Abstract
Morphological features and Inter Simple Sequence Repeat (ISSR) polymorphism were employed to analyse 21 Corynespora cassiicola isolates obtained from a number of Hevea clones grown in rubber plantations in Malaysia. The C. cassiicola isolates used in this study were collected from several states in Malaysia from 1998 to 2005. The morphology of the isolates was characteristic of that previously described for C. cassiicola. Variations in colony and conidial morphology were observed not only among isolates but also within a single isolate with no inclination to either clonal or geographical origin of the isolates. ISSR analysis delineated the isolates into two distinct clusters. The dendrogram created from UPGMA analysis based on Nei and Li’s coefficient (calculated from the binary matrix data of 106 amplified DNA bands generated from 8 ISSR primers) showed that cluster 1 encompasses 12 isolates from the states of Johor and Selangor (this cluster was further split into 2 sub clusters (1A, 1B), sub cluster 1B consists of a unique isolate, CKT05D); while cluster 2 comprises of 9 isolates that were obtained from the other states. Detached leaf assay performed on selected Hevea clones showed that the pathogenicity of representative isolates from cluster 1 (with the exception of CKT05D) resembled that of race 1; and isolates in cluster 2 showed pathogenicity similar to race 2 of the fungus that was previously identified in Malaysia. The isolate CKT05D from sub cluster 1B showed pathogenicity dissimilar to either race 1 or race 2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corynespora cassiicola (Berk. & Curt.) Wei, a plant pathogenic fungus, has been recorded in over 70 countries and more than 280 plant species have been known to host this fungus including fruits, vegetables, grains, perennial crops, forestry and various ornamental plants [1]. On rubber (Hevea brasiliensis), the first record of C. cassiicola was from Sierra Leone in 1936 [2], later followed by India in 1958 [3], Malaysia in 1960 [4], Nigeria in 1969 [5], Indonesia in 1980 [6], Brazil [7], Sri Lanka and Cameroon [8], Thailand in 1985 [9], Bangladesh [10], Vietnam in 1998 [11] and China in 2006 [12]. Initially, C. cassiicola was considered as a weak pathogen on rubber. Since the 1980s however, the severity of Corynespora leaf fall has been on the increasing trend in many rubber growing countries. It has been acknowledged that rubber clones become more susceptible with time and susceptibility to the pathogen varies in different geographical regions [13–18]. These observations inferred to the existence of various physiological races of C. cassiicola infecting rubber tree and/or the evolution of the pathogen that may have generated new races of the fungus. Several races of C. cassiicola infecting rubber have been identified and genetic relationships among isolates have been established [18–23]. Newly bred rubber tree genotypes (clones) should be screened for resistance against the fungus prior to recommendation to growers [24]. Towards this end, detailed information on the inherent variability of the pathogen would prove useful in Hevea breeding programme.
The growing importance of Corynespora leaf fall to the rubber industry in Malaysia has resulted in the undertaking of a number of research projects to elucidate the disease and gain insight on the causal pathogen [25]. This article reports on the morphological and Inter Simple Sequence Repeat (ISSR) markers analyses performed on 21 C. cassiicola isolates that were collected from rubber plantations in Malaysia from 1998 to 2005.
Materials and Methods
Isolates of Corynespora cassiicola
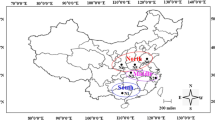
Twenty-one isolates of C. cassiicola studied were obtained from Plant Pathology Laboratory, Crop Improvement and Protection Unit, Rubber Research Institute of Malaysia. These isolates were collected from infected rubber leaves of different clones (series PB, RRIM and BA) and rubber plantations in several states of Malaysia (Johor, Selangor, Perak, Pahang, Kedah, Terengganu and Sarawak) taken from 1998 to 2005 (Table 1, Fig. 1). The isolates were purified using single spore isolation method prior to further analysis.
Map of Malaysia showing the locations of rubber plantations where the Corynespora cassiicola were collected. Numbers indicate the isolates of the fungus listed in Table 1
Colony Morphology
All isolates shown in Table 1 were used for this experiment. A 5 mm mycelial plug was taken from the actively growing margin of 5-day-old culture of C. cassiicola on Potato Sucrose Agar (PSA) (100 g potato, 10 g sucrose, 15 g agar/1 H2O) and was centrally cultured in the Petri dish containing 25 ml of the above medium. Cultures were incubated in the dark at room temperature (28 ± 2°C). The experiment was arranged in completely randomised design (CRD) with 5 replications for each treatment (isolate). Colony characteristics such as colour, shape and structure were recorded. The colony diameter was measured daily by taking the average length of two diameters at right angles for a period of 7 days. Data were subjected to analysis of variance (ANOVA) using SAS software version 8.01.
