Abstract
Dermatophytes are highly specialized pathogenic fungi that exclusively infect the stratum corneum, nails or hair, and it is evident that secreted proteolytic activity is important for their virulence. Endo- and exoproteases-secreted by dermatophytes are similar to those of species of the genus Aspergillus. However, in contrast to Aspergillus spp., dermatophyte-secreted endoproteases are multiple and are members of two large protein families, the subtilisins (serine proteases) and the fungalysins (metalloproteases). In addition, dermatophytes excrete sulphite as a reducing agent. In the presence of sulphite, disulphide bounds of the keratin substrate are directly cleaved to cysteine and S-sulphocysteine, and reduced proteins become accessible for further digestion by various endo- and exoproteases secreted by the fungi. Sulphitolysis is likely to be an essential step in the digestion of compact keratinized tissues which precedes the action of all proteases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dermatophytes are highly specialized pathogenic fungi which are the most common agents of superficial mycoses [1]. They are not opportunists, but true pathogenic fungi infecting stratum corneum, nails or hair of healthy individuals. During infection, hard keratin tissues have to be digested into short peptides and amino acids in order to be assimilated via transporters. The aim of this paper is to briefly review different properties of the proteases secreted by dermatophytes in comparison to other pathogenic fungi, and describe the different steps of keratinized tissue digestion.
Classification of Proteases
The term protease is synonymous with peptidase, proteolytic enzyme and peptide hydrolase. The proteases include all enzymes that catalyse the cleavage of the peptide bonds (CO–NH) of proteins, digesting these proteins into peptides or free amino acids. The classification and the nomenclature of proteases can be found together with information about them in the Handbook of proteolytic enzymes [2] and in the MEROPS database accessible at http://merops.sanger.ac.uk/. The proteases are initially classified following their mode of action and their active sites. Aspartic, cysteine, glutamic, metallo, serine and threonine proteases as well as proteases with unknown catalytic mechanism are recognized. Each protease is then assigned to a family that is a set of homologous enzymes (Table 1). These families are identified by a capital letter representing the catalytic type of the peptidases they contain, together with a unique number. Subfamilies are labelled with a second capital letter (for instance, S9B and S9C in the serine proteases, Table 1). Different families that are thought to have a common origin are grouped into a clan.
The proteases can be further divided into endoproteases (or endopeptidases) and exoproteases (or exopeptidases). The endoproteases cleave peptide bonds internally within a polypeptide. The exoproteases cleave peptide bonds only at the N- or the C-terminus of polypeptide chains.
Many secreted proteases, but not all, are synthesized as precursors in a preproprotein form. The prepeptide or signal peptide [3] is necessary for entering the secretory pathway by transporting the protein across the membrane of the endoplasmic reticulum [4]. Like in bacteria, the further N-terminal extension called the propeptide (30–250 amino acids in length) has been found to be essential and specific in assisting the correct folding and the secretion of its associated protein [5, 6]. Upon completion of folding, the propeptide is removed by an autoproteolytic or an exogenous proteolytic reaction to generate the active enzyme.
Dermatophyte-Secreted Endoproteases
Like many other fungi, all investigated dermatophytes secrete proteolytic activity in vitro when grown in a medium containing protein as sole nitrogen source [7–14]. Small molecules such as ammonium ions and amino acids repress the genes coding for secreted proteases. There are many works reporting the isolation and characterization of one or two secreted proteases from an individual species of dermatophyte [8, 9, 15–28] (Table 2). These enzymes were often described as keratinases without paying attention to the cornified cell envelope made of other components. Keratinized tissues such as the epidermis, nails and hair are not only made of keratins, but also of an insoluble network made of different cross-linked proteins such as involucrin, loricrin and small proline rich proteins which form the cornified cell envelope [29–32]. With the exception of a Microsporum canis 31.5-kDa serine protease [8] belonging to the subtilisin family and a 43.5-kDa metalloprotease [9], there were no reported amino acid sequences of the N-terminus of these proteins to further identify and characterize the isolated extracellular enzymes. The isolated M. canis serine protease and metalloprotease were subsequently called Sub3 (for subtilisin 3) and Mep3 (for metalloprotease 3), respectively, after their genes had been cloned and expressed in Pichia pastoris. The N-terminus sequences of two different peptides of 34 and 48 kDa were reported for one M. canis keratinase called “Ekase” [27]. However, they are identical to the N-terminus sequences of Sub3 and Mep3, respectively, and therefore, the “Ekase” corresponds to a mixture of two proteases.
