Abstract
Automated abnormal brain discovery is an extremely crucial task for clinical diagnosis. Over a decade ago, various techniques had been displayed to improve this technology. This paper presents a hybrid system based on a combination of Gray Wolf Optimizer (GWO) and Support Vector Machine (SVM) with Radial Basis Function (RBF) kernel to classify a given Magnetic Resonance (MR) brain image as benign or malignant. 5-fold cross validation was used to enhance generalization. We applied the hybrid system on 80 images (20 benign and 60 malignant), and found out that the classification accuracy was as high as 98.750%.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Image processing techniques are utilized to improve the quality of images and subsequently to perform features’ extraction and classification. They are effectively used in computer vision, medical imaging, meteorology, astronomy, remote sensing and other related fields [40, 41]. They play an important role in various medical applications to support the computerized disease examination.
Brain diseases constitute one of the major causes of cancer related death among children and adults in the world. Brain diseases like brain tumors are characterized as the growth of abnormal cells inside or around the brain. Detection of the brain tumor in its early stage is the key point of its cure. There are several sorts of brain tumors that make the classification difficult. A good classification process leads to the correct choice of the treatment approach. The human prediction is not always correct. So, we need an automated prediction or classification strategy for tumors. In this paper, a computer aided brain tumor classification system was proposed.
There are various imaging techniques, which were used for brain tumor detection. Among all imaging techniques, MR Imaging (MRI) [14] is widely used to detect a tumor in the brain. The MRI is a safe, fast, and non-invasive imaging technique. The early detection of brain diseases is of primordial importance because it allows the prompt adoption of the proper treatment thus increasing the possibility of a complete cure.
The tumor is essentially an uncontrolled development of cancerous cells in any part of the body, though a brain tumor is an uncontrolled development of cancerous cells in the brain. Brain tumors can be benign or malignant [39]. Benign brain tumors have a consistency in structure and do not contain dynamic cells, while malignant brain tumors have a non-consistency in structure and contain dynamic cells.
Various methods have been identified for classification of brain tumors in MR images such as Fuzzy Clustering Means (FCM) [11], SVM [15, 18, 26], Artificial Neural Network (ANN) [4], Probabilistic Neural Network (PNN) [34, 36], knowledge-based techniques, and Expectation-Maximization (EM) algorithm technique.
The success of any classification technique depends on the correct choice of its parameters. This research concentrates on using SVM for solving the problem of MR classification of brain images. SVM demonstrated its effectiveness as a classification technique. But, SVMs face a few difficulties, when embraced in genuine practical applications. One of these difficulties is setting the optimal parameters of SVM. This research shows a technique for choosing the best parameters for SVM by means of applying an optimization algorithm. Choosing these parameters accurately guarantees to obtain the best classification accuracy. The major parameters that affect the performance of SVM are error penalty parameter C and RBF kernel parameter σ.
In this paper, an automatic brain MRI classification method is proposed. The proposed system combines GWO and SVM so as to achieve enhanced MRI classification accuracy by means of choosing the ideal parameters of SVM.
The rest of this paper is structured as follows. The research work on MR brain image classification is introduced in section 2. Section 3 introduces the proposed system and describes its different phases, namely: preprocessing, feature extraction, and classification phases. Section 4 presents the case study and results. Finally, Section 5 concludes the paper.
2 Related works
There are several studies on the brain MRI Classification. In the most of these algorithms, the input brain MRI is classified as normal or abnormal [3, 5,6,7,8,9, 16, 17, 23, 27,28,29, 31,32,33,34, 38, 42, 44]. In [9], the authors used Berkeley Wavelet Transform (BWT) to segment the MR images and SVM to classify the tumor stage by analyzing feature vectors and areas of the tumor. They segmented the brain tissues into normal tissues such as white matter, gray matter, cerebrospinal fluid (background), and tumor-infected tissues. Fifteen patients infected with a glial tumor, in benign and malignant stages, assisted in this study. This technique obtained an accuracy of 96.51%. Saha and Hossain [32] proposed a scheme for the automatic classification of MR brain images as normal or abnormal using K-Means clustering, Non-Subsampled Contourlet Transform (NSCT) and SVM. In the preprocessing stage of this proposed scheme, median filter was used for removing noise and enhancing resolution of MR brain images. Then K-means clustering was used for segmenting MR brain images because it segments the image faster. NSCT was then applied to the segmented image. Then seven features are extracted from subband coefficients of NSCT and these features are applied to SVM for the classification of MR brain images. This scheme was applied on 88 MR brain images and achieved maximum accuracy for Gaussian Radial Basis (GRB) function kernel of 98.86%.