Conidial Morphology
The conidia of the studied isolates were produced using detached leaf inoculation on appropriate rubber clone from which the isolates were obtained. Detached leaflets at light green stage (10–14 days after budburst) were kept in moist plastic box with abaxial surface facing upward. The leaflets were inoculated by pipetting 20 μl drops of a spore suspension of C. cassiicola (4000 spores/ml) at 10 spots/leaflet. The inoculated leaflets were placed under fluorescent light in air-conditioned room (25°C) for 5 to 10 days. The produced spores were collected and stained using lactophenol blue and fixed onto glass slides for observation. The conidial morphology and dimensions of the isolates were photographed by using Pixelink Megapixel Firewire Camera model PL-A662 fixed on Olympus Microscope BH-2 and measured using VideoTest Size 5 software (100 conidia/isolate, except for isolate CSB16, observation was made on 82 conidia). Data were subjected to unbalance analysis of variance (General Linear Model Analysis, GLM) using SAS software version 8.01.
Fungal DNA Isolation
Pure cultures of the studied isolates were sub-cultured by placing 3 mycelial plugs taken from the actively growing margin of 5-day-old culture into Erlenmeyer flasks containing 200 ml Potato Dextrose Broth (24 g/l). The flasks were shaken at 80 rpm using an orbital shaker at room temperature (28 ± 2°C) for 10 days. The mycelia were filtered through a layer of cheese cloth, and washed with distilled water and immediately stored in a deep freezer at −80°C. The DNA extraction was performed using modified CTAB method adopted from Doyle and Doyle [26] as described by Shaji and Bindu [27]. The purification and the concentration of extracted DNAs were measured using Eppendorf BioPhotometer. The DNA of the studied isolates were then stored in the refrigerator at 4°C and used as mother stock to prepare template DNA (10 ng/μl) right before running Polymerase Chain Reaction (PCR).
Inter Simple Sequence Repeat (ISSR) PCR Analysis
The ISSR-PCR was performed using 8 ISSR primers (UBC826, UBC828, UBC835, UBC840, UBC850, Mj3, Mj4 and Mj5; Table 2) that were selected based on the results of initial screening of the 2 groups of primers against a set of representative studied isolates. One group includes 8 UBC primers (designed from University of British Columbia, Vancouver, Canada) which were successfully applied on Fusarium graminearum [28]. Another group includes 8 Mj primers which were designed from the microsatellite regions of Ganoderma spp. (Puad, unpublished). The primers were synthesized at First BASE Laboratories Sdn Bhd, Malaysia. PCR amplification of ISSR loci was carried out in a 20 μl reaction volume containing 1 × PCR buffer (Finnzymes, Finland), 1.5 mM MgCl2, 0.2 mM each dNTP, 0.5 μM of ISSR primer, 2 U DyNAzymeTMII DNA Polymerase (Finnzymes, Finland) and 20 ng of template DNA. The amplification was performed in a thermocycler (DNA Engine® Peltier Thermal Cycler PTC-200, MJ research, USA) with initial denaturation at 94°C for 5 min, followed by 35 cycles at denaturing temperature 94°C for 30 s, annealing temperature from 51 to 55°C (subject to primer, Table 2) for 1 min, extension temperature 72°C for 2 min and the final extension at 72°C for 10 min. All PCR reactions were performed in 3 replications to confirm the consistency of amplification. The amplified products were size-separated in 1.2% agarose gel under 1× TAE buffer (40 mM Tris, 20 mM Acetic acid and 1 mM EDTA) at 70 V for 45 min at room temperature. Gels were stained with ethidium bromide, visualized under UV light and photographed using a gel documentation system (GeneSnap, Ver 6.03, Syngene Laboratories). The sizes of amplified DNA fragments were estimated using GeneTools (Ver 3.00.13, Syngene Laboratories) by comparison with a 2-Log DNA Ladder (0.1–10 kb) marker (BioLabs).
