Abstract
The accurate separation of ECG signals has become crucial to identify heart diseases. Machine learning methods are widely used to separate ECG signals. The aim of this study was to obtain optimal number of hidden neurons of the Extreme Learning Machine (ELM) using the differential evolution algorithm (DEA) and increase the accuracy rate of ECG classification. In this study, a public database on PhysioNet was used for ECG signal classification. A deep feature method using convolutional neural network was used to extract the major features of the ECG samples. Then, a conventional ELM was applied to the ECG signals. Subsequently, the ECG signals with deep properties were shared with the MATLAB classifier toolbox (k-NN, SVM, Decision Trees). In addition, the ECG signals in the dataset were tested using the Genetic Algorithm Wavelet Kernel-ELM (GAWK-ELM). Finally, the DEA-ELM was improved for the determination of the number of hidden neurons. This study optimized the hidden neuron numbers of traditional ELM with DEA using deep learning capabilities in the feature extraction. The aim of was to maximize the best cost of the DEA and achieve the optimal number of hidden neurons in ELM. Accuracy (Acc), sensitivity (Se), specificity (Spe) and F-measure were used as the performance metrics for the classifier performances. The classification results were 80.60%, 81.50%, and 83.12% with SVM, ELM and DEA-ELM, respectively. Thus, the best classification scores were accomplished with an accuracy of 83.12% with the algorithm supported by the DEA.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
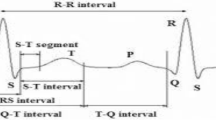
Electrocardiogram (ECG) is the signal that represents the activity of the human heart. ECGs are comprised of various important sections. Figure 1 shows a typical ECG signal [17, 47, 54, 58]. During atrial depolarization, the P wave occurs. Subsequently, the QRS wave occurs in the heart [17, 58]. The movement of the ECG signal has become a significant parameter for defining heart diseases. [22, 38]. The diagnosis of heart diseases at the right time is a very important issue in treating cardiovascular diseases [43]. In particular, attempts have been made to develop powerful computer based algorithms to identify heart disorders using ECG recording classification [50]. Scientists have developed many methods to classify ECG signals. Some of these methods include wavelet transform [52], radial basis function [37], multi-layer perceptron (MLP), support vector machine (SVM) [35], extreme learning machine (ELM) [33] and deep feature learning based method [55]. Deep learning is a new method that has been used in many studies in the literature for Image classification [41], biomedicine applications [8, 20] and agriculture [30]. Deep learning structures are multi-layered and include many parameters. The most commonly used architectures in the literature are automatic encoders and convolutional neural networks (CNNs) [48]. CNNs are well-known deep learning structures and are comprise of two parts. In the initial section of CNN, feature subtraction is applied to the input data [48, 19]. CNN is principally successful in image processing applications [25, 59, 62].
ELM’s have been used in many areas including ECG signal classification and EEG signal classification owing to their impeccable properties such as quick learning and generalization [6, 28, 29, 57]. However, they have several problems including the presence of local minima’s, indefinite learning rate, selection of the number of hidden neurons (NHN) and overfitting [56]. To solve these problems diverse methods with global search capabilities such as differential evolutional (DE) [26, 61], PSO algorithm (classification performance of ECG signals accuracy of 89.72%) [44], GA algorithm (classification performance of radar signals accuracy of 95.50%) [4], ACO algorithm [9] and ABC algorithm [23] have been developed. Clustering Algorithm [11], Intuitionistic Fuzzy Rough Set Technique [10], Segmentation and Feature Extraction method ın medical imaging [13], Intuitionistic Possibilistic Fuzzy C-Mean Clustering Algorithm [12], Digital Mammography [14], Heart Disease Diagnosis Using Rough Set and Cuckoo Search Technique [1], Formal Concept Analysis [60] can be added to these publications. ECG signals, which can be normal or abnormal, are very critical in making an accurate diagnosis. Thereby, computer-aided ECG analysis has a considerable role in detecting heart disorders. In this study, the impact of the differential evolutional extreme learning machine based (DEA-ELM) technique was examined to determine ECG signals as normal and abnormal.