The growth of dermatophytes is rather slow and it is difficult to obtain enough material for the purification of native proteins in sufficient quantities for further characterization. Therefore, a reverse genetic approach (from genes to proteins), which allowed the production of individual proteases in substantial amounts, was chosen to investigate proteolytic activity of different dermatophyte species. This experimental approach also avoided the problem of purification since there would be no contamination from other proteases present in the culture supernatant, as it happens when these fungi are grown in a protein medium. Briefly, dermatophytes were shown to possess a genome encoding a battery of secreted proteases similar to that of Aspergillus species (Table 1). However, five MEP and seven SUB genes isolated in Trichophyton rubrum and in other dermatophytes contrast with the small number of genes encoding neutral- and alkaline-secreted endoproteases in Aspergillus spp. Aspergillus fumigatus has only one gene (MEP) encoding a secreted fungalysin (MEROPS > M36.001), and two (ALP1 and ALP2) encoding secreted subtilisins [alkaline proteases 1 (MEROPS > S08.053) and 2 (MEROPS > S08.052)] [33–36; see also the A. fumigatus Af293 genome (http://www.tigr.org/tdb/e2k1/afu1/)]. Alp1 mep mutants are deficient in proteolytic activity at neutral pH [33]. ALP2 encodes the vacuolar subtilisin which is the orthologue of the proteinase B in the yeast Saccharomyces cerevisiae (MEROPS > S08.052) [36].
The genome of dermatophyte species so far encodes the same set of secreted proteases, and the protein sequence of each orthologue is highly conserved across species [12, 13, 37]. A phylogenetic analysis of genomic and protein sequences of metalloproteases of the fungalysin family revealed a robust tree consisting of five main clades, each of them including a MEP sequence type from each dermatophyte species [12]. The tree topology clearly indicated that the multiplication of MEP genes in dermatophytes occurred prior to species divergence. Likewise, five genes encoding T. rubrum Subs revealed a specific intron–exon structure [13]. Based on sequence data and intron–exon structure, a phylogenetic analysis of subtilisin genes from T. rubrum and other dermatophyte species revealed a presumably ancestral lineage comprising SUB2 orthologues and Aspergillus ALP1. All other genes encoding secreted subtilisins (SUB1, SUB3–SUB7) are dermatophyte specific and seem to have emerged more recently, through successive gene duplication events [13].
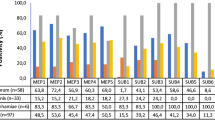
The availability of gene sequences allowed the identification of proteases on SDS-PAGE gels by mass spectrometry (MS) [12, 13, 37]. Stained bands were excised and in-gel digested with trypsin. The digestion products were analysed by reversed-phase liquid chromatography-MS/MS. Alternatively, proteases were identified by Western blotting using specific antibodies raised against individual recombinant proteases produced in Pichia pastoris or large peptides produced in Escherichia coli [37]. Using both identification methods, several Meps and Subs were shown to be secreted by dermatophytes in a protein medium [12, 13, 37]. However, their levels of secretion differ significantly from one species to another [37]. For instance, Sub7 is the major protease secreted by Trichophyton tonsurans. Sub7 is also secreted in different Trichophyton species, but was not detected in Trichophyton soudanense culture supernatants. In contrast Sub6 was found in T. soudanense culture supernatants, together with other Subs, but was not detected in culture supernatants from other species.
Dermatophyte Subs show an apparent molecular mass of 30–37 kDa and are not glycosylated (Fig. 1). As an exception mature Sub6 appears as a 20-kDa polypeptide chain on SDS-PAGE gels [37]. Dermatophyte Subs are made as preproproteins with a prosequence of about 100 amino acids which is removed from the mature catalytic domain of the protease by autoproteolytic activity. In contrast to metalloproteases of the fungalysin family (see below), but like other subtilisins, they very efficiently cleave the synthetic substrate N-Suc-Ala-Ala-Pro-Phe-p-nitroanilide (pNA) [8, 23]. They are alkaline proteases with optimal activity at pH 7.0–9.0, and are strongly inhibited by phenyl methyl sulphonyl fluoride (PMSF) and chymostatin.