Data Analysis
The stable amplified DNA fragments were scored as 1 for presence and 0 for absence of DNA band. The binary matrix data were then used for analysis of genetic similarity index. Cluster analyses were performed from Nei and Li’s similarity matrix [29] using unweighted paired group method with arithmetic mean (UPGMA) with 1000 bootstraped replications, with FreeTree software version 0.9.1.50 [30]. A dendrogram was produced using Treeview software version 1.6.6 [31].
Pathogenesis of the Representative Corynespora cassiicola Isolates on Detached Leaves of Different Rubber Clones
Three isolates (CKT05B, CKT05D and CLN 16) representing the 3 groups in subcluster 1A, 1B and cluster 2 respectively, were selected to test for their pathogenicity on detached leaves of 6 rubber clones (RRIM 600, RRIM 928, KT 39/35, PB 260, RRIM 2020 and RRIM 2014) and later used to distinguish the race of the isolates. The experiments were arranged in randomised complete block design (RCBD) in 3 replications. Each replicate consisted of six detached leaflets. Detached leaflets of about 10–14 days after budburst (brownish to limp green) were floated on water in large Petri dishes with the abaxial surface facing upward. The inoculation was done by dropping 10 μl of spore suspension of C. cassiicola (2000 spores/ml) containing 0.05% of Tween 20 onto the leaflets at 8 spots/leaflet. The leaflets were then placed under fluorescent light in air-conditioned room (25°C).
The development of the infections was assessed 6 days after inoculation using the assessment method adopted from Ismail and Jeyanayagi [18] with modification (Ismail, personal communication). An infection was considered to have occurred when the leaf tissues at the site of inoculation turned dark. The level of the infection was scored as: 0, no visible lesion; 1, small dark discolouration below droplet; 2, prominent large lesion without extended blackish veins, 3, prominent large lesion with extended blackish veins; 4, prominent large lesion with mycelium. For statistical analysis, percent disease intensity (PDI) in each replication was calculated using the following formula:
where 6 is the number of leaves observed in each replicate and 4 is the highest level of infection. Data were transformed to arcsine square-root and subjected to analysis of variance (ANOVA) using SAS software version 8.01.
Results
Colony Morphology
All isolates tested showed similar growth pattern on PSA, one of the best culture media for this fungus [32, 33]. The mycelia grew uniformly in all directions, and produced colonies with concentric growth rings with abundant aerial mycelia. The colonies were dark coloured in the middle and appeared brighter at the edge either with thin or thickly haired, effused mycelia, which were mostly branched, septate, subhyaline to pale brown with smooth walled. The colony morphology of the fungus showed variation among the different isolates. The differences were found either in mycelium colour (pale brown to dark grey), texture (thin to thick hairy, observed from the top) or in colony colour (pale brown to dark grey or black, observed from the bottom of the Petri dishes) as well as the shape of the cultures (round to slightly polygonal; Table 3, Fig. 2).
The mean growth rates calculated over 7 days and the colony sizes at 7 days after incubation differed significantly among isolates (Table 3). The means separation analysis using Duncan multiple range test divided the isolates into 12 groups for these parameters. The isolate CKT05D which was collected from Johor on RRIM 928 in 2005 showed the fastest growth rate (10.0 mm/day) and has the largest colony size (70.8 mm in diameter). On the contrary, the isolate CSB 6 collected from Selangor on RRIM 600 in 1998 showed the slowest growth rate (5.7 mm/day) and smallest colony size (40.3 mm). These two isolates were distinct from the others in Duncan analysis.
Conidial Morphology
The conidial descriptions of the 21 isolates concurred with the description in the C.M.I. notes on C. cassiicola [34]. A high degree of variation in conidial morphology has been observed not only among isolates but also within each isolate of the fungus (Fig. 3). In general, differences were observed in shape (obclavate, cylindrical or oval, straight or curved; Fig. 4), size (17.6–942.3 μm long; 3.1–16.8 μm wide) and the number of pseudoseptate (0–70; Table 4). The variation in conidial shape within isolate was observed and the ratios of these shapes differed significantly among isolates. The obclavate shaped conidia was dominant in some isolates but was minor in others. The oval shape also appeared in several isolates. A similar observation was noted in contour shape (straight or curved) of conidia (Fig. 4) as well. In spite of the variability within isolate, the distinct characteristics of each isolate enabled differentiation of the isolates analysed. The longest spore and the highest number of pseudoseptate were observed in isolate CKT 1 obtained from Johor on RRIM 600 in 1998 (942.3 μm and 70 respectively) and the widest spore found in isolate CLKK 1 obtained from Perak on RRIM 2025 in 2005 (16.8 μm). Isolate CKT1 contained the longest average conidial length (395.5 μm) and the highest average number of pseudoseptate (24.2), whereas the widest average conidial width (13.0 μm) was observed in isolate CLKK1. Based on the means separation analysis, the isolates were separated into 10 groups for spore length; 11 groups for number of pseudoseptate, with CKT1 as the stand alone isolate; and 9 groups for spore width, with CLKK1 as the stand alone isolate (Table 4).