The novelty of the technique proposed in this study is that it uses the most appropriate NHN values, which are determined using the DEA optimization technique to overcome the setbacks of ECG classification (e.g. election of the NHN) with ELM. In addition, as accurate attribute extraction is required to determine the signals as normal and abnormal. In this study, CNN, which is based on the deep learning techniques of the ECG samples, was used for attribute extraction. Moreover, in this study, the efficiencies of several machine learning techniques were examined. The principal objective of this study was to ensure the enhancement of classification efficiency and decrease the mentioned problems of ELM to accurately separate the ECG signals as normal and abnormal using DEA. In this study, a method offering the following advantages was proposed to be used instead of traditional ELM:
-
a)
The DEA-ELM, which is used in a process to separate normal and abnormal samples, has a better classification accuracy than the ELM owing to the improved NHN.
-
b)
This process, which is carried out using DEA, is used to adjust the most optimal efficiency separating the samples as normal and abnormal.
The paper is organized as follows: in Section 1 the collection of the ECG signals is presented. In Section 3 the results of the ELM and DEA-ELM studies used in ECG classification are given. In Sections 4 and 5 the discussions and conclusion are presented.
2 Data collection and techniques
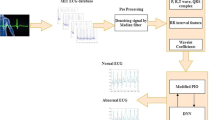
The diagram of the proposed classification method is presented in Fig. 2. This technique can be separated into four parts. Primarily, ECG samples are attained through the PhysioNet. This step is consists of two parts, as baseline-wandering and remove 60 Hz interference. After preprocessing steps, ECG signals which are acquired from Physionet were converted to image using a spectrogram. The next process of the schema is attribute extraction that deep features are attained from CNN. The other step of the recommended system is an improvement that is optimized of a NHN of ELM and, is the last stage which is samples separated as normal and abnormal.
2.1 Dataset
In this study, to determine the adequacy of ELM, PTBDB heart records, which were available on the Physionet, were utilized [24]. A total of 549 ECG samples and samples of signals records were collected from 294 contributors. Roughly 20% of the contributors comprised the healthy samples. The record samples include healthy and different abnormal varieties as indicated in Fig. 3.
A preliminary study was conducted on healthy and MI signals. As a result, 112 MI and 48 healthy signals were selected. Furthermore, in this study, all of the ECG samples were achieved through lead II on the database as shown in Fig. 4.
2.2 Pre-processing
Noise and interference, which increase waves, are unwanted in applications. ECGs have a significant number of interferences, thus the preprocessing stage is important for the classification operation in computer-aided methods [15, 18]. In addition, a normal analysis is carried out with many applications by achieving noiseless clean samples. ECGs contain different kinds of disorderliness including a fundament origin period and device parasites. Using the symlet filter wavelet transformation and denoising operation baseline navigation was adjusted. On the other hand, line parasites and system parasites were decreased in this step. Then, the parasites proceedings on the ECG samples were appeased by employing the Savitzky-Golay filter, which is given in Fig. 5.
2.3 Feature extraction
Feature extraction is a major operation in the classification process. After the pre-processing, it is required to take out the attributes of the ECG samples. In this study, the deep features were obtained by using CNN. Within this context, AlexNet, a CNN structure, was used.
2.3.1 Convolutional neural network structure
CNNs are type of multi-layer sensors [48]. CNN, which is considered to operate data in multiple arrays, has become a popular technique because of its working dexterity in image attribute subtraction [49]. CNNs are comprised of a lot of layers as shown in Fig. 6. All layers consisted of three layers, namely characteristic pooling layer, the filter bank layer, and the non-linear layer. There are a lot of kernels in the filter bank on account of other attribute subtractions.
In the pooling layer, all attribute maps are operated one by one. Every map ensures the mean or maximum value of the contiguous value [48].
2.3.2 Layers used in CNN structure
In this study, AlexNet, which is a CNN architecture, was used to take out the features. The convolution layers in the AlexNet embody the Input-Convolution-ReLu-Pooling-Fully connected layers. The Rectified linear unit (ReLU) used in the AlexNet architecture is the activation function that has a very fast learning speed. ReLU has a higher performance than sigmoid and tanh, both of which are other activation functions. The ReLU activation function fulfills a threshold process to each input element where values less than zero are set to zero thus the ReLU is calculated using Eqs. 1.
ReLU verifies the values of inputs less than zero and forces them to zero and eliminates the vanishing gradient disadvantage observed in previous activation function types [45, 49]. The CNN structure used to take out the attributes of the ECG samples achieved from the ECG spectrograms is presented in Fig. 7.