An efficient detection algorithm to detect tumor in MRIs was introduced by Halder and Dobe [16]. This technique utilized FCM and SVM for feature reduction and classification, respectively. SVM was used to classify the scaned images into two groups, namely, tumor-free and tumor affected. The proposed algorithm was established to give encouraging results than the other existing brain tumor detection algorithms.
Selvapandian and Manivannan [33] proposed a methodology to detect and segment the Glioma tumors in brain MR images. NSCT was used to enhance the brain image and then texture features are extracted from the enhanced brain image. These extracted features are trained and classified using Adaptive Neuro Fuzzy Inference System (ANFIS) approach to classify the brain image into normal and Glioma brain images. As an average, the proposed brain tumor segmentation methodology achieves 92.3% of sensitivity, 96.2% of specificity and 95.9% of accuracy.
In [6] Deep Neural Networks (DNN) based architecture is proposed for brain tumor detection. In the proposed model, seven layers are used for classification that consist of three convolutional, three ReLU and a softmax layer. First the input MR image is divided into multiple patches and then the center pixel value of each patch is supplied to the DNN. DNN assign labels according to center pixels and perform segmentation. Proposed method results on BRATS 2015 dataset are 95% of sensitivity, 95.2% of specificity and 95.1% of accuracy.
Convolutional neural networks (CNN) classifier based brain tumor detection and segmentation methodology was proposed in [7]. This proposed methodology consists of image fusion, feature extraction, classification, and segmentation. Discrete Wavelet Transform (DWT) was used for image fusion and enhanced brain image was obtained by fusing the coefficients of the DWT. Further, Gray Level Co-occurrence Matrix (GLCM) features are extracted and fed to the CNN classifier for glioma image classifications. The methodology proposed in this paper achieves 97.3% of sensitivity, 98.1% of specificity, 98.7% of accuracy, and 0.96 of Dice Similarity Coefficient (DSC).
Amin et al. [5] introduced an automated method to easily differentiate between cancerous and non-cancerous MR brain images. Different techniques were applied for the segmentation of candidate lesion. Then the features set was chosen for every applicant lesion using shape, texture, and intensity. SVM was applied with different cross validations on the features set to compare the precision of the proposed framework. The method achieved average 97.1% accuracy, 0.98 area under curve, 91.9% sensitivity and 98.0% specificity.
A novel methodology based on meta-heuristic optimization approach was proposed in [31] to assist the brain MRI examination. In this study, a novel two stage approach by integrating the Meta heuristic multi-thresholding and the segmentation approach was utilized to extract the tumor region from the well-known brain MRI dataset. This study confirmed that, Shannon’s approach is efficient in providing better image quality. Kharrat et al. [8] presented a novel approach for automatic classification to distinguish between normal and abnormal MRIs of the brain. 2D Wavelet Transform and Spatial Gray Level Dependence Matrix (SGLDM) were used for feature extraction. For feature selection, Simulated Annealing (SA) was applied to reduce features size. Genetic Algorithm and SVM (GA-SVM) model was used to optimize SVM parameters. Stratified K-fold Cross Validation was used to avoid overfitting. An intelligent classification rate of 95.6522% was achieved using the SVM.
In [42], a wavelet-energy based approach was used for the classification of MR brain images. The approach has a three-stage system, including wavelet decomposition, energy extraction, and K-Nearest Neighbors (KNN) algorithm. This approach achieved excellent performance with a sensitivity of 93.75%, a specificity of 100%, and an accuracy of 95.45%. Our group proposed a hybrid optimized classification method to classify the brain tumor by classifying the given MR brain image as normal or abnormal [3]. This hybrid system based on a combination of GWO and ANN classifier. The GWO–ANN classification system performance was compared to the traditional NN classifier using receiver operating characteristic (ROC) analysis. Experimental results obviously indicate that the presented system achieves a high classification rate and perform much better than the traditional NN classifier. An accuracy of 98.91% was obtained using this classification algorithm.
A novel pathological brain detection technique mainly uses the aid of three components: Wavelet Packet Tsallis Entropy (WPTE), Real-Coded Biogeography Based Optimization (RCBBO), and Feed Forward Neural Network (FFNN) [38]. WPTE was introduced to extract global features from brain MR images. RCBBO was introduced to train feed forward neural network. With the use of a 255-image dataset, an average accuracy of 99.49% was accomplished.