Cluster Analysis of Corynespora cassiicola Isolates Based on ISSR Markers
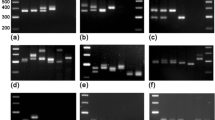
A total of 106 consistently amplified DNA bands were generated from 8 ISSR primers, in which 91.5% were polymorphic (Table 2, Fig. 5). Dendrogram produced from UPGMA analysis based on Nei and Li’s coefficient divided the 21 studied isolates into 2 main clusters (Fig. 6). Cluster 1 includes 12 isolates from Johor and Selangor. This cluster was split into 2 sub cluster (1A, 1B). Sub cluster 1B contained unique isolate CKT05D. Cluster 2 comprised of 9 isolates that were from the other states (Perak, Pahang, Kedah, Terengganu and Sarawak). The bootstrap values for the cluster 1 and 2 were 93% and 100%, respectively. The similarity index was calculated at 41% between the two clusters. The isolates in cluster 1 showed 53% similarity (it must be emphasised that the lower overall similarity in cluster 1 resulted from the inclusion of isolate (CKT05D). The isolates in sub cluster 1A showed 89% similarity and also had a high bootstrap value (100%). The isolates in cluster 2 exhibited 93% of similarity.
Gel electrophoresis of amplification products obtained by 4 ISSR primers (UBC 826, UBC 835, Mj3 and Mj5) using ISSR-PCR technique. The 1st and the last lane (MW) are 10 Kb 2-Log DNA Ladder. Lanes 1 to 12 represent the isolates from Johor and Selangor. Lanes 13 to 21 represent the isolates from other states
Pathogenesis of the Representative Corynespora cassiicola Isolates on Detached Leaves of Different Rubber Clones
The infection level of representative C. cassiicola isolates on different rubber clones ranged from no visible lesion to small dark discolouration below droplet and then to fully prominent large lesion with or without mycelium growth (Fig. 7). The average infection scores and percent disease intensity (PDI) on different clones inoculated with 3 representative isolates are shown in Table 5. Isolate CKT05B caused severe infection on clones RRIM 600 and RRIM 928 (2.9 and 2.3, respectively) but showed only slight infection on clones KT 39/35, PB 260, RRIM 2020 and RRIM 2014 (varied from 0.3 to 0.6). The means separation analysis using Duncan multiple range test separated the clones into 2 groups; group 1 consisted of RRIM 600 and RRIM 928 with PDI values of 72.2% and 58.3% respectively, and group 2 consisted of clones KT 39/35, PB 260, RRIM 2020 and RRIM 2014 with PDI values ranged from 8.3% to 19.4%. Isolate CLN16 caused moderate infection on RRIM 600 (1.4) but caused severe infection on rest of the clones (ranged from 2.6 to 3.1). The Duncan multiple range test also separated the clones into 2 groups based on the PDI values in which, RRIM 600 is a stand alone in a group with lowest PDI value of 34.7%, and the other clones in another group with PDI values ranged from 65.3% to 77.8%. Isolate CKT05D gave a mild infection on RRIM 600 (0.9), severe infection on RRIM 928 (2.4) and moderate infection on the other clones (ranged from 1.5 to 1.8). The PDI values among the clones tested were not significantly different.