CNN structure used in the study was comprised five convolutional strata (C1 to C5 scheme) that included two fully connected strata (FC6 and FC7). The output attributes achieved from the FC7 layer were given as input to the ELM and DEA-ELM classifiers.
2.4 Extreme learning machine
It has been suggested by ref. [28] that ELM can only determine the expiration step weights in the course of training [6, 28, 29]. ELM is a universalized SLFN and as can be seen in Fig. 8 it is not essential to regulate the hidden layer.
For N casually distinct instances (xi, ti), ELM with H hidden nodes and the activation coefficient h(x) is designated by xi = [xi1, xi2, …. . xiD]T ∈ RDand ti = [ti1, xi2, …. . xiK]T ∈ RK. SLFNs are described as shown in Eq. 2:
The ELM is shown in Fig. 9.
Different from standard learning procedures the ELM technique can casually produce hidden node parameters. wi = [wi1, wi2, …. . wiD]T, is the weight vector connected to the hidden node and the input nodes, βi = [βi1, βi2, …. . wiK]K is the weight vector and bi is the ith node. The h(x) activation function and hidden nodes L are shown in Eqs. 3, 4 and 5.
Besides, ELM has represented as in Eq. 6, conversely SLFNs.
H† characterize Moore-Penrose, the universalize reverse of H matrix to attain a least-square solve [28], [7, 27, 36, 42].
2.5 Differential evolution algorithm
The DEA, which is an advanced technique used for working out the problems of constant parameters, was put forward by Price and Storm [34]. DEA is an inhabitants-dependent empirical procedure that is inspired by GA related processing and operators. In addition, DEA can particularly ensure effective results that are the disadvantages of constant data [21, 32, 51, 63]. Incidentally, DEA is exerted investigative on a lot of areas. Through the repetitions in the algorithm, superior consequences for the resolution of the disadvantage by the aid of the operators are explored. Furthermore, the goal of the algorithm is to advance the remedy efficiency of the disadvantages in which coding by constant scores is used, by certain alters in genetic operators. The crossing, mutation and election operators in GA are also used in DEA. However, in DEA not all of the operators are carried out to the all of the inhabitant in the order. Chromosomes are handled one by one and obtained by a new individual through the randım election of three other chromosomes. In the course of this procedure, mutation and crossover processors are used. The compatibility of the recent chromosome obtained using the existing chromosome are compared and the one that is more suitable is transferred to next population as a new individual. As a result, the election operative is also used. The quality of the solutions created is evaluated according to the value they come up with for the goal function [34]. The major benefits and significant parameters of the DEA procedure are presented in Table 1.
The D factor can be characterized by a D vector. In this algorithm, firstly an inhabitant of NP inhabitant vector is selected. This inhabitant is achieved advanced with execution mutation, crossover, and selection operators [21]. The primary steps of DEA are given in Fig. 10. The number of variables of the problem determines the number of genes for each chromosome. The population size (number of chromosomes) in DEA should always be greater than three because three chromosomes are required, excluding the existing chromosome, for the production of new chromosomes.
Mutation is to perform alters in casually decisive amounts on various sections of the genes of the actual chromosome. Three extra chromosomes are selected for mutation within the DEA. The difference of the first two selected chromosomes is obtained. Afterward, this chromosome is multiplied by the scaling parameter. This parameter mostly receives scores among 0–2. The attained weighted difference chromosome is added to the third chromosome. As a result, the result of the mutation chromosome is used in the crossover [34]. The aim is to reduce costs and obtain the optimal number of hidden neurons of ELM using DEA. Table 2 presents the DEA pseudo code used in this study.
3 Classification results
In this section the classification scores for healthy and abnormal samples are presented. To determine the scores a total of 160 ECG samples comprised of 80 abnormal and 80 normal signals were selected from a public dataset. The confusion matrix was employed to appraise classifier efficacy. The accuracy, sensitivity, specificity, quality index, and F-measure of the classifiers were determined using Eqs. 7–10. The true positive (TP) and true negative (TN) characterized the quantity of the normal and abnormal ECG samples, while the false positive (FP) and false-negative (FN) represented the quantity of the normal and abnormal ECG samples [16, 21].
Firstly, the preprocessing step was performed to remove the 60 Hz interference and the baseline wander of the ECG samples obtained from the PTB dataset. Then, the spectrograms to represent the frequency domain the ECG signals were obtained as can be seen in Fig. 11.