A new system for abnormal brain detection was presented by Nayak et al. [27]. This system employed DWT and Kernel Principal Component Analysis (KPCA) for feature extraction and reduction, respectively. Least Squares SVM (LS-SVM) are utilized to classify brain MR images as normal or abnormal. The proposed scheme was validated on a dataset of 90 images and the results show the efficacy of the suggested scheme with considerably a smaller number of features as compared to other schemes.
Zhang et al. [44] introduced a new diagnosis system for the detection of the pathological brain in MRI scanning based on Hu Moment Invariants (HMIs), and a combination of two classifiers—Twin SVM (TSVM) and Generalized Eigenvalue Proximal SVM (GEPSVM). First, HMIs were extracted from a specific MR brain image; then, seven HMIs features were fed into the two classifiers. Then, a 5 × 5-fold cross validation on a data set containing 90 MR brain images, demonstrated that the proposed method accomplished classification accuracy of 98.89%.
In [34], textural features were extracted from GLCM followed by a morphological operation and the PNN classifier was used for the classification of tumors from brain MR images. To remove the noise and smoothen the image, preprocessing was used which also results in the improvement of signal-to-noise ratio. From the observation results, it can be clearly expressed that the detection of brain tumor was fast and accurate when compared to the manual detection carried out by clinical experts. The performance factors evaluated also shows that it gives better outcome by improving Peak signal to noise ratio (PSNR) and Mean Square Error (MSE) parameters.
Nayak et al. [28] presented a new automatic Computer-Aided Diagnosis (CAD) system for brain MR image classification. This method utilized DWT to extract features from the MR images. A combined PCA and Linear Discriminant Analysis (LDA) approach was employed to select the most significant features from the high-dimensional normalized features. Finally, a random forests classifier has been used to determine the normal or pathological brain. The classification accuracy and the area under the curve (AUC) on ‘Dataset-255’ are 99.22% and 0.996, respectively. The results show again the efficacy of the scheme with considerably a smaller number of features as compared to other schemes.
Jahanavi and Kurup [17] introduced a hybrid procedure consisting of SVM with two combined clustering methods, i.e., K-Means and FCM. The feature extraction isperformed by using gray level run length matrix (GLRLM). Finally, SVM helped to classify the image and also grade the location of the tumor. The hybrid technique accomplished an accuracy of 0.99862, a specificity of 0.999894, and a sensitivity of 1.
Parveen and Singh [29] proposed a new hybrid technique based on a combination of SVM and FCM for brain tumor classification. FCM clustering was utilized for the segmentation of the image to detect the suspicious region in brain MRI. GLRLM was used for extraction of features from the brain image, after which SVM technique was applied to classify the brain MR images. Real data set of 120 patients’ MR brain images was used to detect ‘tumor’ and ‘non-tumor’ MR images. The classification results are accurate for large data sets.
Authors in [23], presented an intellectual classification system to recognize normal and abnormal MR brain images. A combination of three classifiers- SVM, KNN and Hybrid Classifier (SVM-KNN) were used to classify 50 images. Experimental outcomes show the effectiveness of the two models. SVM with Quadratic kernel achieves maximum of 96% classification accuracy and the hybrid classifier SVM-KNN demonstrated the highest classification accuracy rate of 98%.
There are few studies on determining the types of the disease.
A hybrid Approach for Classification of Brain MR Tumor Images either as benign or malignant was introduced by Kumar et al. [20]. This hybrid approach includes DWT to be used for extraction of features, GA for diminishing the number of features and SVM for brain tumor classification. The proposed scheme was validated on a dataset of 25 MR brain images (20 benign, 5 malign images). Parameters used for analyzing the images are given as: entropy, smoothness, root mean square error (RMS), kurtosis and correlation. RMS value was found to be near 0.1.
Devkota et al. [13] presented a computer aided detection approach to diagnose brain tumor in its early stage using Mathematical Morphological Reconstruction (MMR). A large number of textural and statistical features are extracted from the segmented image to classify whether the brain tumor in the image is benign or malignant. The study shows that the proposed solution can be used to diagnose brain tumor in patients with a high success rate.