Pictures show necrotic lesions that reflect on the levels of infection of the representative Corynespora cassiicola isolates from subcluster 1A, 1B and cluster 2 on detached leaves of different rubber clones. Note on the top of each picture is the name of the isolate and notes at the bottom are the names of the rubber clones
Discussion
Morphological characteristics play an important role in fungal taxonomy that could assist in the identification of the species of the pathogenic fungus [35]. The similarities in morphological characteristics of 21 isolates to the CMI description [34] indicated that the isolates studied belong to C. cassiicola species. To confirm this identification, the ITS1-5.8S-ITS2 region of 21 studied isolates was amplified and sequenced. BLAST [36] searches of their DNA sequences showed 94% to 100% identity with the deposited C. cassiicola sequences in database (GenBank; data not shown). The 21 sequence data from this study were also deposited in GenBank; their accession numbers are from EU364535 to EU364555. Earlier works have described the variation in colour, texture of the fungal colonies; size and shape of the conidia not only among the isolates obtained from different hosts and geographical regions but also within a single isolate [19, 33, 34, 37–41]. These results indicated the high variability in morphological characteristics of this fungus. The variation in colony and conidial morphology observed in this study concurred with the previous studies. The differences in morphology of the studied isolates however could not be correlated with either clonal or geographical origins of the isolates. On the other hand, Ismail and Jeyanayagi [18] have identified 2 races of C. cassiicola that infect rubber trees in Malaysia. Race 1 isolates severely infect the earlier clones (e.g. RRIM 600, GT1 and IAN 873) and have milder infection on the newer clones (e.g. RRIM 2020 and PB 260); while race 2 isolates severely infect these newer clones. Six isolates i.e. CKS1, CLN16, CLN23, CKK1, CBN5 and CBS1 obtained from clone RRIM 2020 which were expected to be race 2 or at least not to be race 1 did not have any distinguishable morphological characteristic that could separate them into specific groups. This finding warranted further work in order to differentiate the races of the C. cassiicola.
Molecular markers have been widely adopted to determine the genetic characteristics of fungi, plants and animals. The invention of PCR led to the development of faster and inexpensive molecular markers. Several successful uses of PCR-based techniques have been reported for the identification of fungal pathogen at taxonomic level lower than the species [42]. The ITS markers and rDNA-RFLP have been used to study the genetic variability of C. cassiicola but they could not find the polymorphism among the studied isolates [22, 41]. Randomly Amplified Polymorphic DNA (RAPD) has also been employed and revealed significant polymorphism among groups of C. cassiicola isolates. Several earlier workers have reported correlation between the RAPD groups and the features of the isolates e.g. pathogenicity, geographical origin, host plant genotype from which the isolate was collected [20–22, 41] but some others have found no correlation [19, 23]. A methodological disadvantage of RAPD technique is the low reproducibility especially overtime, not only among laboratory but also within laboratory [43]. Inter Simple Sequence Repeat (ISSR) [44], is another PCR-based technique that is similar to RAPD technique except that the ISSR primer sequences are longer and are designed from microsatellite regions. Therefore the annealing temperatures used are higher than those used for RAPD markers which could lead to higher consistency of the PCR products. The advantage of the ISSR technique lies in the effective multilocus markers used for diversity analysis, fingerprinting and genome mapping. They are easy to employ and are highly reproducible compared with other techniques such as RAPD, and no prior sequence knowledge is required [45]. The ISSR have been successfully applied to study the genetic diversity of pathogenic fungi e.g. Fusarium graminearum [28], Sphaeropsis sapinea [46], Phialophora gregata [47], Cryphonectria cubensis [48], Trichaptum abietinum [49], Ustilago spp. [50], Serpula lacrymans [51], Beauveria bassiana [52]. To the best of our knowledge, this is the first report on the use of ISSR markers to analyse isolates of C. cassiicola. In this study, 91.5% of the bands generated using ISSR were polymorphic which reflects the high level of genetic variation that exists among the different isolates. Two clusters derived from the dendrogram created from UPGMA analysis based on Nei and Li’s coefficient correlated to the geographical locations of the isolates. All six isolates that infected rubber clones RRIM 2020 which were mentioned in morphological section were grouped in cluster 2. The low similarity index value (41%) between the two clusters showed the high genetic variability between the isolates from the Johor and Selangor states with the isolates from the other states. The relatively low value of the similarity index obtained in cluster 1 (53%) was mainly due to isolate CKT05D. This isolate was clearly distinct from the rest of the isolates from Johor and Selangor. The high values of similarity index of the isolates in sub cluster 1A (89%) as well as cluster 2 (93%) implied that each group of isolates belongs to the same physiological group. To distinguish the race of the isolates in the study, detached leaf assay using 3 representative isolates was performed on six rubber clones including RRIM 600, RRIM 2020 and PB 260, the clones that were routinely used in the analyses to differentiate the two races of C. cassiicola that infect rubber trees in Malaysia [18]. The isolate CKT05B (a typical representative of the isolates in sub cluster 1A) severely infected clone RRIM 600, RRIM 928; and mildly infected RRIM 2020, PB 260 and the other new clones. The isolate CLN16 (a representative of the isolates in sub cluster 2) showed a severe infection on RRIM 2020, PB 260 and other new clones but moderate infection on RRIM 600. The isolate CKT05D (a unique isolate in sub cluster 1B) caused severe infection on RRIM 928, slight infection on RRIM 600 and moderate infection on other clones. Based on the characteristics of the two races of C. cassiicola described by Ismail and Jeyanayagi [18], the isolates in sub cluster 1A could be grouped into race 1, the isolates in cluster 2 could be grouped into race 2. For the isolate CKT5D, although the PDI values were not significantly different, its pathogenesis results were dissimilar to either race 1 or race 2. Further work is required to include more isolates from the region close to where CKT05D was collected to characterise the isolate and similar member into a new separate physiological group or subgroup.