After obtaining the spectrograms of the ECG signals to extract the features of the ECG signals AlexNet, which is a CNN architecture, was used. As a result, a feature vector of 160 × 4097 size was obtained to be given to the classifiers.
3.1 Comparison of classifiers efficiency
In this section, the performance of the ELM, DEA-ELM, k-NN, SVM, and decision trees algorithms were compared. Within this scope, achievement consequences were achieved via the ELM’s Radbas, Sin, Sig, Hardlim and Tribas activation functions (AC). All of the ECG samples were separated into two sets as training and test sets. Moreover, the ECG samples was tested with a 10-fold cross-validation method. Classifier consequences were attained by tuning the NHN of ELM to 40, 60, 80, 100, and 120 as can be seen in Figs. 12, 13, 14 and 15.
The proposed method was provided by 10-fold cross-validation via the MATLAB Classification Learner Toolbox. The results of the classifiers used in this toolbox are presented in Fig. 16.
In addition, the confusion matrix and receive operating curve (ROC) of the classifiers, namely k-NN, SVM and decision trees, are shown in Fig. 17.
The main aim of this study was to analyze the performance of the model by improving the NHN of ELM via DEA. Therefore, the classification parameters were examined via the NHN of the ELM as seen in Figs. 12, 13, 14 and 15. The classification efficiency of the ELM can be enhanced by NHN. For this, the DEA, which is a powerful meta-heuristic improvement method, was applied. The DEA was used to boost the consequences which were attained by the ECG classification using ELM. This method was carried out as can be seen from Fig. 18. The best costs as efficiency consequences were attained with 250, 500, 750, 1000, 1500, 1600, 1700, 1800, 1900, and 2000 repetitions with DEA-ELM (Fig. 18). Moreover, the factors attained from the DEA-ELM are presented in Table 3.
The efficiency scores for five diverse AC were attained by DEA-ELM. The NHN that improved via DEA-ELM is given in Table 3. Furthermore, the factors used in the DEA method are presented in Table 4.
In the NHN improvement applied via the DEA-ELM method the best accuracy score of 83.12% was achieved with Sigmoid AC and 89 NHN. When the highest best cost value was achieved, the number of repetitions was increased to verify this result. In addition, this dataset was tested using the Genetic Algorithm Wavelet Kernel ELM (GAWK-ELM) [5], which is another meta-heuristic optimization method. The parameters used in the GAWK-ELM method are given in Table 5 and the results of the method are given in Table 6.
As can be understood from the results of the GAWK-ELM method, the DEA-ELM method had higher classification accuracy. This is due to the fact that the DEA method is an iterative solution search-based heuristic search algorithm.
Whether the best cost value changed with excessive repetitions was evaluated. As a result, the best cost score accomplished was left consistent at 1900 and 2000 repetition numerals as referred in Table 3.
4 Discussion
Heart failure is a fatal disease that affects both the heart and the heart vessels and is the most important cause of death worldwide. Therefore, the early detection of heart diseases is very important. As a result, ECG classification has become a critical area of study over the last 20 years [2]. In this section a benchmark regarding automated ECG analysis systems, which is shown in Table 7, among the recommended technique and the ultimately concerned works considering the prominent parameters, such as techniques, databases, classes and attributes type is discussed.
The offered DEA-ELM procedure for the improved NHN of the ELM is completely unique compared to former techniques and is an innovative method in this area. In this study, low accuracy performance was achieved due to the low number of signals used. The sensitivity and specificity values in ELM can be misleading. Therefore, accuracy should be taken into consideration in the system evaluation. Consequently, some studies have reported low accuracy values when using the DEA-ELM. When a collation is accomplished taking notice of the coefficients in Table 4, the classification performance of DEA-ELM based on deep feature may be defined promising. In Ref [65], two deep neural network architecture for the classification of arbitrary-length ECG samples were proposed and evaluated according to the atrial fibrillation (AF) classification data set provided by the PhysioNet/CinC Challenge 2017. Consequently, was found to outperform the first one, obtaining an F1 score of 82.1% on the hidden challenge testing set. In another ref. [46] a novel approach to automatically detect the MI using ECG signals was used. In the present study, a CNN algorithm was implemented for the automated detection of normal and MI ECG beats (with and without noise) and average accuracy of 93.53% and 95.22% using ECG beat with noise and noise removal respectively was obtained, respectively.
