Abstract
This study first report to identify the mating type (−)-specific gene of alpha-box and the mating type (+)-specific gene of the high-mobility-group (HMG) DNA-binding domain in zoophilic dermatophytes of Arthroderma benhamiae in an effort to understand the epidemiological characteristics of Trichophyton mentagrophytes. The sequence of the alpha-box gene (1,387 bp) was found to contain two exons, from 184 to 475 bp and from 525 to 1,387 bp, coding a protein of 384 amino acids, beginning with a putative initiating methionine (ATG). The sequence of the HMG gene (1,910 bp) contained two exons, from 234 to 415 bp and from 479 to 1,457 bp, coding a protein of 386 amino acids, beginning with a putative initiating methionine (ATG).
PCR analysis detected the alpha-box gene in A. benhamiae (−) mating type strains but not in (+) mating type strains. On the other hand, the HMG gene was detected in A. benhamiae (+) mating type strains but not in (−) mating type strains. These findings suggest that the HMG and alpha-box genes could be specific to the (+) and (−) mating types, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In filamentous ascomycetes, the mating type genes are found on a MAT locus [1, 2]. The MAT locus is a region of low sequence similarity between two opposite mating types of fungi and is different in the idiomorph in each mating type [1, 2]. The MAT locus usually contains genes for one or more transcription factors with structural motifs such as alpha-box (in MAT1-1) and the high-mobility-group (HMG) DNA-binding domain (in MAT1-2).
The dermatophytes are members of the genera Trichophyton, Microsporum, and Epidermophyton. Their sexual states are classified in the genus Arthroderma. Many but not all members of Arthroderma frequently cause infections in keratinized tissues, that is, the epidermis and hair and nails, in humans and animals [3]. Full genome sequences of different dermatophyte species (Microsporum gypseum, M. canis, Trichophyton rubrum, T equinum, and T. tosurans) have become available for the public (Broad Institute; http://broad.mit.edu/science/data#). Wenjun et al. identified the MAT locus of four dermatophytes (Microsporum gypseum, M. canis, Trichophyton rubrum, and T. tosurans) but did not examine that of Trichophyton mentagrophytes [4].
T. mentagrophytes is one of the causative agents of both human and animal dermatophytoses and divided into two distinct forms; zoophilic and anthropophilic [3]. The zoophilic isolates of T. mentagrophytes have been generally identified by morphological and biochemical examination as well as through mating experiments. The confirmed teleomorphs of the zoophilic isolates of the T. mentagrophytes complex are A. benhamiae, A. simii, and A. vanbreuseghemii [5–7]. On the other hand, no teleomorph has been identified in T. mentagrophytes var. interdigitale as in the other anthropophilic strains like T. rubrum. The mating ability of dermatophytes appears to be related to their pathogenicity in humans and animals [3].
The aim of this study was to confirm the existence of the alpha-box and HMG genes in genomic DNA from A. benhamiae by PCR analysis and to examine their specificity to the mating types of A. benhamiae, in order to clarify the epidemiological characteristics of T. mentagrophytes.
Materials and Methods
Strains
The type strains of A. benhamiae, (+) mating type strain, VUT-77011, and (−) mating type strain, VUT-77012, were used in this study. The type strains and the other strains of A. benhamiae used are listed in Table 1.
Preparation of Genomic DNA
Mycelia of strains were obtained by culturing cells in Sabouraud’s dextrose broth at 27°C for 7 days [3]. The mycelial cells of dermatophytes were collected by centrifugation at 1,500 ×g for 5 min and then frozen in liquid nitrogen and homogenized. They were then lysed in a lysis buffer containing 1 mg of zymolyase-100T/ml (Takara Bio Company, Kyoto, Japan), 0.1 mM EDTA, 0.1% sodium dodecyl sulfate (SDS), 10 mM Tris hydrochloride (pH 8.0), and 0.3% 2-mercaptoethanol, at 37°C for 16 h. High molecular weight DNA was extracted from the mycelial cells by the phenol/chloroform method. DNA samples dissolved in TE buffer (10 mM Tris–HCl, pH 8.0, and 1 mM EDTA) were used for further analysis.