Protein electrophoretic profiles from culture supernatant of Trichophyton rubrum (Tru) and Arthroderma benhamiae (Abe). Both dermatophytes were grown in a medium containing soy proteins as the sole nitrogen and carbon source. The proteins of 2 ml of culture supernatant were precipitated using trichloroacetic acid, and resuspended in a total volume of 10-μl loading buffer. Major secreted peptidases are annotated. For comparison, T. rubrum recombinant Sub3 (rSub3) produced in P. pastoris is shown in this figure. Ten microlitres of P. pastoris culture supernatant were loaded. The 9% polyacrylamide SDS-PAGE gel was stained with Coomassie brilliant blue. Molecular mass markers (M) are shown on the leftmost lane
Dermatophyte fungalysins are glycoproteins with an apparent molecular mass of 40–48 kDa (Fig. 1). They are synthesized as preproproteins with a prosequence of about 240 amino acids which is removed from the mature catalytic domain of the protease by autoproteolytic activity as in Subs. The fungalysins are Zn metalloproteases with a HEXXE motif in their amino acid sequence [38]. Their optimum pH of activity is between 7.0 and 8.0. These neutral proteases are totally inhibited by phosphoramidon and other chelating agents such as EDTA and 1–10 phenanthroline, and are insensitive to PMSF, pepstatin and iodoacetamide.
Dermatophyte-Secreted Exopeptidases
Genes encoding exopeptidases were not expanded to form gene families in dermatophytes. Like Aspergillus spp., T. rubrum secretes two leucine aminopeptidases, Lap1 and Lap2, and two dipeptidyl-peptidases, DppIV and DppV [14]. The intron–exon structures of the genes encoding these proteases are similar to the genes coding for orthologues in A. fumigatus and Aspergillus oryzae. Lap2 is a major peptidase secreted by dermatophytes (Fig. 1).
Both T. rubrum Lap1 and Lap2 were called leucine aminopeptidases like Aspergillus spp. orthologues because of their preference for leucine-7-amido-4-methylcoumarin (Leu-AMC) as a substrate. However, these enzymes are able to remove any amino acid from the N-terminus of a peptide provided that a proline is not in second position. Lap1 and Lap2 are metalloproteases which are active between pH 6.5 and 10.5 with a broad optimum peak between pH 7.0 and 9.0 [14]. Lap1 structurally belongs to the M28E subfamily as do Vibrio and Aeromonas leucyl aminopeptidases (MEROPS > M28.002; MEROPS > M28.004, respectively). Lap2 structurally belongs to the M28A subfamily like the vacuolar protease Y of S. cerevisiae (MEROPS > M28.001) and the Streptomyces griseus-secreted aminopeptidase (MEROPS > M28.003). Members of the M28A and M28E subfamilies share low sequence similarity. However, the amino acid sequences of two Zn++ binding sites are conserved. Like other fungal-secreted aminopeptidases, T. rubrum Lap1 and Lap2 were found to be sensitive to different ions. Like S. griseus aminopeptidase, Lap2 is highly activated by Co++ [14].
Tricophyton rubrum DppIV and DppV, like orthologues from Aspergillus spp., are serine proteases with a Ser, Asp, His catalytic triad, and are members of the S9 family [14]. Both enzymes are glycoproteins of approximately 90 kDa with about 10 kDa of N-linked carbohydrates. Recombinant T. rubrum DppIV and DppV are active between pH 6.5 and 10.5 with a broad optimum peak between pH 7.0 and 9.0. Recombinant DppIV efficiently hydrolyses X-Pro and X-Ala substrates (X-Pro-AMC, X-Pro-pNA, X-Ala-AMC and X-Ala-pNA, for instance). Like human CD26 and A. fumigatus DppIV, T. rubrum DppIV is inhibited by lys-[Z(NO2)]-pyrolidide and lys-[Z(NO2)]-thiazolidide. Recombinant T. rubrum DppV efficiently hydrolyses X-Ala substrates, but is not capable of removing Gly-Pro from Gly-Pro substrates. Amino acids and tripeptides are not removed from amino acid and tripeptide substrates by either DppIV or DppV. No specific inhibitor has been found for fungal DppVs.
Dermatophytes were also found to secrete a metallocarboxypeptidase (McpA) of the M14A subfamily in the MEROPS data library which is homologous to the human pancreatic carboxypeptidase A (Table 1, unpublished results). In addition to McpA, T. rubrum produces two serine carboxypeptidases of the S10 family, ScpA and ScpB, related to the previously characterized carboxypeptidase S1 secreted by A. oryzae in culture supernatant (Table 1). However, these two carboxypeptidases are glycophosphatidylinositol-anchored (unpublished results).