In [22], the features extracted from GLCM were fed to PNN to classify MR brain images as benign or malignant or normal images. Image recognition and image compression was done by using the PCA method and large dimensionality of the data is reduced. Segmentation process is done by using K-means clustering algorithm and detects the brain tumor spread region. On comparison between PNN and CNN, PNN is considered to have major advantages. As PNN has speed of learning capability, it can adapt its learning in real time.
Kumar and Chatterjee [19] proposed a diagnosis technique consisting of three parts. Firstly, a technique for segmenting the tumor; secondly, optimize the data set and use GLCM for texture analysis is proposed and lastly classify the tumor using SVM classifier into Benign or Malignant one is proposed. The classifier accuracy was obtained by using 3 kernels method that are RBF, Linear and Polynomial kernel. From the observation of the results it can be seen that the RBF kernel yields good performance for the classification of both benign and malignant brain tumors.
A two-stage CAD system was developed by Abd-Ellah et al. [1] for automatic detection and classification of brain tumor through MRIs. In the first stage, the system classifies brain tumor MR into normal and abnormal images. In the second stage, the type of tumor was classified as benign Noncancerous or Cancerous from the abnormal MRIs. The proposed CAD employs K-means, DWT, PCA, and kernel SVM (KSVM) methods for segmentation, feature detection, feature reduction, and MRI classification, respectively. Performance evaluation of the proposed CAD has achieved promising results using a non-standard MRIs database.
In [21], a computer aided brain tumor detection and segmentation methodology was proposed. In this method, ANFIS classifier was used to classify the brain tumor regions as benign or malignant. The performance of tumor segmentation was analyzed with the factors of Similarity Index (SI), Extra Fraction (EF), Overlap Fraction (OF) and accuracy, whose values obtained were 0.78, 0.0098, 0.723 and 99.4% respectively.
Preethi and Sornagopal [30] introduced an efficient method of classifying MR Brain Images as normal, benign and malignant using a PNN with a radial basis function. A set of 15 images were taken as training images and Fast Discrete Curvelet Transform (FDCT) was used to decompose the image and their features (contrast, correlation, energy, homogeneity) were extracted using GLCM. The proposed method, with the help of the texture statistics obtained from Low High (LH) and High Low (HL) sub bands, is able to categorize brain tumor into benign tumor images and malignant tumor images. A new approach for brain tumor detection and classification based on Soft Computing Techniques was presented in [2]. Two types of segmentation (global threshold and watershed segmentation) were used in this proposed approach. ANN was utilized to classify MR images as normal, benign or malignant.
The aim of this paper is to determine the type of brain disease with high accuracy.
3 Proposed method
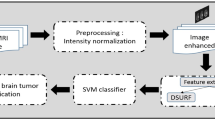
The proposed approach consists of three phases; namely: preprocessing, feature extraction, and GWO-SVMs classification. Figure 1 shows the flow diagram for the proposed method.
3.1 Preprocessing phase
3.1.1 Histogram equalization
The first step in the system is to isolate the tumor area from what remains in the image. Various image processing methods are utilized to isolate the tumor area. Image pre-processing involves basically histogram equalization. The major problem in the process of the edge detection of tumor is that the tumor seems extremely dark on the image, which is very confusing. To overcome this problem, histogram equalization is performed.
3.1.2 Threshold based segmentation
Segmentation partitions an image into its constituent parts. Thresholding is utilized for segmentation so as to get a binarized image with gray level 1 representing the tumor and gray level 0 representing the background. Segmentation is controlled by a parameter known as the intensity threshold. Every pixel in the image is compared with this threshold. If the pixel intensity is larger than the threshold, the pixel is established to white. If it is less than the threshold, it is established to black in the output. The threshold is calculated by the following equation.
With this equation, we can simply evaluate the threshold value. After computing the threshold value, we get the segmented image [24, 35].
3.2 Feature extraction phase
The feature extraction is the procedure to represent a raw image in order to facilitate decision making such as pattern classification. Features will be extracted from the tumor areas from MR brain images. Feature extraction includes decreasing the amount of data required to describe a large set of data correctly. Features are utilized as inputs to classifiers which assign them to the class which they represent. The following features are extracted.
-
Statistical features
-
Texture features
1) Statistical features: Statistical features that are extracted are the mean, variance, standard deviation, skewness and kurtosis.
2) Texture features: GLCM is applied to find the textures which describe the spatial relationship between pixels of different gray levels. This method follows two stages for the extraction of features from the medical images. In the first stage, the GLCM is calculated, and in the next stage, the texture features based on the GLCM [10] are calculated. Some of the texture features are shown below.
