K, Schmidt NO, Jin L, Laterra J, Zagzag D, Way D, Witte M, Weinand M, Goldberg ID, Westphal M, Rosen EM (1998) Scatter factor promotes motility of human glioma and neuromicrovascular endothelial cells. Int J Cancer 75:19–28
Naka T, Boltze C, Samii A, Samii M, Herold C, Ostertag H, Iwamoto Y, Oda Y, Tsuneyoshi M, Kuester D, Roessner A (2009) Expression of c-MET, low-molecular-weight cytokeratin, matrix metalloproteinases-1 and -2 in spinal chordoma. Histopathology 54:607–613
Hecht M, Papoutsi M, Tran HD, Wilting J, Schweigerer L (2004) Hepatocyte growth factor/c-Met signaling promotes the progression of experimental human neuroblastomas. Cancer Res 64:6109–6118
Mazzone M, Comoglio PM (2006) The Met pathway: master switch and drug target in cancer progression. Faseb J 20:1611–1621
Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, Ludovini V, Magrini E, Gregorc V, Doglioni C, Sidoni A, Tonato M, Franklin WA, Crino L, Bunn PA Jr, Varella-Garcia M (2005) Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 97:643–655
Salomon DS, Brandt R, Ciardiello F, Normanno N (1995) Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol 19:183–232
Weinberger PM, Yu Z, Kowalski D, Joe J, Manger P, Psyrri A, Sasaki CT (2005) Differential expression of epidermal growth factor receptor, c-Met, and HER2/neu in chordoma compared with 17 other malignancies. Arch Otolaryngol Head Neck Surg 131:707–711
Fasig JH, Dupont WD, LaFleur BJ, Olson SJ, Cates JM (2008) Immunohistochemical analysis of receptor tyrosine kinase signal transduction activity in chordoma. Neuropathol Appl Neurobiol 34:95–104
Ostroumov E, Hunter CJ (2008) Identifying mechanisms for therapeutic intervention in chordoma: c-Met oncoprotein. Spine 33:2774–2780
Naka T, Kuester D, Boltze C, Scheil-Bertram S, Samii A, Herold C, Ostertag H, Krueger S, Roessner A (2008) Expression of hepatocyte growth factor and c-MET in skull base chordoma. Cancer 112:104–110
Almefty KK, Pravdenkova S, Sawyer JR, Al-Mefty O (2009) Impact of cytogenetic abnormalities on the management of skull base chordomas. J Neurosurg 110:715–724
Brandal P, Bjerkehagen B, Danielsen H, Heim S (2005) Chromosome 7 abnormalities are common in chordomas. Cancer Genet Cytogenet 160:15–21
Scheil S, Bruderlein S, Liehr T, Starke H, Herms J, Schulte M, Moller P (2001) Genome-wide analysis of sixteen chordomas by comparative genomic hybridization and cytogenetics of the first human chordoma cell line, U-CH1. Genes Chromosom Cancer 32:203–211
Glukhova L, Lavialle C, Fauvet D, Chudoba I, Danglot G, Angevin E, Bernheim A, Goguel AF (2000) Mapping of the 7q31 subregion common to the small chromosome 7 derivatives from two sporadic papillary renal cell carcinomas: increased copy number and overexpression of the MET proto-oncogene. Oncogene 19:754–761
Tamayama C, Maruyama K (1990) Expression of EGF receptor and c-neu oncogene product in chordomas. Gan no rinsho 36:773–776
Leo C, Horn LC, Einenkel J, Hentschel B, Hockel M (2007) Tumor hypoxia and expression of c-met in cervical cancer. Gynecol Oncol 104:181–185
Cruz J, Reis-Filho JS, Silva P, Lopes JM (2003) Expression of c-met tyrosine kinase receptor is biologically and prognostically relevant for primary cutaneous malignant melanomas. Oncology 65:72–82
Kelley MJ, Korczak JF, Sheridan E, Yang X, Goldstein AM, Parry DM (2001) Familial chordoma, a tumor of notochordal remnants, is linked to chromosome 7q33. Am J Hum Genet 69:454–460
Sawyer JR, Husain M, Al-Mefty O (2001) Identification of isochromosome 1q as a recurring chromosome aberration in skull base chordomas: a new marker for aggressive tumors? Neurosurg Focus 10:E6
Hallor KH, Staaf J, Jonsson G, Heidenblad M, Vult von Steyern F, Bauer HC, Ijszenga M, Hogendoorn PC, Mandahl N, Szuhai K, Mertens F (2008) Frequent deletion of the CDKN2A locus in chordoma: analysis of chromosomal imbalances using array comparative genomic hybridisation. Br J Cancer 98:434–442
Sandberg AA, Bridge JA (2003) Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: osteosarcoma and related tumors. Cancer Genet Cytogenet 145:1–30
Kuzniacka A, Mertens F, Strombeck B, Wiegant J, Mandahl N (2004) Combined binary ratio labeling fluorescence in situ hybridization analysis of chordoma. Cancer Genet Cytogenet 151:178–181
Bayrakli F, Guney I, Kilic T, Ozek M, Pamir MN (2007) New candidate chromosomal regions for chordoma development. Surg Neurol 68:425–430 discussion 430
Casali PG, Stacchiotti S, Sangalli C, Olmi P, Gronchi A (2007) Chordoma. Curr Opin Oncol 19:367–370
Zhang YW, Vande Woude GF (2003) HGF/SF-met signaling in the control of branching morphogenesis and invasion. J Cell Biochem 88:408–417
Acknowledgment
Maria Begnami work was supported by a grant 453399/2008-8 from CNPQ/Brazil.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Walter, B.A., Begnami, M., Valera, V.A. et al. Gain of chromosome 7 by chromogenic in situ hybridization (CISH) in chordomas is correlated to c-MET expression. J Neurooncol 101, 199–206 (2011). https://doi.org/10.1007/s11060-010-0250-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-010-0250-5