In vivo Dermatophyte Protease Detection and Antigenic Properties
In summary, not less than 20 genes encoding secreted endo- and exopeptidases were identified in dermatophytes (Table 1), and 15 encoded proteases were found to be secreted in vitro during the growth of dermatophytes in a protein medium. However, data are still lacking on proteases like on other hydrolases (e.g. ceramidases and lipases) secreted during the infectious process, and on the kinetics of the individual protease gene expression. Only M. canis Sub3 was identified by a clear immunohistochemical signal in the intradermal and the intraepidermal portions of infected hair structures in cats using specific anti-Sub3 IgG [39]. In addition, M. canis SUB1, SUB2, SUB3, MEP2 and MEP3 mRNA were detected by reverse transcriptase-nested polymerase chain reaction in hair from experimentally infected guinea pigs [10, 11]. Since dermatophytes grow exclusively in the stratum corneum, nails or hair utilizing them as sole nitrogen and carbon sources, it is reasonable to postulate that during infection the dermatophytes behave like in a protein medium, and also secrete different exoproteases. Synergistic action of endo- and exoproteases allows the degradation of the keratinized tissues into amino acids and short peptides that are assimilable via dermatophyte transporters (see below).
Secreted Subs and Meps are antigenic and induce both cell-mediated and humoral immune reactions. Specific lymphoproliferative response towards recombinant M. canis Sub3 and Mep3 was observed in guinea pigs experimentally infected by M. canis [40, 41]. The response was only transient with Mep3. Antibody response was observed towards Mep3, but not towards Sub3 except for one out of fourteen animals. Although being immunogenic, both proteases were not protective against M. canis experimental infection in guinea pigs in a vaccination trial [42, 43].
Two individual protease antigens, Tri r2 and Tri t4, like Trichophyton extracts, elicit either immediate (IH) or delayed-type (DTH) hypersensitivity skin test reactions in different individuals [44]. IH is associated with chronic, low-grade infections and the presence of IgE antibodies to purified Trichophyton antigens. In contrast, DTH is associated with highly inflamed lesions, which are acute and resolve spontaneously. Thus, it has been proposed that cell-mediated immune responses to Trichophyton antigens confer protection. Tri r2 is the protease Sub6 [13, 37], and was identified by screening a phage display library with sera from individuals with high IgE antibody titers and IH skin test reactions to a Trichophyton extract [45]. Tri t4 was purified from T. tonsurans mycelium extract [46], and is the orthologue of A. fumigatus and T. rubrum DppV [14, 45]. Skin tests and in vitro T cell proliferation assays were used to monitor this antigen in the isolated fractions during the different steps of its purification [46].
Protein Digestion by Endo- and Exoproteases
The endo- and exoproteases of many microorganisms cooperate very efficiently in protein digestion. During this process, the main function of the former is to produce a large number of free ends on which the latter may act. The high keratinolytic activity of T. rubrum Sub3 and Sub4 suggests that the dermatophyte endoproteases play an important role in virulence. In comparison to proteinase K and subtilisin Carlsberg, Sub3 and Sub4 were more active on keratin azure, but less active on other protein sources, especially elastin, suggesting a specificity towards hard keratin substrates [13].
The four aminopeptidases (T. rubrum Lap1, Lap2, DppIV and DppV) showed activities similar to those of A. fumigatus orthologues [14]. Large peptides generated by endoprotease digestion can subsequently be digested into amino acids and X-pro dipeptides in a way similar to that utilized by A. oryzae and Lactobacillus spp. [47, 48]. Laps degrade peptides from their N-terminus; however, X-Pro acts as a stop sequence. In a complementary manner, these X-Pro sequences can be removed by DppIV, thus allowing Laps access to the next residue. Synergistic action of Lap and DppIV to degrade large peptides seems to be common to many ascomycete fungi. In conclusion, dermatophytes like Aspergillus spp. secrete a battery of endo- and exoproteases which allow the digestion of a protein into oligopeptides and free amino acids which are then assimilable via transporters.