3.3 GWO-SVMs classification phase
3.3.1 SVMs
The SVMs represent a new type of learning machines which are used in pattern classification. To improve the Quality of Service (QoS) and the security, many researchers use these techniques. SVMs can solve any classification problem by means of attempting to find out an optimal isolating hyper plane between two classes. The main objective of the SVMs is that it simultaneously maximizes the geometric margin around the isolating hyper plane between a positive and a negative class. Consider the training dataset is in the form of {(x1, y1), (x2, y2), ……, (xn, yn)} with n samples, where xi is an n-dimensional input vector and yi = {−1, 1} corresponding to the class 1 or 2. The construction of an optimal hyper plane with the maximal geometric margin to isolate two classes needs to solve the optimization problem, as shown in Eqs. (12) and (13).
Where, αi is the weight given to every training point xi .Those points are called support vectors for αi > 0 .C is known as error penalty parameter. K is a kernel function. In this paper, the widely used Gaussian RBF kernel is assumed, that is:
3.3.2 GWO
The GWO algorithm was proposed by Mirjalili et al. [25]. It simulates the hunting behavior and social leadership of gray wolves in life. In this algorithm the population is organized into four groups: alpha (α), beta (β), delta (δ), and omega (ω). The first three best gray wolves are considered as α, β, and δ. The rest of wolves are assumed as omega (ω) and required to encircle α, β, and δ as follows:
where t indicates the current iteration,\( \overrightarrow{\ A}=2\overrightarrow{a}.\overrightarrow{r_1}-\overrightarrow{a}\kern1.25em \),\( \overrightarrow{\ C}=2.\overrightarrow{r_2}\kern1em \)\( , \overrightarrow{X} \) indicates the position vector of a gray wolf, \( \overrightarrow{X_p}\kern0.5em \) is the position vector of the prey, \( \overrightarrow{a} \)is linearly decreased from 2 to 0 and \( \overrightarrow{r_1} \), \( \overrightarrow{r_2} \) are random vectors in (0, 1).
During optimization, the three best solutions obtained so far are assumed as α, β, and δ respectively are saved and each ω wolf is required to update its position with respect to α, β, and δ simultaneously as follows:
Figure 2 shows the flow chart of GWO.
3.3.3 Cross-validation
In this paper the 5-fold cross validation is applied because of its properties as straightforward, simple, and utilizing all the data for training and validation. The dataset is arbitrarily divided into 5 commonly exclusively subsets of roughly equal size, in which 4 subsets are utilized as training set, and the latest subset is utilized as validation set. The previously mentioned technique repeated 5 times, so every subset is utilized once for approval. The fitness function of GWO selected the classification accuracy of the 5-fold cross validation:
where ys is the number of succeeded classification and ym is the number of misclassifications. GWO is implemented to maximize the fitness function (classification accuracy).
3.3.4 GWO- SVMs classifier
Here, we describe the proposed GWO-SVMs system to obtain the ideal parameters of SVM which result in a superior classification accuracy rate. The outline of the proposed GWO-SVMs algorithm is as follows:
-
1.
Initialize the gray wolf population.
-
2.
Training the SVM classifier, evaluate the fitness of each gray wolf according to (20).
-
3.
Determine whether the gray wolf’s fitness reach the best classification accuracy, if reached, go to step 5, otherwise the next step into the next generation iterative process.
-
4.
Update the position of the gray wolves in the population according to (19), then return to step 2.
-
5.
Get the best classification accuracy.
-
6.
Training data set to get learning model, utilize the model to predict the test data set to get the prediction accuracy of the test data set.
3.3.5 Pseudocode of our method
In total, our method can be described as shown in fig. 3:
4 Experiments and discussions
We succeeded in classifying the MR brain images as normal and abnormal images [3] with very high accuracy, compared to other published work, using hybrid GWO- ANN classification technique. In this work, we further classify MR abnormal brain images as benign or malignant.
4.1 Database
Two datasets are used for the performance evaluation of the proposed system. The first dataset contains 80 T2-weighted MR brain images: 20 are benign tumors experiencing a low-grade glioma, meningioma and 60 are malignant tumors experiencing a Bronchogenic Carcinoma, Glioblastoma Multiform, and Sarcoma. These images were taken from Harvard Medical School Website (http://www.med.harvard.edu/AANLIB/). The second dataset is the BRATS 2015 [12] dataset which consists of 60 High Grade Glioma (HGG) images and 20 Low Grade Glioma (LGG) images.