It should be emphasized that in this study only DEA-ELM was used for ECG classification yet the primary aim of this study was to advance the classification efficiency and improve NHN. Furthermore, it was determined that the classification efficiency of the ECG sample was increased via the DEA-ELM procedure as is shown in Figure 18. It must realized that system efficiency can be increased via different attributes taken by various domains such as temporal and deep learning. The experimental consequences showed that NHN is beneficial for ECG classification efficiency.
5 Conclusion
In this study, DEA-ELM, a recently improved procedure, was used for ECG classification. The proposed system was tested by operating the open access PTB Diagnostic database. Moreover, a comparisonwas achieved by considering major parameters from the point of the ECG analysis. The best classification scores were achieved with an accuracy of 81.50% via ELM. The best cost of 83.12% was achieved with the Sigmoid function, while the best classification results of 89 NHN were obtained with the DEA-ELM approach. The main aim of this study was to classify ECG with the DEA procedure that was optimized with ELM.
References
Acharjya DP (2020) A hybrid scheme for heart disease diagnosis using rough set and cuckoo search technique. J Med Syst 44(1):27
Amato F, López A, Peña-Méndez EM, Vaňhara P, Hampl A, Havel J (2013) Artificial neural networks in medical diagnosis. J Appl Biomed 11(2):47–58
Andersen RS, Peimankar A, Puthusserypady S (2019) A deep learning approach for real-time detection of atrial fibrillation. Expert Syst Appl 115:465–473
Avci E (2013) A new method for expert target recognition system: Genetic Wavelet Extreme Learning Machine (GAWELM). Expert Syst Appl
Avci D (2016) An automatic diagnosis system for hepatitis diseases based on genetic wavelet kernel extreme learning machine. J Electr Eng Technol 11(4):993
Avci E, Coteli R (2012) A new automatic target recognition system based on wavelet extreme learning machine. Expert Syst Appl 39(16):12340–12348
Bin Huang G, Chen L, Siew CK (2006) Universal approximation using incremental constructive feedforward networks with random hidden nodes. IEEE Trans Neural Netw 17(4):879–892
Cao C et al (2018) Deep learning and its applications in biomedicine. Genomics, Proteomics Bioinforma 16(1):17–32
Chen X, Zhang P, Du G, Li F (2018) Ant Colony optimization based Memetic algorithm to solve bi-objective multiple traveling salesmen problem for multi-robot systems. IEEE Access 6:21745–21757
Chowdhary CL, Acharjya DP (2016) A hybrid scheme for breast cancer detection using intuitionistic fuzzy rough set technique. Int J Healthc Inform Syst Inf (IJHISI) 11(2):38–61
Chowdhary CL, Acharjya DP (2017) Clustering algorithm in possibilistic exponential fuzzy c-mean segmenting medical images. In: Journal of Biomimetics, biomaterials and biomedical engineering, vol 30. Trans Tech Publications Ltd, pp 12–23
Chowdhary CL, Acharjya DP (2018) Segmentation of mammograms using a novel intuitionistic possibilistic fuzzy C-mean clustering algorithm. In: Nature inspired computing. Springer, Singapore, pp 75–82
Chowdhary CL, Acharjya DP (2020) Segmentation and feature extraction in medical imaging: a systematic review. Procedia Comput Sci 167:26–36
Chowdhary CL, Sai GVK, Acharjya DP (2016) Decrease in false assumption for detection using digital mammography. In: Computational intelligence in data mining—, vol 2. Springer, New Delhi, pp 325–333
Cömert Z, Fatih Kocamaz A (2015) Determination of QT interval on synthetic electrocardiogram In Signal processing and communications applications conference (SIU), 2015 23th, pp 1–4
Cömert Z, Kocamaz AF (2017) Using wavelet transform for Cardiotocography signals classification 25th signal process. Commun Appl Conf:1–4
Diker A, Avcı E, Gedikpinar M (2017) Determination of R-peaks in ECG signal using Hilbert Transform and Pan-Tompkins Algorithms Signal Process. Commun Appl Conf (SIU)
Diker A, Cömert Z, Avcı E (2017) A diagnostic model for identification of myocardial infarction from electrocardiography signals. Bitlis Eren Univ J Sci Technol 7(2):132–139