Alpha-Box Gene Sequence
To clone the alpha-box gene from 4 (−) mating type strains (VUT-77012, VUT-97010, 4–23, and 4–40), primers were prepared based on the conserved sequence of the alpha-box gene of Microsporum gypseum MAT1-1 and T. rubrum MAT 1-1 [4]. The sense primers were ABalpha1S (1–19 bp): 5′-GCAAGTTCACCTCCCAGCC, Abalpha2S (400–409 bp): ACTCGACCTGCGTCACGCAG, and Abalpha3S (736–754 bp): CCTTGATACCATGGGTTTG. The antisense primers were ABalpha1R (1–19 bp): 5′-GCAAGTTCACCTCCCAGCC, Abalpha2R (400–409 bp): ACTCGACCTGCGTCACGCAG, and Abalpha3R (1,368–1,387 bp): CCTTGATACCATGGGTTTG.
Thirty-five cycles of PCR amplification were performed under the following conditions: denaturation for 1 min at 94°C, primer annealing for 1 min at 55°C, and polymerization for 2 min at 72°C in the total reaction volume of 30 μl of amplification mixture containing 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 200 mM each deoxynucleoside triphosphate, 1.0 U Taq polymerase (Takara, Kyoto, Japan), and 0.5 μg of each primer.
The PCR products were purified by gel electrophoresis and cloned into the pCRII vector (Invitrogen, San Diego, CA, USA). The plasmid DNA from more than three clones of each species was extracted using the QIAGEN plasmid kit (QIAGEN, Vallencia, CA, USA) and separately sequenced by the dideoxy chain termination method using an ABI PRISM 310 Genetic Analyzer (PerkinElmer, Inc., Foster City, CA, USA).
HMG Gene Sequence
To clone the HMG gene 3 (+) mating type strains (VUT-77011, 4–13 and 4–18), degenerate primer sequences were prepared based on the conserved amino acid sequence of the HMG box transcription factor of Aspergillus nidulans MAT1-2-1 [2]. The sense primer (HsHMG1S) was 5′-CCI CGI CCI CCI AAT(C) GCI TT-3′ (amino acid sequence, PRPPNAF), and the antisense primer (HsHMG1R) was 5′-CGI CGT(C) TTT(C) TTT(C) TCI GAI GG-3′ (amino acid sequence, PSEKKRR).
The gene-specific primers for GenomeWalker™ were designed from the sequences of progressively amplified products beginning with the sequences of the MAT1-2 gene fragment of A. benhamiae. GenomeWalker™ procedures were carried out according to the GenomeWalker™ Universal user manual (Takara). The PCR products obtained were sequenced using the techniques mentioned previously.
PCR Analysis of the Alpha-Box and HMG Genes Specific to the Genomic DNA of the A. benhamiae (+) and (−) Mating Type Strains
The primers Abalpha3S and Abalpha3R amplified a 650-bp fragment of the A. benhamiae alpha-box gene. The primers used for amplification of the HMG fragment were 5′-CTGTATCGCCAACATCACCA-3′ (primer ABHMG1S; nucleotides [nt.] 714–733 in the A. benhamiae HMG sequence DDBJ - AB542198) and 5′-AGCCTCACTGGGCATCATCA-3′ (primer ABHMG1R; nt. 1,455–1,474 in the same sequence).
The genomic DNA samples (100 ng) were amplified by PCR in a volume of 30 μl, using a reaction mixture containing 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 200 mM each deoxynucleoside triphosphate, 1.0 unit of Taq polymerase (Takara), and 0.5 μg of a pair of primers. Amplification was carried out over 35 cycles consisting of template denaturation (1 min, 94°C), primer annealing (1 min, 55°C), and polymerization (2 min, 72°C). The PCR products were analyzed on a 2% agarose gel, stained with ethidium bromide, and visualized with UV light.
