Abstract
Extensive survey was carried out in the state of Maharashtra, India, as part of a 3-year project to explore keratinophilic fungal diversity for conservation and biotechnological potential. A total of 578 soil samples were collected from keratin-rich habitats across 24 districts of Maharashtra State. Hair-baiting technique and micro-dilution drop-trail method were employed for isolation and purification of keratinophilic fungi from soil. A total of 66 species belonging to 17 genera of order Onygenales were recorded in hair baits. Eleven taxa were found to be new to science, most of which were rare as they were recorded in only one sample out of the > 500 samples analyzed. Three novel taxa have been characterized at morphological and molecular level and described here as new to science. These taxa include Currahmyces indicus gen. et sp. nov., Canomyces reticulatus gen. et sp. nov., Ctenomyces indicus sp. nov. All these novel taxa are morphologically and phylogenetically distinct from known taxa of order Onygenales. The study indicates that systematic sampling of a larger area is needed to uncover the hidden (unknown) diversity of keratinophilic fungi which is overlooked in sporadic samplings as evident from previous studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Keratinophilic fungi are specialized group of fungi which have affinity towards animal protein keratin which is produced by all higher vertebrates as outer covering. Most of the keratinophilic fungi are restricted to single order Onygenales [1] due to their ability to produce keratin-degrading enzymes (proteases) which help them utilize keratin as sole source of carbon and nitrogen. Phylogenetically, order Onygenales is a monophyletic group comprising around 375 species dispersed in 55 genera [1,2,3,4,5,6,7,8,9,10,11]. There have been regular reports of keratinophilic fungi from various parts of the world suggestive of their cosmopolitan nature. However, not all fungi are found uniformly across the globe (restricted distribution) due to limited dispersal which is mostly carried out by small animals, insects and birds. Furthermore, there are several Onygenalean species described long back but still not reported from any other locality indicating their rarity. Except for migratory birds which help intercontinental dispersal of keratinophilic fungi, the only other source of dispersal is by humans (mostly dermatophytes) due to frequent travel across globe. Most of the dermatophytes which earlier had specific distribution pattern [12] are now being reported from newer areas of the world due to frequent international travel and material transfer. But still there are numerous Onygenalean fungi which are missing in our records that need discovery and formal description.
Since Currah’s revision of the order Onygenales more than three decades ago [1], several genera and species have been added to this interesting group of ascomycetes (Ascomycota). These additions have been mostly carried out on the basis of molecular phylogenetic studies, and hence, their positions are fairly stable in the phylogenetic tree of life. Nineteen new genera have been added to the Onygenales after Currah revised the order, viz, Amauroascopsis [13], Aphanoascella [3], Auxarthronopsis [4], Castanedomyces [14], Chlamydosauromyces [10], Emergomyces [5], Emmonsiellopsis [6], Harorepupu [7], Helicocarpus [6], Kraurogymnocarpa [15], Lophophyton [8], Myotisia [9], Ophidiomyces [16], Paranannizziopsis [17], Paraphyton [8], Pseudoamauroascus [18], Pseudospiromastix [11], Sigleria [11] and Testudomyces [19]. Most of these fungi have been isolated from either soil or from clinical material of human or animal origin. Except for those that are based on clinical samples, most of the genera are monotypic in nature and more studies are needed to explore other species (if they exist). Some species that form extremely minute ascomata after extended periods of incubations (> 2 months) in hair baits are often overlooked in routine investigations for, e.g. Bifidocarpus cubensis Cano, Guarro & Castaneda [20] and Gymnoascus verrucosus Rahul Sharma & SK Singh [21]. The former species was first described in 1994 by Cano et al. from Cuba [20] and was known only from type locality until recently when it was observed on goat dung samples in India [22].
In the present study, we surveyed a large area encompassing Maharashtra State of India in which we sampled 24 districts for collection of soil from keratin-rich habitats. The present paper reports the distribution of keratinophilic fungi in soil and provides description of two new genera and one new species of order Onygenales using morphology and molecular phylogeny.
Materials and Methods
Collection of Soil and Isolation of Fungi
Soil samples were collected from keratin-rich habitats, viz, vicinity of barbers shop, entrances of animal burrows, hen frequented areas, under trees along roadsides. Subsurface soil were collected after removing surface soil in zip lock polythene bags using sterile spatula (metal spoon) and stored at room temperature until processed for fungal isolation. For isolation of Onygenalean fungi, Vanbreuseghem’s hair-baiting technique was used with slight modifications [23, 24]. Further, when ascomata or fungal growth developed in hair baits, they were transferred to artificial medium (Sabouraud Dextrose Agar with Chloramphenicol 250 mg L−1) using micro-dilution drop-trail method [25] to obtain pure cultures. In case of fragile ascomata, ascospore mass was picked up using fine needle moistened with sterile distilled water to make ascospore suspension for drop-trail method. Ex-type cultures were submitted to National Centre for Microbial Resource (NCMR), Pune, India, and Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands.