The setting of the training images and validation images was shown in Fig. 4. The data set is divided into 5 equally distributed groups; each groups contain 4 benign brains and 12 malignant brains. Since 5-fold cross-validation was used, we would perform 5 experiments.
In each experiment, 4 groups were used for training, and the left group was used for validation. Each group was used once for validation. In total, in this cross-validation way, 320 images were used for training, and 80 images were used for validation.
4.2 Classification accuracy
The Confusion Matrices of our proposed method taken from benchmarkdatasets are mentioned in Tables 1 and 2. The element of the ith row and the jth column characterizes the classification accuracy belonging to class i and is allocated to class j after the supervised classification. The results show that our proposed method just misclassified 1 image of the total 80 MR brain images. The whole classification accuracy was (20 + 59)/80 = 98.75% using Harvard data set and (19 + 60)/80 = 98.75% using BRATS 2015 dataset. Moreover, our proposed method was compared with the recently published results in the literature that used the same MRI datasets, as shown in Tables 3 and 4. It is clear that the introduced classification method provides the highest classification accuracy.
(O stand for output, T stand for Target).
(O stand for output, T stand for Target).
4.3 Parameter selection
The last parameters achieved by GWO were C = 959.784 and σ = 28. 571. Where C is the error penalty parameter, σ is RBF kernel parameter. We compared this case with a randomly selected technique, which arbitrarily produced the values of C in the range of (1, 1000) and σ in the range of (1, 100). Then we compared them with the optimized values by GWO (C = 959.784 and σ = 28.571). The obtained results achieved by random selection technique are shown in Table 5. We saw that the classification accuracy varied with the change of parameters σ and C, so it was important to determine the optimal values before constructing the classifier. It is noticed from Table 5 that by trying different values for 휎 and C parameters, the accuracy change significantly. Accuracy changed from 93.75% to 97.50% without optimization, while it has increased to 98.75% using GWO-SVMs proposed approach.
The random selection technique was hard to run over the best values, this proved that the GWO was an effective method for this problem compared to the random selection technique.
5 Conclusion
In this study, a hybrid classifier has been developed to distinguish between benign and malignant MR brain images. This hybrid technique combined GWO and SVM in order to achieve enhanced MRI classification accuracy by means of choosing the ideal parameters of SVM. The hybrid system just misclassified 1 image of the total 80 MR brain images and the whole accuracy was 98.75%. A comparison between the introduced approaches with previously published ones shows that the proposed one gives the highest classification accuracy. However, this paper only evaluates the proposed method on T2-weighted images. When facing with the T1-weighted images or other types of medical image (e.g. three dimensional image, CT image, and so on), more experiments are needed to be done to prove the effectiveness of the proposed method. Moreover, another major limitation is that the proposed method was tested on a small dataset hence in the future work we suggest employment of advanced machine learning methods like deep neural network models with large dataset for further improvement.