Reduction of Cystine Disulphide Bridges Preceding Proteolytic Activity
Fungal-secreted proteases are incapable of dissolving by themselves compact keratinous tissues. Proteins of the cornified cell envelope, in particular loricrin and small proline rich proteins, contain numerous cysteine residues that form disulphide bridges. This results in an insoluble proteinaceous complex made of a network of different cross-linked proteins. Efficient protein degradation of keratinous tissues by hydrolytic enzymes has to be accompanied by a simultaneous reduction of the cystine disulphide bridges, which are an important structural feature of keratin complexes [13, 49, 50]. Efficient in vitro degradation of the hair structure of keratin azure by hydrolytic enzymes was only possible in the presence of a reducing agent, such as 1% β-mercaptoethanol or dithiothreitol (DTT).
During infection, dermatophytes and filamentous fungi were shown to excrete sulphite as a reducing agent [50–52]. In the presence of sulphite, disulphide bonds of the keratin substrate are directly cleaved to cysteine and S-sulphocysteine (Fig. 2). The presence of sulphocysteine was confirmed in human hair attacked by Microsporum gypseum [53]. This compound was found both in the free form and in oligopeptides with a molecular mass of about 700–2,500 Da, the primary products of keratinolysis [52, 54]. As highly specialized fungi, the dermatophytes are able to utilize free cystine added to a nutrient broth and excrete sulphite which is a product of the metabolism of cysteine compounds. In presence of disulphide bridges, sulphite is immediately bound in the form of S-sulphocysteine (Fig. 2). Although it totally inhibits the activity of subtilisins in vitro, sulphite should not interfere with the activity of these enzymes secreted by the fungus in vivo. In fact, sulphite is only detectable in vitro when its amount exceeds that of cystine [50, 55].
Genes encoding T. rubrum, Arthroderma benhamiae and A. fumigatus sulphite efflux pumps (TruSSU1, AbeSSU1 and AfuSSU1, respectively) were cloned and expressed in S. cerevisiae [56]. These transporters like the S. cerevisiae sulphite transporter SceSsu1p belong to the Tellurite-resistance/Dicarboxylate Transporter (TDT) family including the Escherichia coli tellurite efflux pump and the Schizosaccharomyces pombe malate transporter encoded by the genes TEHA and MAEI, respectively. TruSSU1, AbeSSU1 and AfuSSU1 were predicted to encode proteins of 375, 375 and 396 amino acid residues with molecular masses of approximately 42, 42 and 44 kDa, respectively. Like the S. pombe malate transporter encoded by the gene MAEI [57], the deduced amino acid sequences revealed proteins with hydrophilic N- and C-termini and 10 putative membrane-spanning helices which are typical of membrane transport proteins [56]. The relatively high expression of TruSSU1 and AbeSSU1 compared to that of AfuSSU1 likely reflects a property of dermatophytes, which renders these fungi pathogenic in the stratum corneum, hair and nails.
Conclusion
One of the remarkable properties of dermatophytes is their ability to decompose keratin. However, dermatophyte-secreted proteases are similar to those secreted by other fungi, which are not pathogenic agents in keratinized tissues. The hypothesis that one keratinase, alone or with further proteases, decomposes hard keratin is currently abandoned [49]. By excreting large quantities of sulphite, rather than converting sulphite into sulphate, dermatophytes are capable of cleaving disulphide bridges, so that reduced proteins become accessible for further digestion by various endo- and exoproteases. A schema of the keratinized tissue degradation pathway by the dermatophytes is given in Fig. 3.
The activities of several dermatophyte-secreted endoproteases are redundant [13]. However, there are possible bottlenecks in the degradation process which cannot be circumvented by another mechanism. For instance, sulphitolysis is so far the sole known dermatophyte mechanism that allows the reduction of disulphide bridges, an absolute necessity in the process of degradation of compact keratinized tissues. As in other fungi, there apparently is a single gene (SSU1) encoding a sulphite efflux pump in dermatophytes [56]. The secreted DppIV (X-prolyl peptidase) may also be important for virulence being necessary for degradation of peptides derived from loricrin and small proline-rich proteins of the cornified cell envelope. DppIV is the only enzyme permitting the removal of the N-terminal X-Pro sequence during the aminopeptidase degradation of large peptides.