Morphological Study
Cultural characteristics were noted after 14 days to 30 days of incubation at 28 °C and 37 °C on Sabouraud Dextrose Agar (SDA; HiMedia Laboratories, Mumbai, India), Cornmeal Agar (CMA; HiMedia Laboratories, Mumbai, India), Potato Dextrose Agar (PDA; HiMedia Laboratories, Mumbai, India) and Oatmeal Agar (OA; HiMedia Laboratories, Mumbai, India). For microscopic features, a slight modification of Riddle’s slide culture technique was used [26]. Instead of agar block, a drop of molten agar medium was placed on sterilized slide. Cultures were grown on small drop of solidified SDA on sterile glass slides which were covered with autoclaved cover glass and incubated for few days or 1–2 weeks, and photographs were taken thereof.
Light Microscopy (LM)
For light microscopy, fungal material (mostly ascomata) was picked up from hair baits using fine needle and mounted in lactophenol (HiMedia Laboratories, Mumbai, India) and photomicrographs were taken by Olympus BX53 research microscope fitted with ProgRes digital camera. For asexual morphs, slide cultures were mounted in lactophenol and observed under microscope. Measurements of microscopic features at various magnifications were taken using calibrated ProgRes software of the microscope.
Scanning Electron Microscopy (SEM)
For scanning electron microscopy, air-dried fungal material was mounted on stubs using double-sided adhesive tapes and observed directly on EVO-LS10 (Zeiss, Jena) or after sputter coating with palladium using JFC 1600 auto fine coater and observed on JEOL-JSM 6360A (Jeol,Tokyo) analytical scanning electron microscopes.
Molecular Study
DNA Extraction, PCR and Sequencing
Genomic DNA was extracted from freshly grown fungal cultures using method of Edwards et al. [27, 28] with slight modifications. Briefly, 300 mg of mycelia mat was scraped from a 7–10 days old colony and placed in 1.5-mL Eppendorf tube. Care was taken to remove agar medium attached to the mycelia mat by using sterile tooth pick. Three hundred micro-litre of Edwards buffer (200 mM Tris.HCl, pH 8.0; 200 mM NaCl, 25 mM EDTA; 0.5% SDS) was added to the Eppendorf tube containing mycelia mat. The mycelia were crushed with the help of autoclaved plastic pestle (Tarson, Kolkata) for 5 min until a fine suspension was obtained. Additional 300 µL of Edwards buffer was added to the micro-tube, and the process of crushing was repeated for another 5 min. The finely crushed mycelia in suspension were briefly vortexed for 10–20 s and then kept at 100 °C for 15–20 min. The suspension was centrifuged for 10 min at 14,000 rpm, and supernatant was transferred to a fresh tube. Equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) (Sigma, St Louis, MO, USA) was added and briefly vortexed before keeping at room temperature for 10 min. The mixture was centrifuged for 15 min at 14,000 rpm. Upper aqueous layer was gently transferred to fresh tube. Equal volume of ice-cold absolute ethanol was added and mixed by inversion until precipitate appeared. The tubes were incubated overnight at − 20 °C. Next day, the precipitated DNA was centrifuged at 14,000 for 15 min. The pellet was washed with 70% ethanol. Air-dried precipitate was suspended in 100 µL of TE buffer, and 1 µL of RNase (100 µg/mL final conc.) (Sigma, St Louis, MO, USA) was added and mixed with the help of pipette and incubated at 37 °C for 15–20 min. RNase was deactivated by incubating at 65 °C for 10 min. Quantification of DNA was carried out by NanoDrop spectrophotometer (NanoDrop, USA) and by running along with samples a standard of lambda DNA of known concentration on a 0.8% agarose gel.
Polymerase chain reaction was carried out on an ABI 2720 thermocycler (Applied Biosystems, Singapore) with the following reaction conditions. Reaction mixture contained 16.8 µL-H2O, 2.5 µL-10X buffer, 1 µL-dNTP’s (200 µM), 1.5 µL-MgCl2 (25 mM), 1 µL-ITS1 primer, 1 µL-ITS4 primer, Taq polymerase-0.2 µL (New England Biolabs) and 0.25 to 1 µL-template DNA. Amplification conditions in PCR machine consist of 1 cycle of 94 °C for 5 min; 30 cycles of 94 °C for 1 min; 55 °C for 1 min; 72 °C for 1 min; and a final extension of 72 °C for 7 min. Five micro-litre of PCR product was run on 1.2% agarose gel to check amplification. Successfully, amplified products were cleaned using FavoPrep PCR purification mini kit (Favorgen Biotech Corp., Taiwan) as per manufacturer’s instructions. Sequencing was carried out using Big Dye Terminator Cycle Sequencing kit as per manufacturer’s instruction on an ABI 3730 xl automated DNA sequencer (Applied Biosystems Inc, Japan). Sequences obtained from sequencer were manually checked for inconsistencies using Chromas Lite software v.2.1.1 (http://www.technelysium.com.au).
Phylogeny Analysis
A BLASTn search was carried out with NCBI and CBS databases to check for the closest taxon hits. Based on the identities received from BLASTn results, phylogeny analysis was carried out using closely related sequences from GenBank. Sequence alignment was carried out by CLUSTAL W in MEGA v.5.05 software (MEGA release# 5110426) in which sequences from both ends are trimmed. Phylogenetic trees were constructed in MEGA v.5.05 using neighbour-joining, maximum parsimony and maximum likelihood methods and bootstrap analysis of 1000 replicates.