Dogan RO, Kayikcioglu T (2018) R-peaks detection with convolutional neural network in electrocardiogram signal in 2018 26th. Signal Processing and Communications Applications Conference (SIU) 2018(2):1–4
Donida Labati R, Muñoz E, Piuri V, Sassi R, Scotti F (2018) Deep-ECG: convolutional neural networks for ECG biometric recognition. Pattern Recogn Lett 0:1–8
Engelbrecht A (2007) An introduction differential evolution. Comput Intell
Escalona-Morán MA, Soriano MC, Fischer I, Mirasso CR (2015) Electrocardiogram classification using reservoir computing with logistic regression. IEEE J Biomed Heal Informatics 19(3):892–898
Farahani HF, Rashidi F (2017) Optimal allocation of plug-in electric vehicle capacity to produce active, reactive and distorted powers using differential evolution based artificial bee colony algorithm. IET Sci Meas Technol 11(8):1058–1070
Goldberger AL et al (2000) PhysioBank, PhysioToolkit, and PhysioNet components of a new research resource for complex. Physiologic Signals
Harangi B (2018) Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform 86(January):25–32
Hu J, Wang C, Liu C, Ye Z (2008) Fly optimization and differential evolution no. ICCSE:464–467
Huang G, Zhu Q, Siew C (2004) Extreme learning machine: a new learning scheme of feedforward neural networks. Neural Netw 2:985–990
Huang G-B, Zhu Q-Y, Siew C-K (2006) Extreme learning machine: theory and applications. Neurocomputing 70(1):489–501
Huang G-B, Zhou H, Ding X, Zhang R (2012) Extreme learning machine for regression and multiclass classification. IEEE Trans Syst Man Cybern B Cybern 42(2):513–529
Kamilaris A, Prenafeta-Boldú FX (2018) deep learning in agriculture: a survey. Comput Electron Agric 147(July 2017):70–90
Karaboğa N, Koyuncu CA (2005) Diferansiyel Gelişim Algoritmasi Kullanılarak Adaptif Lineer Toplayıcı Tasarımı, in EMO, III. Otomasyon Sempozyumu, Denizli, pp 216–220
Karci A (2017) Differential evolution algorithm and its variants Anatol. J Comput Sci 2(1):10–14
Karpagachelvi S (2011) Classification of electrocardiogram signals with extreme learning machine and relevance vector machine. J Comput Sci 8(1)
Keskintürk T (2006) Differential evolution algorithm İstanbul Ticaret Üniversitesi Fen Bilim. Derg. 5(9):85–99
Khorrami H, Moavenian M (2010) A comparative study of DWT, CWT and DCT transformations in ECG arrhythmias classification. Expert Syst Appl 37(8):5751–5757
Kim J, Shin H, Shin K, Lee M (2009) Robust algorithm for arrhythmia classification in ECG using extreme learning machine. Biomed Eng Online 8(1):31
Koruürek M, Doğan B (2010) ECG beat classification using particle swarm optimization and radial basis function neural network. Expert Syst Appl 37(12):7563–7569
Kulkarni SP (2015) DWT and ANN based heart arrhythmia disease Diagnosis from MIT-BIH ECG signal data. Int J Recent Innov Trends Comput Commun (January):276–279
Limam M, Precioso F (2017) AF detection and ECG classification based on convolutional recurrent neural network in Computing in Cardiology 44:1–4
Lin C-L, Hsieh S-T (2017) A beat-to-beat atrial fibrillation detecting system. J Cardiovasc Dis Diagn
Liu D, Jiang Y, Pei M, Liu S (2018) Emotional image color transfer via deep learning. Pattern Recogn Lett 110:16–22
Liu H, Zhang Y, Zhang H (2020) Prediction of effluent quality in papermaking wastewater treatment processes using dynamic kernel-based extreme learning machine. Process Biochem
Mannurmath JC, Raveendra M (2014) MATLAB based ECG signal classification. Int J Sci Eng Technol Res 3(7):1946–1951
Melgani F, Bazi Y (2008) Classification of electrocardiogram signals with support vector machines and particle swarm optimization. IEEE Trans Inf Technol Biomed 12(5):667–677
Nwankpa CE, Ijomah W, Gachagan A, Marshall S (2018) Activation functions : comparison of trends in practice and research for deep learning. arXiv:1–20
Oh SL, Ng EYK, Tan RS, Acharya UR (2018) Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats. Comput Biol Med
Ojha DK, Subashini M (2014) Analysis of electrocardiograph ( ECG ) signal for the detection of abnormalities using MATLAB. Int J Medical, Heal Biomed Pharm Eng 8(2):114–117