Although the protein sequence of each protease is highly conserved across species, their level of secretion was shown to be variable depending on the species [37]. These results suggest that a switch of habitat and different inflammatory reactions could be related to a differential expression of genes encoding homologous secreted proteins. The proteins which are secreted in a medium containing proteins as the sole carbon and nitrogen source (most of them being proteases) likely represent the spectrum of enzymes that allow the degradation of keratinized tissues into assimilable compounds during infection, and are the best vaccine candidate antigens since they are also involved in immediate and delayed-type hypersensitivity reactions in infected hosts.
Abbreviations
- AMC:
-
7-Amido-4-methylcoumarin
- Dpp:
-
Dipeptidyl peptidase
- DTH:
-
Delayed-type hypersensitivity
- IH:
-
Immediate hypersensitivity
- Lap:
-
Leucine aminopeptidase
- Mcp:
-
Metallocarboxypeptidase
- Mep:
-
Metalloprotease
- MM:
-
Molecular mass
- MS:
-
Mass spectrometry
- pNA:
-
p-Nitroanilide
- PMSF:
-
Phenyl methyl sulphonyl fluoride
- Scp:
-
Serine carboxypeptidase
- Sub:
-
Subtilisin
References
Weitzman I, Summerbell RC. The dermatophytes. Clin Microbiol Rev. 1995;8:240–59.
Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of proteolytic enzymes. 2nd ed. London: Elsevier; 2004.
Von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–90.
Pfeffer SR, Rothman JE. Biosynthetic protein transport and sorting by the endoplasmic reticulum and Golgi. Ann Rev Biochem. 1987;56:829–52.
Eder J, Fersht AR. Pro-sequence-assisted protein folding. Mol Microbiol. 1995;16:609–14.
Beggah S, Léchenne B, Reichard U, Foundling S, Monod M. Intra- and intermolecular events directing maturation of the Candida albicans secreted aspartic proteinase Sap1p. Microbiology. 2000;146:2765–73.
Mignon B, Nikkels A, Pierard G, Losson B. The in vitro and in vivo production of a 31.5 kDa keratinolytic subtilase from Microsporum canis and the clinical status in naturally infected cats. Dermatology. 1998;196:438–41.
Mignon B, Swinnen M, Bouchara JP, Hofinger M, Nikkels A, Pierard G, Gerday C, Losson B. Purification and characterization of a 31.5 kDa keratinolytic subtilisin-like serine protease from Microsporum canis and evidence of its secretion in naturally infected cats. Med Mycol. 1998;36:395–404.
Brouta F, Descamps F, Fett T, Losson B, Gerday C, Mignon B. Purification and characterization of a 43.5 kDa keratinolytic metalloprotease from Microsporum canis. Med Mycol. 2001; 39:269–75.
Brouta F, Descamps F, Monod M, Vermout S, Losson B, Mignon B. Secreted metalloprotease gene family of Microsporum canis. Infect Immun. 2002;70:5676–83.
Descamps F, Brouta F, Monod M, Zaugg C, Baar D, Losson B, Mignon B. Isolation of a Microsporum canis gene family encoding three subtilisin-like proteases expressed in vivo. J Invest Dermatol. 2002;11:830–5.
Jousson O, Léchenne B, Bontems O, Capoccia S, Mignon B, Barblan J, Quadroni M, Monod M. Multiplication of an ancestral gene encoding secreted fungalysin preceded species differentiation in the dermatophytes Trichophyton and Microsporum. Microbiology. 2004;150:301–10.
Jousson O, Léchenne B, Bontems O, Mignon B, Reichard U, Barblan J, Quadroni M, Monod M. Secreted subtilisin gene family in Trichophyton rubrum. Gene. 2004;339:79–88.
Monod M, Léchenne B, Jousson O, Grand D, Zaugg C, Stocklin R, Grouzmann E. Aminopeptidases and dipeptidyl-peptidases secreted by the dermatophyte Trichophyton rubrum. Microbiology. 2005;151:145–55.
Sanyal AK, Das SK, Banerjee AB. Purification and partial characterization of an extracellular proteinase from Trichophyton rubrum. Sabouraudia. 1985;23:165–78.
Asahi M, Lindquist R, Fukuyama K, Apodaca G, Epstein WL, McKerrow JH. Purification and characterization of major extracellular proteinases from Trichophyton rubrum. Biochem J. 1989;232:139–44.
Apodaca G, McKerrow JH. Purification and characterization of a 27,000-Mr extracellular proteinase from Trichophyton rubrum. Infect Immun. 1989;57:3072–80.