Results
During the soil survey, a total of 578 soil samples were collected from 24 districts of Maharashtra State of India. The percentage distribution of samples from various habitats is as follows: animal frequented—27.7%, barber shop—18%, chicken shop—11%, public place—9%, roadside—7.5%, burrow—7.4%, under tree—6.7%, garbage—5.5%, drainage—3% and others—3.5%. Of the sampled habitats, barber shop, animal burrows, animal resting grounds, chicken shops have nearly all samples positive for keratinophilic fungi (> 95% frequency of occurrence). Table 1 shows habitat-wise frequency of occurrence of Onygenalean fungi in soil samples. A total of 66 species belonging to 17 genera of order Onygenales were recorded in hair baits. District-wise occurrence of various Onygenalean species is shown in Supplementary Table 1. An unassigned species of Nannizzia was most frequently occurring species in various districts (83% of districts), while other species with decreasing order of frequency are as follows: N. gypsea—75%, G. petalosporus—66%, N. corniculata—62%, N. incurvata—62%, Chrysosporium tropicum—50%, N. fulva—46%, C. linfenense—41%, Ctenomyces serratus—41% and Aphanoascus orissae—37%. The remainder of the species was even less frequent in the 24 district samples. Of the 66 Onygenalean species recorded, 23 species were represented by only one district which is suggestive of their limited distribution in Maharashtra.
Phylogenetic and Molecular Analysis
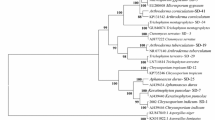
The complete ITS1, ITS2 and 5.8S regions along with partial 18S and 28S rRNA genes were sequenced for MCC 1548 (newly designated as Currahmyces indicus) and the sequences deposited in GenBank with accession nos. MK340498 (ITS), MK340499 (LSU), MK340500 (SSU). Sequencing of the partial 28S rRNA gene of MCC 1548 resulted in 841 bp nucleotide sequence of the 5′ end. A nucleotide BLAST search of nrLSU showed a maximum similarity of 96% with Amauroascus volatilis-patellis (CBS 633.72, MH872297.1), Auxarthronopsis guizhouensis (LC5705, KU746714.1), Amauroascus purpureus (IFO 32622, AY176707.1). Sequencing of partial 28S rRNA gene of MCC 1486 resulted in 841 bp nucleotide sequence of the 5′ end. A nucleotide BLAST search of nrLSU showed a maximum similarity of 97% with Amauroascus volatilis-patellis (CBS 633.72, MH872297.1), 96% with Am. purpureus (IFO 32622, AY176707.1). Additionally, nrLSU sequences of onygenaceous (17 taxa), gymnoascaceous (5) and arthrodermataceous (4) from previous studies were retrieved from GenBank and aligned with MCC 1548 and MCC 1486. A neighbour-joining (NJ) tree was constructed using Kimura-2 parameter nucleotide substitution model. It showed distinct clades for members of three families of Onygenales (Fig. 1). The new isolates were placed within the Onygenaceae clade along with Renispora flavissima, Neogymnomyces demonbreunii and Auxarthronopsis bandhavgarhensis. Sequencing of the partial 18S rRNA gene of MCC 1548 resulted in 1332 bp nucleotide sequence of the 5′ end. A nucleotide BLAST search with 1332 bp sequence showed maximum similarity of 99% with Chlamydosauromyces punctatus (UAMH9990, NG_061019.1), Renispora flavissima (CBS 708.79, AB015784.1), Auxarthronopsis bandhavgarhensis (NFCC 2185, NG_061122.1). Sequencing of partial 18S rRNA gene of MCC 1486 resulted in 1675 bp nucleotide sequence of the 5′ end. A nucleotide BLAST search with 1675 bp sequence showed maximum similarity of 99% with Chlamydosauromyces punctatus (UAMH 9990, NG_061019.1), Renispora flavissima (CBS 708.79, AB015784.1), Aphanoascus mephitalis (CBS 453.75, AB015779.1), Uncinocarpus reesii (UAMH 160, L27991.1), A. fulvescens (FMR 2179, AJ315172.1), A. verrucosus (NBRC 32382, JN941595.1). The nrSSU sequences from previous studies, including 17 Onygenaceous, 3 Gymnoascaceous and 3 Arthrodermataceous taxa, were aligned with MCC 1548 and MCC 1486. Incomplete portions from both ends were excluded from the analysis. The neighbour-joining SSU tree resolved the strains into distinct families of Onygenales (Fig. 2). The new fungi formed a distinct clade, comprised of Chlamydosauromyces punctatus, Auxarthronopsis bandhavgarhensis and Renispora flavissima. Other related species of this clade, such as Am. purpureus, Am. volatilis-patellis, Nannizziopsis albicans and Neogymnomyces demonbreunii could not be included in the analysis as their SSU sequences were not available in GenBank. The high sequence similarity of LSU and SSU of MCC 1548 and MCC 1486 to other members of Onygenales suggests that they are members of this order and can be placed within Onygenaceae.