In conclusion, morphological analysis served to identify but could not differentiate the isolates of C. cassiicola; the ISSR markers proved useful to differentiate the isolates of this fungus. This study also confirmed that there are at least 2 distinct groups of C. cassiicola that infect rubber trees in Malaysia. The data from the markers could also be expanded for a wider genetic diversity study of C. cassiicola from different geographical regions on different host plants.
Abbreviations
- PDI:
-
Percent disease intensity
- CMI:
-
Commonwealth Mycology Institute
- UBC:
-
University of British Colombia
References
Farr DF, Rossman AY, Palm ME, McCray EB. Fungal databases. Systematic Botany & Mycology Laboratory, ARS, USDA. Retrieved Dec 13, 2007, from http://nt.ars-grin.gov/fungaldatabases/.
Deighton FC. Preliminary list of fungi and diseases of plants in Sierra Leone. Kew Bull. 1936;7:397–424.
Ramakrishnan TS, Pillay R. Leaf spot of rubber caused by Corynespora cassiicola (Berk. & Curt.). Wei Rubber Bd Bull. 1961;5:32–5.
Newsam S. Pathology division report. Rubber Res Inst Malaysia 1961;63–70.
Awoderu VA. New leaf spot of para rubber (Hevea brasiliensis) in Nigeria. Plant Dis Reptr. 1969;53(5):406–8.
Teoh CH. Corynespora leaf fall of Hevea in West Java. Malaysian Plant Protection Society. Newsletter. 1983;7(2):12–3.
Junqeuira NTV, Gasparotto L, Moraes VHF, Silva HM, Lim TM. New diseases caused by virus, fungi and also bacterium on rubber from Brazil and their impact on international quarantine. In: Proceeding of the regional conference on plant quarantine support for agricultural development, Kuala Lumpur, Malaysia, 10–12 December 1985. p. 253–60.
Liyanage AS, Jayasinghe CK, Liyanage NIS, Jayaratne AHR. Corynespora leaf spot disease of rubber (Hevea brasiliensis)––A new report. J Rubber Res Inst Sri Lanka. 1986;65:47–50.
Pongthep K. Corynespora disease of Hevea in Thailand. IRRDB’s symposium on pathology of Hevea in Chieng Mai, Thailand, 2–3rd Nov. 1987.
Rahman MA. Diseases of Hevea brasiliensis in Bangladesh. Bano Biggyyan Patrika. 1988;17:73–9.
Dung PT, Hoan NT. Corynespora leaf fall on rubber in Vietnam, a New record. In: Proceeding of IRRDB Symposium 1999. Hainan Publishing House. p. 273–5.
Jinji P, Zhang X, Qi Y, Xie Y, Zhang H, Zhang H. First record of Corynespora leaf fall disease of Hevea rubber tree in China. Aust Plant Dis Notes. 2007;2:35–6. doi:10.1071/DN07017.
Tan AM, Lo TP, Vadivel G, Bachik MS, Yoon KP. Survey of major leaf diseases of rubber in Peninsular Malaysia. RRIM Planters’ Bull. 1992;211:51–62.
Jayashinghe CK, Silva WPK. Current status of Corynespora leaf fall in Sri Lanka. In: Proceeding workshop on Corynespora leaf fall disease of Hevea rubber, 16–17 December 1996, Medan, Indonesia. p. 15–9.