Yu RJ, Harmon SR, Blank F. Isolation and purification of an extracellular keratinase from Trichophyton mentagrophytes. J Bacteriol. 1968;96:1435–6.
Yu RJ, Harmon SR, Grappel SF, Blank F. Two cell-bound keratinases of Trichophyton mentagrophytes. J Invest Dermatol. 1971;56:27–32.
Tsuboi R, Ko IJ, Takamori K, Ogawa H. Isolation of a keratinolytic proteinase from Trichophyton mentagrophytes with enzymatic activity at acidic pH. Infect Immun. 1989;57:3479–83.
Aubaid AH, Muhsin TM. Partial purification and kinetic studies of an extracellular proteinase from Trichophyton mentagrophytes var. erinacei. Mycoses. 1998;41:163–8.
Siesenop U, Böhm KH. Comparative studies on keratinase production of Trichophyton mentagrophytes strains of animal origin. Mycoses. 1995;38:205–9.
Moallaei H, Zaini F, Larcher G, Beucher B, Bouchara JP. Partial purification and characterization of a 37 kDa extracellular proteinase from Trichophyton vanbreuseghemii. Mycopathologia. 2006;161:369–75.
Takiuchi I, Higuchi D, Sei Y, Koga M. Isolation of an extracellular proteinase (keratinase) from Microsporum canis. Sabouraudia. 1982;20:281–8.
Takiuchi I, Higuchi D, Sei Y, Koga M. Immunological studies of an extracellular keratinase. J Dermatol. 1983;10:327–30.
Takiuchi I, Sei Y, Tagaki H, Negi M. Partial characterization of the extracellular keratinase from Microsporum canis. Sabouraudia. 1984;22:219–24.
Hamaguchi T, Morishita N, Usui R, Takiuchi I. Characterization of an extracellular keratinase from Microsporum canis. Jpn J Med Mycol. 2000;41:257–62.
Lee KH, Park KK, Park SH, Lee JB. Isolation, purification and characterization of keratinolytic proteinase from Microsporum canis. Yonsei Med J. 1987;28:131–8.
Simon M, Green H. Enzymatic cross-linking of involucrin and other proteins by keratinocytes particulates in vitro. Cell. 1985;40:687–3.
Hohl D, de Viragh PA, Amiguet-Barras F, Gibbs S, Backendorf C, Huber M. The small proline-rich proteins constitute a multigene family of differentially regulated cornified cell envelope precursors proteins. J Invest Dermatol. 1995;104:902–9.
Steinert PM, Marekow LN. The proteins elafin, filagrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human cornified cell envelope. J Biol Chem. 1995;270:17702–11.
Kalinin AE, Kajava AV, Steinert PM. Epithelial barrier function: assembly and structural features of the cornified cell envelope. BioEssays. 2000;24:789–800.
Jaton-Ogay K, Paris S, Huerre M, Quadroni M, Falchetto R, Togni G, Latgé JP, Monod M. Cloning and disruption of the gene encoding an extracellular metalloprotease of Aspergillus fumigatus. Mol Microbiol. 1994;14:917–28.
Reichard U, Büttner S, Eiffert H, Staib F, Rüchel R. Purification and characterisation of an extracellular serine proteinase from Aspergillus fumigatus and its detection in tissue. J. Med Microbiol. 1990;33:243–51.
Jaton-Ogay K, Suter M, Crameri R, Falchetto R, Fatih A, Monod M. Nucleotide sequence of a genomic and a cDNA clone encoding an extracellular alkaline protease of Aspergillus fumigatus. FEMS Microbiol Lett. 1992;92:163–8.
Reichard U, Cole GT, Hill TW, Rüchel R, Monod M. Molecular characterization and influence on fungal development of ALP2, a novel serine proteinase from Aspergillus fumigatus. Int J Med Microbiol. 2000;290:549–58.
Giddey K, Favre B, Quadroni M, Monod M. Closely related dermatophyte species produce different patterns of secreted proteins. FEMS Microbiol Lett. 2007;267:95–101.
Jongeneel CV, Bouvier J, Bairoch A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989;242:211–4.
Mignon BR, Coignoul F, Leclipteux T, Focant C, Losson BJ. Histopathological pattern and humoral immune response to a crude exo-antigen and purified keratinase of Microsporum canis in symptomatic and asymptomatic infected cats. Med Mycol. 1999;37:1–9.