Neighbour-joining tree based on nucleotide sequence of 28S rRNA gene of the 26 strains of Arthrodermataceae, Gymnoascaceae and Onygenaceae listed in Table 2 along with Canomyces reticulatus MCC 1486T and Currahmyces indicus MCC 1548T. Three strains belong to Trichocomaceae, Byssochlamys nivea (AY176750.1), Eurotium herbariorum (AY176751.1), and Petromyces alliaceus (AY176752.1) served as out-group. The branch lengths are propagated to the distance values calculated in MEGA5 and values at nodes represent bootstrap percentage at 1000 replicates. Bootstrap values above 50% are shown
Neighbour-joining tree based on nucleotide sequences of 18S rRNA gene of the 23 strains of Arthrodermataceae, Gymnoascaceae, and Onygenaceae listed in Table 2 along with Canomyces reticulatus MCC 1486T and Currahmyces indicus MCC 1548T. Three strains belonging to Trichocomaceae, Byssochamys nivea (GU733368.1), Eurotium herbariorum (GU733351.1), and Petromyces alliaceus (AB002071.1) served as out-group. The branch lengths are proportional to distance values calculated in MEGA5, and values at nodes represent bootstrap percentage of 1000 replicates. Bootstrap values above 50% are shown
Sequencing of ITS region of MCC 1548 resulted in 563 bp long nucleotide sequence that included 225 bp of ITS1, 150 bp of 5.8S and 183 bp of ITS2. A nucleotide BLAST search of 563 bp ITS sequence showed maximum similarity of 86% with Chrysosporium speluncrum (CCF 3760, NR_13753719), 85% with Auxarthronopsis guizhouensis (LC5705, KU746668.1), A. bandhavgarhensis (NFCCI 2185, NR_153515.1), Am. purpureus (IFO 32622, AJ271564.1), all with less than 85% query coverage. In comparison with LSU and SSU, the ITS sequence showed more difference, minimum 14% with other closest related species. Sequencing of ITS region of MCC 1486 resulted in 631 bp long nucleotide sequence that included 261 bp of ITS1, 150 bp of 5.8S and 203 bp of ITS2. A nucleotide BLAST search of 631 bp of ITS sequence showed maximum similarity of 84% with Auxarthronopsis guizhouensis (LC5705, KU746668.1), Chrysosporium speluncrum (CCF3760, NR_137537.1), Am. volatilis-patellis (CBS 249.72, MH860467.1), 82% with Nannizziopsis albicans (IMI 155645, AJ271432.1), all with very less query coverage. The remaining sequences were even more distant. The ITS sequences of MCC 1548 and MCC 1486 were compared with sequences of 30 other species from ten genera of Onygenaceae (Table 2), and a phylogenetic tree (NJ) was constructed using Kimura-2 parameter model (Fig. 3). Both the new isolates MCC 1548 and MCC 1486 were placed in a clade comprising of Neogymnomyces demonbreunii and Renispora flavissima.
Neighbour-joining tree based on nucleotide sequences of internal transcribed spacer (ITS) region, 5.8S rRNA gene of 31 strains listed in Table 2 which includes reference strains of species belonging to genera in Onygenaceae and Arthrodermataceae along with Canomyces reticulatus (MCC 1486T) and Currahmyces indicus (MCC 1548T). The unrooted tree was drawn using 736 nucleotides of ITS 1, 2 and 5.8S rRNA gene using MEGA5 software. The branch lengths are proportional to distance calculated in MEGA5, and values at nodes represent bootstrap percentages of 1000 replicates. Bootstrap values above 50% are shown
Sequencing of the 28S rRNA gene of CBS 143635 resulted in 852 bp long sequence of the 5′ end. A nucleotide BLAST search of nrLSU showed maximum similarity of 99% with Ctenomyces serratus (CBS 491.63, MH896995.1; CBS 187.61, NG_058765.1), 97% with Chrysosporium vallenarense (UAMH 6914, AY176732.1). Sequencing of 18S rRNA gene of CBS 143635 resulted in 1024 bp long sequence of the 5′ end. A BLAST search showed maximum similarity of 99% with Ctenomyces serratus (CBS 187.61, NG_062605.1), Arthroderma curreyi (CBS 138.26, AJ315165.1), Nannizzia gypsea (CBS 118893, XR_001951136.1). Sequencing of ITS region of CBS 143635 resulted in 593 bp long sequence of the 5′ end. A nucleotide BLAST search of ITS sequence of CBS 143635 showed maximum similarity of 96% with type strain of Ctenomyces serratus (CBS 187.61, NR_144890.1) and 97% with Ctenomyces vellereus (CBS 478.76, HQ871796.1). However, two sequences deposited as Ctenomyces serratus, i.e. CBS 223.64 (MH858426.1) and CBS 222.64 (MH858425.1) showed 99% and 98% similarity, respectively. It seems that these two strains belong to the new species as they are distant from the type strain of Ct. serratus. An NJ phylogenetic tree constructed using ITS sequences of CBS 143635 along with nine sequences belonging to eight species of Arthrodermataceae shows Ct. indicus clustering with Ct. serratus and Ct. vellereus with high bootstrap support (Fig. 4).