Sinulingga W, Suwarto, Soepena H. Current status of Corynespora leaf fall in Indonesia. In: Proceeding workshop on Corynespora leaf fall disease of Hevea rubber, 16–17 December 1996, Medan, Indonesia. p. 29–36.
Breton F, D’Auzac J, Garcia D, Sanier C, Eschbach JM. Recent research on Corynespora cassiicola/Hevea brasiliensis interaction. In: Proceeding workshop on Corynespora leaf fall disease of Hevea rubber, 16–17 December 1996, Medan, Indonesia. p. 49–78.
Shamsul Kamar AS, Shamsuri MH. Current status of Corynespora leaf fall in Malaysia. In: Proceeding workshop on Corynespora leaf fall disease of Hevea rubber, 16–17 December 1996, Medan, Indonesia. p. 21–8.
Ismail H, Jeyanayagi I. Occurrence and identification of physiological races of Corynespora cassiicola of Hevea. In: Proceeding of IRRDB Symposium 1999. Hainan Publishing House. p. 263–72.
Darmono TW, Darussamin A, Pawirosoemardjo S. Variation among isolates of Corynespora cassiicola associated with Hevea brasiliensis in Indonesia. In: Proceeding workshop on Corynespora leaf fall disease of Hevea rubber, 16–17 December 1996, Medan, Indonesia. p. 79–91.
Silva WPK, Karunanayake EH, Wijesundera RLC, Priyanka UMS. Genetic variation in Corynespora cassiicola: a possible relationship between host origin and virulence. Mycol Res. 2003;107(5):567–71. doi:10.1017/S0953756203007755.
Saha T, Arun Kumar A, Sreena S, Joseph A, Kuruvilla Jacob C, Kothandaraman R, Nazeer MA. Genetic variability of Corynespora cassiicola infecting Hevea brasiliensis isolated from the traditional rubber growing areas in India. Indian J Nat Rubber Res. 2000;13(1&2):1–10.
Atan S, Hamid NH. Differentiating races of Corynespora cassiicola using RAPD and internal transcribed spacer markers. J Rubber Res. 2003;6(1):58–64.
Romruensukharom P, Tragoonrung S, Vanavichit A, Toojinda T. Genetic variability of Corynespora cassiicola population in Thailand. J Rubber Res. 2005;8(1):38–49.
Othman R, Benong M, Ong SH, Ismail H. Strategies and development of resistant Hevea clones against Corynespora leaf fall. In: Proceeding workshop on Corynespora leaf fall disease of Hevea rubber, 16–17 December 1996, Medan, Indonesia. p. 177–94.
RRIM Malaysian Country Report, IRRDB Workshop on Corynespora Leaf Fall of Rubber, 6–9 June 2000, Malaysia and Indonesia 2000.
Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–5.
Philip S, Roy B. Molecular characterization of Corynespora cassiicola. In: A laboratory manual for international training on strategies for management of Corynespora leaf fall disease of H. brasiliensis. 18–29 April 2006, Rubber Research Institute of India. p. 8–16.
Mishra PK, Tewari JP, Clear RM, Turkington TK. Molecular genetic variation and geographical structuring in Fusarium graminearum. Ann Appl Biol. 2004;145:299–307. doi:10.1111/j.1744-7348.2004.tb00387.x.
Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–73. doi:10.1073/pnas.76.10.5269.
Pavlíček A, Hrdá Š, Flegr J. FreeTree––Treeware program for construction of phylogenetic trees on the basis of distance data and for bootstrap/jackknife analysis of the trees robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biologica (Praha). 1999;45:97–9.
Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8.
Liyanage AS, Jayasinghe CK, Liyanage NIS, Jayaratne AHR. Corynespora leaf spot disease of rubber (Hevea brasiliensis)––A new report. J Rubber Res Inst Sri Lanka. 1986;65:47–50.
Chee KH. Studies on sporulation, pathogenicity and epidemiology of Corynespora. J Nat Rubber Res. 1988;1(3):21–9.
Ellis MB, Holliday P. Corynespora cassiicola. C. M. I. description of pathogenic fungi and bacteria. No. 303. 1971.
Mushrif SK. Morphology, physiology and survival of Corynespora cassiicola (Bert.&Curt.) Wei. In: K Jacob, editor. Corynespora leaf disease of Hevea brasiliensis. Strategy for management. Rubber Research Institute of India; 2006. p. 26–32.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10.