References
Abd-Ellah MK, Awad AI, Khalaf AAM, Hamed HFA (2016) Design and implementation of a computer-aided diagnosis system for brain tumor classification. 28th IEEE Int Conf Microelectron (ICM), Giza Egypt 17-20:73–76
Abdullah HN, Habtr MA (2015) Brain tumor extraction approach in MRI images based on soft computing techniques. 8th IEEE Int Conf Intell Netw Intell Syst (ICINIS), Tianjin China 1-3:21–24
Ahmed HM, Youssef BAB, Elkorany AS, Saleeb AA, Abd El-Samie F (2018) Hybrid gray wolf optimizer–artificial neural network classification approach for magnetic resonance brain images. Appl Opt 57(7):B25–B31
Ahmmed R, Swakshar AS, Hossain F, Rafiq A (2017) Classification of tumors and it stages in brain MRI using support vector machine and artificial neural network. IEEE Int Conf Electric Comput Commun Eng (ECCE), Cox's Bazar, Bangladesh 16-18:229–234
Amin J, Sharif M, Yasmin M, Fernandes SL (2017) A distinctive approach in brain tumor detection and classification using MRI. Pattern Recogn Lett
Amin J, Sharif M, Yasmin M, Fernandes SL (2018) Big data analysis for brain tumor detection : deep convolutional neural networks. Futur Gener Comput Syst 87:290–297
Anitha R, Raja SS (2017) Segmentation of glioma tumors using convolutional neural networks. Int J Imaging Syst Technol 27(4):354–360
Ayed MB, Kharrat A, Halima MB (2015) MRI brain tumor classification using support vector machines and Meta-heuristic method, 15th IEEE international conference on intelligent systems design and applications (ISDA). Marrakech, Morocco: 446–451
Bahadure NB, Ray AK, Thethi HP (2017) Image analysis for MRI based brain tumor detection and feature extraction using biologically inspired BWT and SVM. Int J Biomed Imag 2017
Bahadure NB, Ray AK, Thethi HP (2017) Feature extraction and selection with optimization technique for brain tumor detection from MR images. IEEE Int Conf Comput Intell Data Sci (ICCIDS), Chennai, India 2-3:1–7
Benson CC, Deepa V, Lajish VL, Rajamani K (2016) Brain tumor segmentation from MR brain images using improved fuzzy c-means clustering and watershed algorithm. IEEE International Conference on Advances in Computing, Communications and Informatics (ICACCI), Jaipur, India: 187–192
BRATS database (2015). https://www.smir.ch/BRATS/Start
Devkota B, Alsadoon A, Prasad PWC, Singh AK, Elchouemi A (2018) Image segmentation for early stage brain tumor detection using mathematical morphological reconstruction. Proc Comput Sci 125:115–123
Fink JR, Muzi M, Peck M, Krohn KA (2015) Continuing education: multi-modality brain tumor imaging: MRI, PET, and PET/MRI. J Nucl Med 56(10)
Gandhi T, Panigrahi BK, Gupta T (2016) Classification of post contrast T1 weighted MRI brain images using support vector machine,” 3rd IEEE international conference on computing for sustainable global development (INDIACom), New Delhi, India: 2560–2563
Halder A, Dobe O (2016) Detection of tumor in brain MRI using fuzzy feature selection and support vector machine. IEEE Int Conf Adv Comput Commun Inform (ICACCI), Jaipur, India 21-24:1919–1923
Jahanavi MS, Kurup S (2016) A novel approach to detect brain tumour in MRI images using hybrid technique with SVM classifiers. IEEE Int Conf Recent Trends Electron Inform Commun Technol (RTEICT), Bangalore, India 20-21:546–549
Kharrat A, Gasmi K, Messaoud MB, Benamrane N, Abid M (2010) A hybrid approach for automatic classification of brain MRI using genetic algorithm and support vector machine. Leonardo J Sci 9(17):71–82
Kumar PMS, Chatterjee S (2016) Computer aided diagnostic for Cancer detection using MRI images of brain (brain tumor detection and classification system). IEEE Annual India Conf (INDICON), Bangalore, India 16-18:1–6
Kumar S, Dabas C, Godara S (2017) Classification of brain MRI tumor images: a hybrid approach. Proc Comput Sci 122:510–517
Lakshmi A, Arivoli T (2014) Computer aided diagnosis system for brain tumor detection and segmentation. J Theor Appl Inf Technol 64(2):561–567
Lavanyadevi R, Machakowsalya M, Nivethitha J, Kumar AN (2017) Brain tumor classification and segmentation in MRI images using PNN. IEEE Int Conf Electric Instrum Commun Eng (ICEICE), Karur, India 27-28:1–6
Machhale K, Nandpuru HB, Kapur V, Kosta L (2015) MRI brain Cancer classification using hybrid classifier ( SVM- KNN ). IEEE international conference on industrial instrumentation and control (ICIC), Pune, India: 60–65
Madhusudhanareddy P, Prabha IS (2013) Novel approach in brain tumor classification using artificial neural networks. Int J Eng Res Applic (IJERA) 3(4):2378–2381
Mirjalili S, Mirjalili SM, Lewis A (2014) Grey wolf optimizer. Adv Eng Softw 69:46–61
Nandpuru HB, Salankar SS, Bora VR (2014) MRI brain Cancer classification using support vector machine. IEEE Stud Conf Electric Electron Comput Sci (SCEECS), Bhopal, India 1-2:1–6
Nayak DR, Dash R, Majhi B (2015) Least squares SVM approach for abnormal brain detection in MRI using multiresolution analysis. IEEE Int Conf Comput Commun Sec (ICCCS), Pamplemousses, Mauritius 4-5:1–6
Nayak DR, Dash R, Majhi B (2015) Classification of brain MR images using discrete wavelet transform and random forests. Fifth National IEEE Conf Comput Vision Pattern Recogn Image Process Graph (NCVPRIPG), Patna, India 16-19:1–4
Parveen, Singh A (2015) Detection of brain tumor in MRI images, using combination of fuzzy C-means and SVM. 2nd IEEE international conference on signal processing and integrated networks (SPIN), Noida, India, pp. 98–102, .