Neighbour-joining tree based on nucleotide sequence of internal transcribed spacer 1, 2 and 5.8S rRNA gene of 9 strains of family Arthrodermataceae which includes reference strains of seven genera along with Ctenomyces indicus CBS 143635T. Guarromyces ceretanicus was used as out-group. The phylogenetic tree was drawn using 558 nucleotides of ITS 1, 2 and 5.8S rRNA gene using MEGA5 software. The branch lengths are proportional to distance values calculated in MEGA5, and values at nodes represent bootstrap percentage of 1000 replicates. Bootstrap values above 50% are shown
Morphologically, the new taxon designated as Currahmyces indicus (MCC 1548) resembles Amauroascus due to it fragile ascoma (incompositoperidium) and broadly punctate-reticulate ascospores. The new taxon forms crystals on its peridial hyphae that are rich in Ca, C and Cl as revealed by EDAX-SEM analysis (data not shown). The exact composition of these crystals is still not known. However, crystals on peridial hyphae are absent in Amauroascus. The other new taxon Canomyces reticulatus forms an incompositoperidium similar to Neogymnomyces, but both are phylogenetically distinct and also in their anamorphs which is Chrysosporium-like in the latter. The new species Ctenomyces indicus differs from Ct. serratus in the size of the conidia which are smaller in the new species and also the number of swollen pedicels that are more in number in the new species (usually 3–4). Phylogenetically, they have 4% difference in the ITS region.
Taxonomy
Currahmyces Rahul Sharma & Shouche gen. nov.
Mycobank MB829247
Etymology
Named in the honour of Dr. Randolf S. Currah, Canadian Mycologist for his contribution to the taxonomy of order Onygenales.
Description
Ascomata solitary, globose to subglobose, white on the outer periphery and brown in the centre (due to ascospore mass). Peridium made up of loosely arranged thin- to thick-walled asperulate, septate hyphae. Peridial hyphae densely asperulate, towards margin covered with spine-like crystal structures. Asci globose, brown, evanescent, 8-spored. Ascospores unicellular, pale brown globose, with broadly punctate-reticulate walls. Asexual morph: Terminal and intercalary slender arthroconidia.
Type Species
Currahmyces indicus Rahul Sharma & Shouche (2018)
Currahmyces indicus Rahul Sharma & Shouche sp. nov.
Mycobank MB829248
Genbank MK340498 (ITS), MK340499 (LSU), MK340500 (SSU)
Fig. 6 Currahmyces indicus (MCC 1548T). a, b Steriomicroscopic view of ascoma growing on hair-baited soil plate; c Ascoma with Perdial hyphae and ascospores; d Peridial hyphae with developing crystals; e Rough-walled peridial hyphae with spine like crystals; f–h broadly punctate-reticulate ascospores as seen under LM and SEM
Etymology
Indicus—referring to the country of origin India.
Description
Ascomata discrete, globose to subglobose, white, loosely arranged gymnothecium, brown at centre, 450–600 µm diam including peripheral hyphae. Peridial hyphae branched, 1.4–2.8 µm wide, thick-walled hyaline to brown, covered with crystals aggregations 2.7–6.1 µm diam. Asci globose, brown, 7–8 µm, 8 spored, evanescent. Ascospores yellowish to pale brown, globose, 3–3.4 µm diam, punctate. Puncta broad, mostly 4 seen from side view, 0.67–1.2 µm wide.
Asexual Morph
Slender terminal and intercalary arthroconidia. Intercalary conidia are more abundant than terminal, 0.8–1.2 × 2.7–9.3 µm. Vegetative hyphae smooth walled, septate, 0.7–1.1 µm wide.
Cultures
Colonies after 4 week at 28 °C (Fig. 1) on SDA (4.5–4.7 cm) grossly circular, margin lobate, white to cream with pinkish tinge, cottony, slightly raised, reverse reddish pink with cream-coloured margin; on PDA (4.4 cm) circular with lobate tufts of mycelia, white cottony, raised, dense mycelia growth except margin, reverse reddish pink centre, rest yellowish; on OA (4 cm) circular, snowy white, cottony, slightly raised throughout, reverse cream coloured; on CMA (4.6 cm) circular, white, sparse aerial growth, centre with dense mycelia growth, some sectors with little or no aerial growth, reverse yellowish centre, rest uncoloured. At 37 °C after 16 d on SDA (2 cm) circular cottony white with yellowish patches in centre, raised throughout, margin flat, reverse dark reddish brown, margin pale yellow.
Type:
India: Maharashtra: Beed, Nandur Phata. Ascoma developed in soil S385 (hair baited) collected from hen resting area. 04 June 2016, R. Sharma (MCC H1007-holotype); MCC 1548 = MS385-ASCO1-ex-type cultures).
Substratum
Soil. Distribution: Known only from type locality, Nandur Phata, Beed District, Maharashtra, India.
Canomyces Rahul Sharma & Shouche gen. nov.
Mycobank MB829249
Etymology
Named in the honour of Dr. Josep Lira-Cano for his many contribution to the order Onygenales.
Description
Ascomata solitary, globose, cream coloured. Peridium made up of loosely arranged undifferentiated hyphae. Asci globose, pale brown, evanescent, 8-spored. Ascospores unicellular, pale cream to yellowish, regularly punctate-reticulate. Asexual morph Narrow elongate arthroconidia.
Type Species
Canomyces reticulatus Rahul Sharma & Shouche (2018)
Canomyces reticulatus Rahul Sharma & Shouche so. nov.