Spencer JA, Walter HJ. Variation in certain isolates of Corynespora cassiicola. Phytopathology. 1969;59:58–60.
Onesirosan PT, Arny DC, Durbin RD. Host specificity of Nigerian and North American isolates of Corynespora cassiicola. Phytopathology. 1974;64:1364–7.
Duarte MLR, Asano S, Albuquerque FC. Comparative study of the morphological and physical characteristics of two Corynespora cassiicola isolates. Fitopatologia Brasileira. 1983;8:205–14.
Dung Phan Thanh. Studies on Corynespora cassiicola (Bert. & Curt.) Wei on Rubber. Master Thesis, Universiti Pertanian Malaysia (UPM) 1995.
Silva WPK, Deverall BJ, Lyon BR. Molecular, physiological and pathological characterization of Corynespora leaf spot fungi from rubber plantations in Sri Lanka. Plant Pathol. 1998;47(3):267–77. doi:10.1046/j.1365-3059.1998.00245.x.
Gil-Lamaignere C, Roilides E, Hacker J, Muller FMC. Molecular typing for fungi––a critical review of the possibilities and limitation of currently and future methods. Clin Microbiol Infect 2003;9:172–85. doi:10.1046/j.1469-0691.2003.00649.x.
International Atomic Energy Agency. Mutant germplasm characterization using molecular markers: a Manual, IAEA-TCS-19, IAEA. 2002; Introduction 11.
Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20:176–83. doi:10.1006/geno.1994.1151.
Goldwin ID, Aitken AB, Smith LW. Application of Inter simple sequence repeats (ISSR) markers to plant genetics. Electrophoresis. 1997;18:1524–8. doi:10.1002/elps.1150180906.
Burgess T, Wingfield MJ, Wingfield BW. Simple sequence repeat markers distinguish among morphotypes of Sphaeropsis sapinea. Appl Environ Microbiol. 2001;67(1):354–62. doi:10.1128/AEM.67.1.354-362.2001.
Meng X, Chen W. Applications of AFLP and ISSR techniques in detecting genetic diversity in the soybean brown stem rot pathogen Phialophora gregata. Mycol Res. 2001;105(8):936–40. doi:10.1016/S0953-7562(08)61949-8.
Van Der Merwe NA, Wingfield BD, Wingfield MJ. Primers for the amplification of sequence-characterized loci in Cryphonectria cubensis populations. Mol Ecol Notes. 2003;3:494–7. doi:10.1046/j.1471-8286.2003.00508.x.
Kauserud H, Schumacher T. Regional and local population structure of the pioneer wood-decay fungus Trichaptum abietinum. Mycologia. 2003;95(3):416–25.
Menzies JG, Bakkeren G, Matheson F, Procunier JD, Woods S. Use of inter-simple sequence repeats and amplified fragment length polymorphisms to analyze genetic relationships among small grain-infecting species of Ustilago. Phytopathology. 2003;93:167–75. doi:10.1094/PHYTO.2003.93.2.167.
Kauserud H. Widespread vegetative compatibility groups in the dry-rot fungus Serpula lacrymans. Mycologia. 2003;96(2):232–9. doi:10.2307/3762059.
Elena Estrada M, Camacho MV, Benito C. The molecular diversity of different isolates of Beauveria Bassiana (Bals.) Vuill. As assessed using intermicrosatellites (ISSRs). Cell Mol Biol Lett. 2007;12:240–52. doi:10.2478/s11658-006-0069-4.
Acknowledgements
The study presented in this paper was undertaken as part of a doctoral study project which was supported by Universiti Putra Malaysia (UPM) and Malaysian Rubber Board (MRB). Nguyen Anh Nghia, a research officer from Rubber Research Institute of Vietnam (RRIV), was financially sponsored by a Scholarship from the Ministry of Education, Vietnam Government. The authors would like to thank Dr. Shamsul Bahari from the MRB for granting permission to use the facilities for conidial morphology study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nghia, N.A., Kadir, J., Sunderasan, E. et al. Morphological and Inter Simple Sequence Repeat (ISSR) Markers Analyses of Corynespora cassiicola Isolates from Rubber Plantations in Malaysia. Mycopathologia 166, 189–201 (2008). https://doi.org/10.1007/s11046-008-9138-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-008-9138-8