Özyurt, F. (2019). Efficient deep feature selection for remote sensing image recognition with fused deep learning architectures. The Journal of Supercomputing, 1–19
Özyurt, F. (2020). A fused CNN model for WBC detection with MRMR feature selection and extreme learning machine. Soft Computing, 24 (11), 8163–8172
Pasolli E, Melgani F (2015) Genetic algorithm-based method for mitigating label noise issue in ECG signal classification. Biomed Signal Process Control 19:130–136
Qin AK, Huang VL, Suganthan PN (2009) Differential evolution algorithm with strategy adaptation for global numerical optimization. IEEE Trans Evol Comput 13(2):398–417
Rai HM, Trivedi A, Shukla S (2013) ECG signal processing for abnormalities detection using multi-resolution wavelet transform and artificial neural network classifier. Measurement 46(9):3238–3246
Rajendra Acharya U, Fujita H, Oh SL, Hagiwara Y, Tan JH, Adam M (2017) Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inf Sci (Ny) 415416:190–198
Sadhukhan D, Mitra M (2012) Detection of ECG characteristic features using slope thresholding and relative magnitude comparison. 3rd International Conference on Emerging Applications of Information Technology, EAIT 122–126
Sannino G, De Pietro G (2018) A deep learning approach for ECG-based heartbeat classification for arrhythmia detection. Futur Gener Comput Syst 17:32454–32458
Singh R, Balasundaram S (2007) Application of extreme learning machine method for time series analysis. Int J Intell Technol 2(4):256–262
Tavares LD, Saldanha RR, Vieira DaG (2015) Extreme learning machine with parallel layer perceptrons. Neurocomputing 166:164–171
Teodoro FGS, Peres SM, Lima CAM (2017) Feature selection for biometric recognition based on electrocardiogram signals. Int Jt Conf Neural Networks no mV:2911–2920
Traore BB, Kamsu-Foguem B, Tangara F (2018) Deep convolution neural network for image recognition. Ecol Inform 48(September):257–268
Tripathy BK, Acharjya DP, Cynthya V (2013) A framework for intelligent medical diagnosis using rough set with formal concept analysis. arXiv preprint arXiv 1301:–6011
Villarreal-cervantes MG, Rodr A, Garc C (2017) Multi-objective on-line optimization approach for the DC motor controller tuning using differential. Evolution:1–15
Vinícius dos Santos Ferreira M, Oseas de Carvalho Filho A, Dalília de Sousa A, Corrêa Silva A, Gattass M (2018) Convolutional neural network and texture descriptor-based automatic detection and diagnosis of glaucoma. Expert Syst Appl 110:250–263
Wen-An ZQ, K-LT Y (2016) Differential evolution-based feature selection and parameter optimisation for extreme learning machine in tool wear estimation. Int J Prod Res 54(15):4703–4721
Wu JF, Bao YL, Chan SC, Wu HC, Zhang L, Wei XG (2017) Myocardial infarction detection and classification-a new multi-scale deep feature learning approach. Int Conf Digit Signal Process DSP:309–313
Zihlmann M, Perekrestenko D, Tschannen M (2017) Convolutional recurrent neural networks for electrocardiogram. Classification in Computing in Cardiology (CinC), 1–4
Funding
This review was not supported by any external funding sources
Author information
Authors and Affiliations
Contributions
Aykut Diker: Literature analysis, Interpretation of the results, Drawing of the figures, Preparation of the manuscript. Yasin Sönmez: Literature analysis, Interpretation of the results, Preparation of the manuscript. Fatih Özyurt: Literature analysis, Drawing of the figures, Preparation of the manuscript. Engin Avcı: Literature analysis, Interpretation of the results. Derya Avcı: Literature analysis, Interpretation of the results, Preparation of the manuscript.
Corresponding author
Ethics declarations
Conflict interests
The authors declare no conflicts of interest.
Statements of ethical approval
The authors did not carry out any work that involved human or animal participants for this study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Diker, A., Sönmez, Y., Özyurt, F. et al. Examination of the ECG signal classification technique DEA-ELM using deep convolutional neural network features. Multimed Tools Appl 80, 24777–24800 (2021). https://doi.org/10.1007/s11042-021-10517-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-10517-8