Mycobank MB829250
Genbank MK340501 (ITS), MK340502 (LSU), MK340503 (SSU)
Fig. 9 Canomyces reticulatus (MCC 1486T). a Close-up of colony on SDA showing formation of ascomata after 2 months of incubation; b Developing ascomata in between hyphal network on SDA; c, d Peridial hyphae and ascospores; e punctate-reticulate ascospores; f Asci; g Asci and ascospores (most of the ascospores have collapsed due to vacuum pressure except few indicated by arrow; h, j Narrow elongated arthroconidia; i Arthroconidia with swollen ends
Etymology
Reticulatus—referring to the punctate-reticulate ascospores of the fungus.
Description
Ascomata discrete, globose, pale cream, 300–450 µm diam. Peridial hyphae undifferentiated from vegetative hyphae, smooth walled, 1.2–3.6 µm wide. Asci globose pale brown 13–16.7 µm, 8-spored, evanescent. Ascospores unicellular, yellowish to pale brown, globose, regularly punctate-reticulate, 5.6–6.3 µm diam, mostly 18 puncta seen from surface view, 0.8–0.9 µm broad.
Asexual Morph
Narrow elongate arthroconidia.
Cultures
Colonies after 4 week at 28 °C on SDA (5.4–5.5 cm) circular, white, cottony flat, uniform with some sectors with sparse aerial mycelia reverse pale brown fading towards margin; on PDA (4.3 cm) circular, white with sectors having little aerial mycelia giving yellowish brown appearance due to submerged growth, reverse brown at centre fading towards margin; on OA (4.5 cm) circular, snowy white, with irregular patches, flat, reverse cream coloured with central brown spot; on CMA (2.5 cm) grossly circular, lobate, yellowish, sparse aerial mycelia with irregular rings, reverse pale brown with dark centre. At 37 °C after 16 days on SDA (2.8 cm) circular, white, cottony, flat with central raised knob, centre pale coloured, rest white, reverse pale brown, growing margin pale yellow.
Type
India, Maharashtra, Beed, Pithi, Ascomata developed in soil S393 (hair baited) collected under a tree, 04 June 2016, R. Sharma (MCC H1008-holotype: MCC 1486 = MS393-AVP-cultures-extype).
Substratum
Soil
Distribution
Known only from type locality Pithi, Beed District, Maharashtra, India.
Ctenomyces indicus Rahul Sharma & Shouche sp. nov.
Mycobank MB829251
Genbank MK340504 (ITS), MK340505 (LSU), MK340506 (SSU)
Fig. 10
Etymology
Indicus—referring to the country of origin India.
Diagnosis
Conidiophores short swollen pedicels or inflated cells, conidia hyaline to buff rough-walled obovoid to globose that are smaller in size in comparison with Ctenomyces serratus.
Description
Conidiophore absent or made up of one, two or three swollen/inflated vesicular cells. Conidia rough-walled obovoid to globose hyaline when young turning to buff, (6.2) 6.4–8.2 (8.6) × (7.3) 7.9–9.8 (10.4) µm. Swollen conidiogenous cells hyaline, thin walled, 2.2–4.2 × 5.2–5.4 µm. Vegetative hyphae hyaline, thin-walled, smooth, septate, 1.1–1.3 µm wide.
Cultures
After 2 week at 28 °C on SDA (4.2–4.6 cm) circular, powdery uniform, tan coloured dense growth, with or without concentric rings, growing margin white, reverse pale coloured; on PDA (4.9–5.4 cm) circular, pale tan in colour, flat, powdery with patches without mycelia, with 1–2 concentric rings, reverse pale cream. After 16 days at 37 °C (0.6–2 cm) circular, tan, powdery, centre knob-like raised, flat, margin white, reverse centre dark brown rest pale brown.
Sexual Morph
Not seen.
Type
India, Maharashtra, Latur District, Rachanwadi, conidia developed on hair-baited soil S 448, hen frequented area, 29 December 2016, R. Sharma (MCC H1009-holotype: MS448 M = CBS 143635-cultures-extype).
Substratum
Soil
Distribution
Latur, Hingoli and Nanded District of Maharashtra, India.
Other Material examined: MS466 M (MCC 1513 = CBS 143634); MS447M2 (MCC 1514)
Discussion
Survey of soils of Maharashtra State in India showed diverse forms of Onygenalean fungi represented by members of family Arthrodermataceae, Onygenaceae and Gymnoascaceae. The most common genera encountered were Nannizzia and Chrysosporium. Although Chrysosporium is a group of morphologically similar forms that are phylogenetically diverse and may represent distinct genera. A revision of this genus is currently needed taking into account all close relatives as to ascertain the taxonomic boundaries. Although in the last phylogenetic assessment of the genus, 38 Chrysosporium species were included along with only 18 species from other genera [29].
Saprophytic Onygenalean fungi are mostly found in habitats that are rich in keratin as well as dung of various animals, mostly rodents. A fact that might be overlooked in previous studies is their association with insects which help disperse these fungi. In present study, we found some of the Onygenales fungi forming ascomata on insect remains in soil collected from burrows or hen frequented areas. The mesh-like peridial hyphae may help the peridium to cling to insect body and disperse the ascospores to newer locations.
The current study showed high per cent incidence of keratinophilic fungi in almost all habitats sampled in comparison with previous studies [30,31,32,33], mainly due to the fact that we collected soil from locations where there was a clear presence of shed keratin substrate. All burrows sampled were positive for Onygenalean fungi.