Results and Discussion
The sequence of the alpha-box gene (1,387 bp) contained two exons, from 184 to 475 bp and from 525 to 1,387 bp, coding a protein of 384 amino acids, beginning with a putative initiating methionine (ATG).
These two amino acid sequences of the A. benhamiae alpha-box gene shared approximately 76.3% sequence similarity with that of the Microsporum gypseum alpha-box gene in the conserved region (GenBank accession number: FJ798800) (Fig. 1), but only shared 35 and 36% sequence similarity with those of the Coccidioides immitis alpha-box gene (GenBank accession number: EF512009S4) and the Coccidioides posadasii alpha-box gene (GenBank accession number: EF512013), respectively, in the conserved region.
Nucleotide sequence and putative amino acid sequence similarities were 99% and 93–99%, respectively, among the alpha-box genes of the four A. benhamiae strains (VUT-77012, VUT-97010, 4–23, and 4–40). The sequences determined in this study have been deposited in GenBank (Arthroderma benhamiae alpha-box gene for the alpha-box domain protein, complete coding sequences (cds). GenBank accession numbers: AB570254, AB570255, AB570256, and AB570257).
The sequence of the HMG gene (1,910 bp) contained two exons, from 234 to 415 bp and from 479 to 1,457 bp, coding a protein of 386 amino acids, beginning with a putative initiating methionine (ATG).
Nucleotide sequence and putative amino acid sequence similarities were 99% and 99–100%, respectively, among the HMG genes of the three A. benhamiae strains (VUT-77011, 4–13 and 4–18). The sequences determined in this study have been deposited in GenBank (Arthroderma benhamiae HMG gene for the HMG domain protein, complete cds. GenBank accession numbers: AB542198, AB570252, and AB570253).
The amino acid sequences of the A. benhamiae HMG genes and Microsporum gypseum HMG gene shared approximately 80.5% sequence similarity in the conserved region (GenBank accession number: FJ798798) (Fig. 2), but only shared 42 and 43.5% sequence similarity with those of the Ajellomyces capsulatus HMG gene (GenBank accession number: EF472255) and the Coccidioides posadasii HMG gene (GenBank accession number: EF472258), respectively, in the conserved region.
Comparison of the homologous regions of the predicted protein sequences of (+) mating type strains (VUT-77011) of A. benhamiae HMG and M. gypseum HMG (GenBank FJ798798). An asterisk indicates identity with the amino acid found in the A. benhamiae HMG sequence. The box indicates the highly conserved HMG signature sequence [2]
Previous phylogenetic studies of 18S and 25S rDNA sequences indicated that the dermatophytes are closely related to A. capsulatus and C. posadasii [8, 9]. In a previous study, we sequenced the chitin synthase 1 (CHS1) gene of A. benhamiae and found that the amino acid sequence of the A. benhamiae CHS1 gene in the conserved region shared approximately 70% sequence similarity with the Aspergillus nidulans CHSC gene, the Coccidioides immitis CHS1 gene, the Exophiala dermatitidis CHS2 gene, and the Neurospora crassa CHS3 gene [10].
Therefore, the alpha-box and the HMG genes of dermatophytes might be intraspecifically conserved among species and be low sequence sililality from the other MAT genes of filamentous ascomycetes.
In the present study, the sequences of the alpha-box and HMG genes of A. benhamiae were fully determined. The amino acid sequence similarity of the alpha-box genes between A. benhamiae and M. gypseum was relatively high (Fig. 1). However, neither of these two alpha-box genes had the highly conserved alpha-box signature sequences “RPLNSFIAFRSFYS” and “DPFKAKWAIVAKAYS” of filamentous ascomycetes [2].