Descamps F, Brouta F, Vermout S, Monod M, Losson B, Mignon B. Recombinant expression and antigenic properties of a 31.5-kDa keratinolytic subtilisin-like serine protease from Microsporum canis. FEMS Immunol Med Microbiol. 2003;38:29–34.
Brouta F, Descamps F, Vermout S, Monod M, Losson B, Mignon B. Humoral and cellular immune response to a Microsporum canis recombinant keratinolytic metalloprotease (r-MEP3) in experimentally infected guinea pigs. Med Mycol. 2003;41:495–502.
Descamps F, Brouta F, Vermout S, Willame C, Losson B, Mignon B. A recombinant 31.5 kDa keratinase and a crude exo-antigen from Microsporum canis fail to protect against a homologous experimental infection in guinea pigs. Vet Dermatol. 2003;14:305–12.
Vermout S, Brouta F, Descamps F, Losson B, Mignon B. Evaluation of immunogenicity and protective efficacy of a Microsporum canis metalloprotease subunit vaccine in guinea pigs. FEMS Immunol Med Microbiol. 2004;40:75–80.
Woodfolk JA. Allergy and dermatophytes. Clin Microbiol Rev. 2005;18:30–43.
Woodfolk JA, Wheatley LM, Piyasena RV, Benjamin DC, Platts-Mills TAE. Trichophyton antigens associated with IgE antibodies and delayed type hypersensitivity – sequence homology to two families of serine proteinases. J Biol Chem. 1998;273:29489–96.
Woodfolk JA, Slunt JB, Deuell B, Hayden ML, Platts-Mills TAE. Definition of a Trichophyton protein associated with delayed hypersensitivity in humans. Evidence for immediate (IgE and IgG4) and delayed hypersensitivity to a single protein. J Immunol. 1996;156:1695–701.
Byun T, Kofod L, Blinkovsky A. Synergistic action of an X-prolyl dipeptidyl aminopeptidase and a no specific aminopeptidase in protein hydrolysis. J Food Chem. 2001;49:2061–3.
O’Cuinn G, Fitzgerald R, Bouchier P, McDonnell M. Generation of non-bitter casein hydrolysates by using combinations of a proteinase and aminopeptidases. Biochem Soc Trans. 1999;27:730–4.
Kunert J. Effect of reducing agents on proteolytic and keratinolytic activity of enzymes of Microsporum gypseum. Mycoses. 1992;35:343–8.
Kunert J. Physiology of keratinophilic fungi. In: Kushwaha RKS, Guarro J, editors. Biology of dermatophytes and other keratinophilic fungi. Bilbao: Revista Iberoamericana de Micología; 2000. p. 77–85.
Kunert J. Keratin decomposition by dermatophytes: evidence of the sulphitolysis of the protein. Experientia. 1972;28:1025–6.
Kunert J. Keratin decomposition by dermatophytes. II. Presence of S-sulphocysteine and cysteic acid in soluble decomposition products. Z Allg Mikrobiol. 1976;16:97–105.
Kunert J. Thiosulphate esters in keratin attacked by dermatophytes in vitro. Sabouraudia. 1972;10:6–13.
Ruffin P, Andrieu S, Biserte G, Biguet J. Sulphitolysis in keratinolysis. Biochemical proof. Sabouraudia. 1976;14:181–4.
Kunert J. Formation of sulphate, sulphite and S-sulphocysteine by the fungus Microsporum gypseum during growth on cystine. Folia Microbiol (Praha). 1975;20:142–51.
Léchenne B, Reichard U, Zaugg C, Fratti M, Kunert J, Boulat O, Monod M. Sulphite efflux pumps in Aspergillus fumigatus and dermatophytes. Microbiology. 2007;153:905–13.
Grobler J, Bauer F, Subden RE, vanVuuren HJJ. The mae1 gene of Schizosaccharomyces pombe encodes a permease for malate and other C4 dicarboxylic acids. Yeast. 1995;11:1485–91.
Acknowledgements
We thank Dr. Sandy Vermout, Dr. Peter Staib and Dr. Massimo Lurati for their critical review of the manuscript and assistance with the English. This work was supported by the Swiss National Foundation for Scientific Research, grant 3100-105313/1.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Monod, M. Secreted Proteases from Dermatophytes. Mycopathologia 166, 285–294 (2008). https://doi.org/10.1007/s11046-008-9105-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-008-9105-4