The newly isolated Onygenalean fungi represent missing forms indicated by Currah in his seminal paper (1985) [1] as evidenced by sudden discontinuity in the morphological forms. Currahmyces indicus is unusual in Onygenales as it forms crystal-like structures on its peripheral hyphae of its peridium. The gross morphology of the asomata is reminiscent of genus Amauroascus some of whose species forms fragile ascomata with punctate-reticulate ascospores; however, they lack formation of crystalline structures. Phylogenetically, C. indicus is distant from the type species of Amauroascus, i.e. A. niger. The genus which currently comprises 13 species [2] is polyphyletic in nature. The species of Amauroascus forms two groups based on ascospores types, one forms punctate-reticulate ascospores while other forms irregularly verruculose-punctate ascospores, but these groups are not phylogenetically supported. Morphologically, the globose ascospores of C. indicus resemble those of Am. burundensis in having broadly punctate-reticulate wall, but the walls of puncta are broader in this Amauroascus species. Also, the ascospore size of C. indicus is slightly smaller (3–3.4 µm diam) than Am. burundensis (3.5–4 µm). The colony of C. indicus is white as compared to Am. burundensis which is orange (on OA). Phylogenetically, C. indicus is placed in a clade comprising Canomyces reticulatus, Neogymnomyces demonbreunii and Renispora flavissima (Figs. 1, 2 and 3). All these taxa are morphologically quite distinct from the new taxon, and hence, it could not be placed in any of them. Ascospores of N. demonbreunii are discoid-oblate, punctate to smooth (2.5–3 × 2.5–4 µm), while those of Renispora flavissima are reniform and finely pitted (2.5 × 4–5.5 µm) unlike C. indicus which are globose, broadly punctate-reticulate (3–3.4 µm).
The second new taxon Canomyces reticulatus is phylogenetically close to C. indicus, N. demonbreunii and Renispora flavissima. It also could not be placed in any of the phylogenetically close relatives due to morphological differences. Although the gross morphology of the ascomata of C. reticulatus is somewhat similar to N. demonbreunii, it differs in shape and size of the ascospore and the type of anamorph which is arthroconidial in the former and Chrysosporium in the latter. Renispora flavissima also differs from C. reticulatus in having reniform, pitted ascospores and the type of anamorph which are tuberculate aleurioconidia.
Ctenomyces indicus clusters with other Ctenomyces species with high bootstrap value. Morphologically, both the species form rough-walled aleurioconidia borne on swollen cells, the difference lies in the size of conidia (bigger in Ct. serratus) and number of swollen pedicels (which are more in Ct. indicus). The new name Ct. vellereus is used instead of Myceliophthora vellerea [34, 35] in the phylogenetic tree. Figure 12 shows nucleotide differences in the three Ctenomyces species. In Ct. indicus, there are eight loci with unique apomorphies while Ct. vellereus has four and Ct. serratus has three loci with unique apomorphies.
Of the three novel taxa being described here, two (both new genera) were recovered from only one sample each out of the total 578 samples analyzed. Had we missed those two samples during our sampling, we would have failed to recover these two novel forms. The present study indicates that systematic sampling of a large area is essential to recover forms that have very restricted distribution. Selection of keratin-rich habitats is also a key in obtaining rare Onygenalean forms, many of which are still un-described.
References
Currah RS. Taxonomy of the Onygenales: Arthrodermataceae, Gymnoascaceae, Myxotrichaceae and Onygenaceae. Mycotaxon. 1985;24:1–216.
Guarro J, Gene J, Stchigel AM, Figureas MJ. Atlas of soil ascomycetes., CBS Biodiversity Series 10Utrecht: Centraalbureau voor Schimmelcultures; 2012.
Sutton DA, Marin Y, Thompson EH, Wickes BL, et al. Isolation and characterization of a new fungal genus and species, Aphanoascella galapagosensis, from carapace keratitis of a Galapagos tortoise (Chelonoidis nigra microphyes). Med Mycol. 2013;51:113–20.
Sharma R, Graeser Y, Singh SK. Auxarthronopsis, a new genus of Onygenales isolated from the vicinity of Bandhavgarh National Park, India. IMA Fungus. 2013;4(1):89–102.
Dukik K, Munoz J, Jiang Y, Feng P, et al. Novel taxa of thermally dimorphic systemic pathogens in the Ajellomycetaceae (Obygenales). Mycoses. 2017;60(5):296–309.
Marin-Felix Y, Stchigel AM, Cano JF, et al. Emmonsiellopsis, a new genus related to the thermally dimorphic fungi of the family Ajellomycetaceae. Mycoses. 2015;58(8):451–60.
Johnston PR, Nguyen HD, Park D, Hirooka Y. Harorepupu aotearoa (Onygenales) gen. sp. nov.; a threatened fungus from shells of Powelliphanta and Paryphanta snails (Rhytididae). IMA Fungus. 2015;6(1):135–43.
De Hoog GS, Dukik K, Monod M, Packeu A, et al. Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia. 2017;182(1):5–31.
Crous PW, Wingfield MJ, Burgess TL, et al. Fungal planet description sheets: 558–624. Persoonia. 2017;38:240–384.
Sigler L, Hambleton S, Pare JA. Chlamydosauromyces punctatus gen. & sp. nov. (Onygenaceae) from the skin of a lizard. Stud Mycol. 2002;47:123–9.