The amino acid sequence similarity between the A. benhamiae and M. gypseum HMG genes was relatively high, particularly in the highly conserved HMG signature sequences “PRPPNAFILYR” and “PRKPSEKKRR” of filamentous ascomycetes [2] (Fig. 2). Thus, the amino acid sequences of the A. benhamiae HMG gene were examined in further detail.
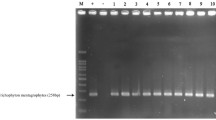
PCR analysis using 35 amplification cycles also detected the alpha-box gene in A. benhamiae (−) mating type strains, but not in A. benhamiae (+) mating type strains (Fig. 3). On the other hand, the HMG gene was detected in the A. benhamiae (+) mating type strains, but not in the A. benhamiae (−) mating type strains (Fig. 3).
Results of PCR of the A. benhamiae HMG gene and the A. benhamiae alpha-box gene. Lane 1, VUT-77011 (+); lane 2, NUBS-09011 (+); lane 3, 4–13 (+); lane 4, 4–18 (+); lane 5, VUT-77012 (−); lane 6, VUT-97010 (−); lane 7, 4–23 (−); and lane 8, 4–40 (−). Chitin synthase gene 1 (CHS1) [10] was the positive control for PCR amplification for sample DNA
PCR analysis indicated that the alpha-box gene is present exclusively in A. benhamiae (−) mating type strains while the HMG gene is present exclusively in A. benhamiae (+) mating type strains (Fig. 3). These results suggest that the alpha-box gene and HMG gene exist in the opposite mating type strain, indicating that A. benhamiae is heterothallic species by this molecular analysis.
References
Casselton LA. Fungal sex genes-searching for the ancestors. BioEssays. 2008;30:711–4.
Bubnick M, Smulian AG. The MAT1 locus of Histoplasma capsulatum is responsive in a mating type-specific manner. Eukaryotic Cell. 2007;6:616–21.
Kwon-Chung KJ, Bennett EJ. Medical mycology. Philadelphia: Lea & Febiger; 1992. pp.136-7 and 816-26.
Li W, Metin B, White TC, Heitman J. Organization and evolutionary trajectory of the mating type (MAT) locus in the dermatophyte and dimorphic pathogen. Eukaryotic Cell. 2009;6:616–21.
Ajello L, Cheng S-L. The perfect state of Trichophyton mentagrophytes. Sabouraudia. 1967;4:230–4.
Stockdale MP, Mackenzie WRD, Austwick KCP. Arthroderma simii sp. nov., the perfect state of Trichophyton simii (Pinoy) comb. nov. Sabouraudia. 1967;4:112–23.
Takashio M. The Trichophyton mentagrophytes complex. In: Iwata K, editor. Recent advances in medical and veterinary mycology. Tokyo: University of Tokyo Press; 1977. p. 271–6.
Bialek R, Ibricevic A, Fothergill A, Begerow D. Small subunit ribosomal DNA sequence shows Paracoccidioides brasiliensis closely related to Blastomyces dermatotidis. J Clin Microbiol. 2000;34:3190–3.
Leclerc CM, Philippe H, Guého E. Phylogeny of dermatophytes and dimorphic fungi based on large subunit ribosomal RNA sequence comparisons. J Med Vet Mycol. 1994;32:331–41.
Kano R, Nakamura Y, Watanabe S, Hasegawa A. Chitin synthase 1 and 2 genes of dermatophytes. Stud Mycol. 2002;47:49–56.
Kawasaki M, Anzawa K, Takeda K, et al. Genetic and phenotypic variation among F1 progenies of Arthroderma benhamiae. Jpn J Med Mycol. 2008;49:103–10.
Acknowledgments
This study was supported by Grants-in-Aid from the Academic Frontier Project of the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Nihon University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kano, R., Yamada, T., Makimura, K. et al. Arthroderma benhamiae (The Teleomorph of Trichophyton mentagrophytes) Mating Type-Specific Genes. Mycopathologia 171, 333–337 (2011). https://doi.org/10.1007/s11046-010-9383-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-010-9383-5