Preethi G, Sornagopal V (2014) MRI image classification using GLCM texture features. IEEE international conference on green computing communication and electrical engineering (ICGCCEE), Coimbatore, India: 1–6
Rajinikanth V, Satapathy SC, Fernandes SL, Nachiappan S (2017) Entropy based segmentation of tumor from brain MR images – a study with teaching learning based optimization. Pattern Recogn Lett 94:87–95
Saha C, Hossain F (2017) MRI brain tumor images classification using K-means clustering, NSCT and SVM. 4th IEEE Uttar Pradesh section international conference on electrical, computer and electronics (UPCON), Mathura, India: 329–333
Selvapandian A, Manivannan K (2018) Fusion based glioma brain tumor detection and segmentation using ANFIS classification. Comput Methods Prog Biomed 166:33–38
Shree NV, Kumar TNR (2018) Identification and classification of brain tumor MRI images with feature extraction using DWT and probabilistic neural network. Brain Inform 5(1):23–30
Syed AQ, Narayanan K (2014) Detection of tumor in MRI images using artificial neural networks. Int J Adv Res Electric Electron Instrum Eng 3(9):11749–11754
Thara KS, Jasmine K (2016) Brain tumour detection in MRI images using PNN and GRNN. IEEE Int Conf Wireless Commun Signal Process Netw (WiSPNET), Chennai, India 23-25:1504–1510
Urban G, Bendszus M, Hamprecht FA, Kleesiek J (2014) Multi-modal brain tumor segmentation using deep convolutional neural networks, MICCAI multimodal brain tumor segmentation challenge (BraTS): 1–5
Wang S, Li P, Chen P, Phillips P, Liu G, Du S, Zhang Y (2017) Pathological brain detection via wavelet packet Tsallis entropy and real-coded biogeography-based optimization. Fundamenta Informaticae 151(1–4):275–291
Wu M, Chin W, Tsan T, Chin C (2016) The benign and malignant recognition system of nasopharynx in MRI image with neural-fuzzy based Adaboost classifier. 2nd IEEE international conference on information management (ICIM), London, UK: 47–51
Yan C, Xie H, Chen J, Zha Z, Hao X, Zhang Y, Dai Q (2018) A fast Uyghur text detector for complex background images. IEEE Trans Multimed 20(12):3389–3398
Yan C, Li L, Zhang C, Liu B, Zhang Y, Dai Q (2019) Cross-modality bridging and knowledge transferring for image understanding. IEEE Transactions on Multimedia
Zhang G, Lu Z, Ji G, Sun P, Yang J, Zhang Y (2015) Automated classification of brain MR images by wavelet-energy and k-nearest neighbors algorithm. Seventh IEEE international symposium on parallel architectures, algorithms and programming (PAAP), Nanjing, China: 87–91
Zhang G, Wang Q, Lee E, Ji G, Wang S, Yan J, Zhang Y (2015) Automated classification of brain MR images using wavelet-energy and support vector machines. International Conference on Mechatronics, Electronic, Industrial and Control Engineering (MEIC), Shenyang, China: 683–686
Zhang Y, Yang J, Wang S, Dong Z, Phillips P (2016) Pathological brain detection in MRI scanning via Hu moment invariants and machine learning. J Experiment Theor Artif Intell 29(2):299–312
Zhou X, Wang S, Xu W, Ji G, Phillips P, Sun P, Zhang Y (2015) Detection of pathological brain in MRI scanning based on wavelet-entropy and naive Bayes classifier. International Work-Conference on Bioinformatics and Biomedical Engineering (IWBBIO), Granada, Spain: 683–686
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ahmed, H.M., Youssef, B.A.B., Elkorany, A.S. et al. Hybridized classification approach for magnetic resonance brain images using gray wolf optimizer and support vector machine. Multimed Tools Appl 78, 27983–28002 (2019). https://doi.org/10.1007/s11042-019-07876-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-019-07876-8