Hirooka Y, Tanney JB, Nguyen HDT, Seifert KA. Sigleria gen. nov. in Spiromastigaceae (Onygenales, Eurotiomycetes). Mycologia. 2016;108(1):135–56.
Ajello L. Natural history of the dermatophytes and related fungi. Mycopath Mycol Appl. 1974;53:93–110.
Guarro J, Gene J, de Vroey Ch. Amauroascopsis, a new genus of Eurotiales. Mycotaxon. 1992;45:171–8.
Cano J, Sole M, Pitrach LB, Guarro J. Castanedomyces australiensis, gen. nov., sp. nov., a keratinophilic fungus from Australian soil. Stud Mycol. 2002;47:165–72.
Udagawa S, Uchiyama S. Taxonomic studies on new or critical fungi of non-pathogenic Onygenales-1. Mycoscience. 1999;40:277–90.
Sigler L. Ophidiomyces Sigler, Hamleton & Paré, gen. nov. IF550166 and IF550167. In: Nomenclatural novelties: Lynne Sigler. Index Fungorum no. 19. 2013; http://www.indexfungorum.org/Publications/Index%20Fungorum%20no.19.pdf.
Sigler L, Hambleton S, Pare JA. Molecular characterization of reptile pathogens currently known as members of the Chrysosporium anamorph of Nannizziopsis vriesii complex and relationship with some human-associated isolates. J Clin Microbiol. 2013;51(10):3338–57.
Cano J, Sole M, Pitarch LB, Guarro J. Pseudoamauroascus, a new genus of the Onygenales (Ascomycota). Stud Mycol. 2002;47:173–9.
Solé M, Cano J, Pitarch LB, Stchigel AM, Guarro J. Molecular phylogeny of Gymnoascus and related genera. Stud Mycol. 2002;47:141–52.
Cano J, Guarro J, Castenada RF. Studies on keratinophilic fungi IV. Bifidocarpus, a new genus of the Eurotiales. Mycotaxon. 1994;52:53–7.
Sharma R, Singh SK. A new species of Gymnoascus with verrucolose ascospores. IMA Fungus. 2013;4:177–86.
Sharma R. Some keratinophilic fungi new to India. J Mycopathol Res. 2016;54:35–9.
Vanbreuseghem R. Technique biologique pour l’isolment des dermatophytes du sol. Annales de la Societe Belge de Medicine Tropicale. 1952;32:173–8.
Sharma R, Rajak RC. Keratinophilic fungi: nature’s keratin degrading machines! Their isolation, identification and ecological role. Resonance. 2003;8:28–40.
Sharma R, Rajak RC, Pandey AK. Teaching technique for mycology 19: a micro-dilution drop-trail method for isolating Onygenalean ascomycetes from hair baits. Mycologist. 2002;16:153–8.
Riddel RW. Permanent stained mycological preparations obtained by slide culture. Mycologia. 1950;42:265–70.
Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349.
Tamari F, Hinkley CS, Ramprasad N. A comparison of DNA extraction methods using Petunia hybrida tissues. J Biomol Tech. 2013;24:113–8.
Vidal P, Vinuesa MA, Sanchez-Puelles JM, Guarro J. Phylogeny of the anamorphic genus Chrysosporium and related taxa based on rDNA internal transcribed spacer sequences. Rev Iberoam Micol. 2000;17:22–9.
Deshmukh SK, Verekar SA. The occurrence of dermatophytes and other keratinophilic fungi from the soils of Himachal Pradesh (India). Czech Mycol. 2006;2006(58):117–24.
Deshmukh SK, Verekar SA, Chavan YG. Incidence of keratinophilic fungi from the selected soils of Kaziranga National Park, Assam, (India). Mycopathologia. 2017;182:371–7.
Kachuei R, Emami M, Naeimi B, et al. Isolation of keratinophilic fungi from soil in Isfahan province, Iran. J Mycol Med. 2012;22:8–13.
Garg AK. Isolation of dermatophytes and other keratinophilic fungi from soils in India. Sabouraudia. 1966;4:259–64.
van den Brink J, Samson RA, Hagen F, Boekhout T, de Vries RP. Phylogeny of the industrial relevant, thermophilic genera Myceliophthora and Corynascus. Fungal Divers. 2012;52:197–207.
Kirk PM. Nomenclatural novelties. Index Fungorum. 2014;120:IF660468.
Acknowledgements
RS thanks Rohit Sharma for extensive help with soil sample collections, Council of Scientific and Industrial Research (CSIR) for Senior Research Associateship (SRA-Pool No. 8766-A). The authors thank Sugat V Shende, Department of Physics, Savitribai Phule Pune University, and Mr Vijay, Indian Institute for Science, Education and Research (IISER), Pune, for scanning electron microscopy. The authors thank Director, National Centre for Cell Science (NCCS), Pune, for laboratory facilities. The work was financially supported by Department of Biotechnology, New Delhi, India (BT/PR10054/NDB/52/94/2007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Handling Editor: Sybren de Hoog.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharma, R., Shouche, Y.S. Diversity of Onygenalean Fungi in Keratin-Rich Habitats of Maharashtra (India) and Description of Three Novel Taxa. Mycopathologia 185, 67–85 (2020). https://doi.org/10.1007/s11046-019-00346-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-019-00346-7